Abstract
A chitinase gene (SmChiC) and its two C-terminal truncated mutants, SmChiCG426 and SmChiCG330 of Serratia marcescens, were constructed and cloned by employing specific polymerase chain reaction (PCR) techniques. SmChiCG426 is derived from SmChiC molecule without its C-terminal chitin-binding domain (ChBD) while SmChiCG330 is truncated from SmChiC by its C-terminal deletion of both ChBD and fibronectin type III domain (FnIII). To study the role of the C-terminal domains of SmChiC on the enzyme properties, SmChiC, SmChiCG426, and SmChiCG330 were expressed in Escherichia coli by using the pET-20b(+) expression system. The His-tag affinity-purified SmChiC, SmChiCG426, and SmChiCG330 enzymes had a calculated molecular mass of 51, 46, and 36 kDa, respectively. Certain biochemical characterizations indicated that the enzymes had similar physicochemical properties, such as the optimum pH (5), temperature (37 °C), thermostability (50 °C), and identical hydrolyzing product (chitobiose) from both the soluble and insoluble chitin substrates. The overall catalytic efficiency k cat /K M was higher for both truncated enzymes toward the insoluble α-chitin, whereas the binding abilities toward the insoluble α-chitin substrate were reduced moderately. The fluorescence and circular dichroism (CD) spectroscopy data suggested that both mutants retained a similar folding conformation as that of the full-length SmChiC enzyme. However, a CD-monitored melting study showed that the SmChiCG330 had no obvious transition temperature, unlike the SmChiC and SmChiCG426.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chitinases (EC 3.2.1.14) hydrolyze the β-1, 4-glycoside linkage between two N-acetylglucosamine residues (GlcNAc) in chitin. Chitin is an abundant biopolymer that is similar to cellulose in nature. Chitinases are found in chitin-producing organisms as well as in plants, bacteria, and vertebrates, where they might play a defensive role against pathogens [1, 2]. The CAZy database (http://www.cazy.org/) classified these chitinases into 18, 19, 23, and 48 glycoside hydrolase families based on their catalytic modules [3]. Family 18 contains chitinases from bacteria, fungi, viruses, and animals, and class III and V chitinases from plants. Family 19 contains class I, II, and IV plant chitinases [4]. Both families differ in their primary sequences, three-dimensional structures, and catalytic mechanisms [5–7]. The catalytic domains of family 18 chitinases have an (α/β)8 (TIM-barrel) fold [8–10]. In addition to the catalytic domain, numerous family 18 chitinases contain one or more domains that could be involved in the substrate binding (chitin-binding domains, ChBD) [11–13]. The architectures of substrate-binding clefts of chitinases differ, in which processive exochitinases are typically deep and tunnel-shaped, whereas nonprocessive endochitinases tend to have shallower and open substrate-binding regions [14, 15]. Family 18 chitinases can be further divided into four subgroups: A, B, C, and D-like [16].
Serratia marcescens is a Gram-negative soil bacterium that produces four chitinases (A, B, C1, and C2) in its efficient chitinolytic machinery. All chitinases, A (ChiA), B (ChiB), as well as C1 and C2 (ChiC), belong to the family 18 chitinase. The crystal structures of ChiA [9, 17] and ChiB [10, 18, 19] have shown that both enzymes have GH18 subfamily-A catalytic domain with retaining mechanism. The crystal structure of ChiC is unknown; yet, its GH18 subfamily-B catalytic domain resembles that of hevamine, a plant endochitinase [20–23]. Both ChiC and hevamine lack the so-called α/β-domain that comprises one of the walls of the deep substrate-binding clefts such as those of ChiA and ChiB. In addition to the GH18 subfamily-B catalytic domain, ChiC1 has one C-terminal ChBD and fibronectin type III-like domain (FnIII-like), whereas ChiC2, a proteolytic derivative of ChiC1, has the catalytic domain only. Consequently, the structure of ChiC1 differs substantially from that of ChiA and ChiB. Many previous studies have observed a substantial synergism on the hydrolysis of powdered chitin for several combinations of Serratia chitinases [14, 24, 25].
The roles of C-terminal regions of family 18 and 19 chitinases have been investigated extensively, and their effects on the chitin binding and hydrolysis of insoluble chitin substrate remain inconsistent [26–29]. Suzuki et al. and Synstak et al. had studied specific biochemical properties of S. marcescens ChiC1 and ChiC2 [21, 23, 24]. Previous research indicated that the C-terminal motif portion of ChiC1 did not affect the hydrolysis of soluble and amorphous chitin substrates, but it is required for efficient hydrolysis of crystalline chitins [21]. To further demonstrate the possible involvement of C-terminal region of ChiC1 on its enzyme intact structure and functional relationships, additional biochemical characterizations, including fluorescence and circular dichroism (CD) spectrometries, were performed in SmChiC and its two C-terminal truncation mutants—SmChiCG426 and SmChiCG330. Comparisons among these three molecules are useful and valuable for elucidating the molecular characteristics of SmChiC.
Materials and Methods
Bacterial Strains, Plasmids, and Culture Conditions
S. marcescens (BCRC15326) was purchased from Bioresource Collection Research Centre, Institute of Food Industry and Development, Hsinchu, Taiwan, and was used as the source of chromosomal DNA for the SmChiC gene cloning. The Escherichia coli hosts, plasmids, and culture conditions were identical to those detailed in the previous research [30].
DNA Manipulations
Chromosomal DNA from S. marcescens was prepared by employing the method detailed by Marmur [31]. Other recombinant DNA techniques were performed according to the standard methods described by Ausubel et al. [32]. The SmChiC gene was cloned by polymerase chain reaction (PCR) from S. marcescens genomic DNA by referring the reported ChiC1 sequence of S. marcescens ATCC13880 under the gene accession number BAA76623 [21]. For SmChiC, the sense primer SmChiC N0 forward: 5′-GGAATTCCATATGAGCACAAATAACATTATTAATG-3′ with an NdeI restriction site (italic) and the antisense primer SmChiC C0 reverse: 5′-CCGCTCGAGGGCGATGAGCTGCCACAGGGTGAAG-3′ with an XhoI restriction site (italic) were designed and used for PCR. The truncated genes, SmChiCG426 (ChBD deletion only) and SmChiCG330 (both ChBD and FnIII-like deletion), were constructed based on the analyzed and reported domain structures of SmChiC for their C-termini predictions and the reported molecular weights of ChiC1 and ChiC2 [21, 33]. The 3′-end primers, SmChiCG426 reverse, 5′-CCGCTCGAGGCCATCGTTCGCGGTTTTCACCGCCAGCGCG-3′ and SmChiCG330 reverse, 5′-CCGCTCGAGGCCGCCCTGAATCAGCGGCG-3′, were therefore designed accordingly. The cloned genes were sequenced to ensure that no point mutations occurred during the PCR cloning.
Protein Expression, Purification, Gel Electrophoresis, and Zymogram Analysis
E. coli BL21(DE3)pLysS containing the recombinant plasmids were cultured in Luria-Bertani broth containing 100 μg ml−1 ampicillin and 34 μg ml−1 chloramphenicol at 37 °C until A600 reached 0.6 for recombinant protein induction by IPTG inducer. Protein expression and purification followed the procedures described in previous studies [27, 28]. Sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12 %) and chitinase zymogram analyses were performed simultaneously as reported previously [27, 34].
Characterizations of Recombinant Chitinases
The chitinase activity was determined with 4-MU-(GlcNAc)2 as the assay substrate according to the procedure described in previous research using purified enzymes [27]. Protein concentrations were determined by Bradford method with bovine serum albumin as the standard [35]. Effects of the pH and temperature on the enzyme activity of SmChiC, SmChiCG426, and SmChiCG330 were investigated by the same procedure as previously described [27].
Hydrolysis of the Polymeric α-Chitin
The reaction mixtures (total volume of 0.5 ml) containing 20 μM for each enzyme and 0.1 g α-chitin (Cat. no. C9752; Sigma, St. Louis, MO, USA) in 20 mM sodium acetate buffer (pH 5.0) were incubated at 37 °C for various periods. Aliquotes of 50-μl reaction mixtures were removed on the 0.5, 1, 2, and 4 days after the hydrolysis reaction started. The identities and amounts of the hydrolysis products were analyzed by multiple reaction monitoring (MRM, LC-MS-MS) using the calibration curve of (GlcNAc)1–5 chitooligo standards.
Kinetics Analysis
The kinetic parameters were measured for the α-chitin substrate. The enzyme activity was measured by the released chitobiose at 37 °C for the time required for initial velocity in a 0.3-ml solution containing 0.5 μM purified enzyme in 20 mM sodium acetate buffer (pH 5.0) and various concentrations (2–20 mg) of α-chitin substrate. The released chitobiose was quantified by MRM. The hydrolysis rates were plotted against the substrate concentration in a Michaelis-Menten plot. The K M and k cat values were obtained after the experimental data were fitted to the Michaelis-Menten equation using nonlinear fitting in Origin 7 software [36].
Chitin-Binding Ability Assay
Different amounts (6–150 μg) of each enzyme and 2 mg α-chitin were mixed in a final volume of 0.25 ml ddH2O (pH 6.5) for 1 h at 4 °C under constant shaking. The supernatant, which contained unadsorbed proteins, was collected by centrifugation at 10,000 × g for 15 min, and the protein concentration was determined. The difference between the added protein and that in the supernatant was counted as the chitin-bound protein [27].
Thin Layer Chromatography Analysis of the Hydrolyzed Chitin Products
The hydrolysis products from various substrates, (GlcNAc)6 (Cat. no. H1396; Sigma) or α-chitin (Cat. no. C9752; Sigma), by same amounts of SmChiC, SmChiCG426, and SmChiCG330 at 37 °C for various time periods were performed, and the products of the reactions were placed in silica gel plates (Kiesel gel 60 F254; Merck, Rahway, NJ). The thin layer chromatography (TLC) profiles were developed with 2-propanol–water–ammonia (6.93:2.94:0.13 by volume) as the mobile phase, and the chitooligo standards, (GlcNAc)1-7, were applied. The plates were sprayed with a color developing reagent comprising 2 g diphenylamine, 2 ml aniline, 10-ml 85 % orthophosphoric acid, and 100 ml isopropanol. The plates were heated at 95 °C for 10 min to observe the carbohydrate spots [37].
Thermostability Analyses
Two micrograms each of SmChiC, SmChiCG426, and SmChiCG330 in 50 μl of 20 mM sodium phosphate buffer (pH 7.0) was heated at 30–99 °C for 30 min. The residual enzyme activity was measured at 37 °C by adding 100 μl of 50 μM 4-MU-(GlcNAc)2 as a substrate in the final reaction volume of 500 μl [27, 28].
Spectrometry
The fluorescence emission spectra of SmChiC, SmChiCG426, and SmChiCG330 were obtained at 25 °C using a Hitachi F-2500 spectrofluorometer, and the excitation spectra at 282 nm and the emission at 300–430 nm were recorded. The 0.03-mg ml−1 protein concentrations were measured in 0.5-ml solution of 20 mM sodium phosphate buffer (pH 7.0). To remove any variations in protein concentrations, the obtained spectra were corrected to ensure that the molar enzyme concentrations were identical. CD spectrometry was performed using an Aviv CD spectrophotometer under constant N2 flush. The CD spectra for the 190–260-nm far-ultraviolet wavelengths were measured at 25 °C. The results of triplicate scans were averaged, and all CD spectra were corrected against their corresponding buffer blanks. The protein concentrations were 0.17 mg ml−1 in 5 mM sodium phosphate buffer (pH 7.0) [27, 28].
Results
Domain Structures of SmChiC
Following the protein homology comparisons, the SmChiC comprised one GH18 subfamily-B catalytic domain, one C-terminal ChBD, and one FnIII-like domain (Fig. 1). The amino acid sequence alignments between the SmChiC from both S. marcescens BCRC15326 and S. marcescens ATCC13880 exhibited identities of 98.5 % (data not shown) [21].
Construction, Expression, and Purification of SmChiC, SmChiCG426, and SmChiCG330
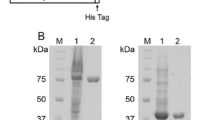
The 1440-bp DNA fragment encoding SmChiC from S. marcescens BCRC15326 chromosomal DNA was cloned by PCR. Two smaller and enzymatically active molecules, SmChiCG426 and SmChiCG330, were obtained from the truncated SmChiC after the deletion of the C-terminal ChBD only or both ChBD and FnIII-like domain, respectively (Fig. 1). SmChiC, SmChiCG426, and SmChiCG330 recombinant proteins were produced from the pET-20b(+) expression system. The homogeneities of these His-tag affinity-purified chitinases were investigated using 12 % SDS-PAGE, and their chitinase activity zymograms are shown with a molecular masses of 51, 46, and 36 kDa, respectively (Fig. 2a, b).
Biochemical and Molecular Characterizations of SmChiC, SmChiCG426, and SmChiCG330
The optimum pH and temperature for SmChiC, SmChiCG426, and SmChiCG330 were 5.0 and 37 °C, respectively, with 4-MU-(GlcNAc)2 as the assay substrate (data not shown). The effects of the C-terminal truncation on the kinetic parameters of the SmChiC, SmChiCG426, and SmChiCG330 were determined. The overall catalytic efficiency (k cat/K M ) of the SmChiCG426 and SmChiCG330 toward the α-chitin was of 1.06 and 2.09 relative fold, respectively, compared to that of the SmChiC. The SmChiCG426 retained approximately 85 % of the relative chitin-binding abilities of the SmChiC toward the insoluble α-chitin substrate. Compared to the SmChiC, the chitin-binding ability of the SmChiCG330 toward the α-chitin was further decreased to 63 % (Fig. 3). The effects of C-terminal truncation on hydrolyzing the soluble (GlcNAc)6 and insoluble α-chitin substrates were investigated by TLC or MRM (LC-MS-MS) methods. All three chitinases produced the chitobiose as the final product (TLC data not shown). The results of time course experiments of substrate hydrolysis indicated that SmChiCG426 and SmChiCG330 had different hydrolyzing abilities compared to SmChiC toward the polymeric α-chitin (Fig. 4). The structural integrities of the SmChiCG426 and SmChiCG330 molecules were further analyzed by fluorescence and CD spectrometry experiments. Similar spectra patterns in fluorescence for the native forms of three recombinant chitinases appeared in a maximum emission peak at 342 nm (data not shown). The CD spectra intensity of three chitinases were not changed substantially, except for a slight decrease at 215–230 nm observed in the SmChiCG330 (Fig. 5). The thermostability analysis indicated that all three chitinases were heat labile and lost approximately 55 % of enzyme activity at 50 °C after 30 min of heat incubation (Fig. 6). The CD-monitored thermounfolding studies of these three chitinases showed that the transition temperatures of SmChiC and SmChiCG426 were 54 and 55 °C, respectively, whereas the SmChiCG330 did not have a distinct transition temperature (Fig. 7).
Time course of hydrolysis of insoluble α-chitin by SmChiC (black circle), SmChiCG426 (black square), and SmChiCG330 (black triangle) enzymes. Reaction mixtures containing 0.1 g α-chitin and 20 μM each enzyme in a final reaction volume of 500 μl, 20 mM NaOAc buffer pH 5.0, were incubated at 37 °C for various time periods as indicated. The amounts of released chitobiose, (GlcNAc)2, were assayed and quantified by multiple reaction monitoring (MRM, LC-MS-MS). The linearity of the standard calibration curve was confirmed by the correlation coefficient value (r 2 = 0.9997)
CD spectra of SmChiC (black circle), SmChiCG426 (black square), and SmChiCG330 (black triangle) in the far UV region. The CD spectra were obtained at 25 °C, and the protein concentration was 0.17 mg ml−1 in 5 mM sodium phosphate buffer (pH7.0) in a 0.1-cm light path CD cell. After background subtraction, the CD data were converted from CD signals into molar ellipticity values (degree∙cm2 dmol−1)
Thermal unfolding of SmChiC (black circle), SmChiCG426 (black square), and SmChiCG330 (black triangle) monitored by CD spectroscopy at 216 nm. The CD melting curves were generated by monitoring the changes in the dichroic intensity at 216 nm as a function of temperature. The protein concentration and buffer were the same as in Fig. 5. Thermal denaturation was measured in the range of 25–95 °C at 1 °C intervals
Discussion
Functional chitinases can be derived from their precursors through protein processes such as proteolytic cleavages at either the N- or C-terminal region. The SmChiCG426 and SmChiCG330 were artificially derived from SmChiC, though they retained their enzyme activities. Evidence including (1) the relatively higher catalytic efficiency, k cat /K M , (2) hydrolyses of the soluble 4MU-(GlcNAc)2, (GlcNAc)6 and insoluble α-chitin substrates, and (3) the moderately decreased substrate-binding abilities all supported the fact that the SmChiCG426 and SmChiCG330 remained active after removing the C-terminal motif either ChBD or both ChBD and FnIII-like domains, respectively (Figs. 2, 3 and 4).
The boundary of the C-terminal region for the functional SmChiC was studied using C-terminal deletion mutants. Currently, the smallest active enzyme molecule, SmChiCG330, which was 150 amino acids shorter than SmChiC, was obtained by removing the presumably entire ChBD and FnIII-like domain region of the V331-A480 residues. This truncation did not abolish the enzyme against the soluble and insoluble chitin substrate hydrolyses. However, the loss of the C-terminal motif portion of the SmChiC rendered that both SmChiCG426 and SmChiCG330 have different hydrolytic activities toward the insoluble chitin polymer (α-chitin). SmChiCG330 has an approximate two-fold activity compared with those of SmChiC and SmChiCG426 (k cat/K M ratio and Fig. 4).
In BcChiA1, the ChBDChiA1 is necessary for binding specifically insoluble chitin and for hydrolyzing it efficiently [13]. The ChBDChiA1 domain itself does not bind with chito-oligosaccharides or soluble derivatives of chitin, although it does bind with insoluble crystalline chitin [11]. Wang et al. also reported that Chi1ΔChBD and Chi1ΔAΔChBD chitinases from Aeromonas caviae CB101 could not bind α-chitin and β-chitin (<5 %) because they had lost their C-terminal ChBDs [37]. The ChBD-deleted mutants of Chi1 retained 15 % hydrolyzing ability against the α-chitin substrate [38]. In contrast, this study shows that the deletion of the 54 C-terminal amino acid residues of SmChiC, the entire putative ChBD, SmChiCG426 retained 85 % relative binding ability and approximately the same catalytic efficiency (k cat/K M ratio,1.06:1) toward the α-chitin. Further C-terminal deletion of the SmChiCG426, SmChiCG330, having relative 63 % substrate-binding ability and almost two-fold catalytic efficiency (k cat/K M ratio, 2.09:1) was observed compared to that of the SmChiC. The binding study shown in Fig. 3 might provide a basis for the possible involvement of the C-terminal ChBD between the insoluble α-chitin substrate binding and the SmChiC enzyme, but other interactions such as the presence of FnIII and other neighboring aromatic amino acid residues could not be totally excluded. These results obtained from this study were not completely consistent with previous findings in binding studies of ChiC1 and ChiC2 of S. marcescens 2170 toward the regenerated chitin, colloidal chitin, and powdered chitin [21, 22, 24, 25].
Many C-terminally truncated chitinases have been reported to lose their chitin-binding abilities and hydrolyzing efficiencies toward various insoluble chitin substrates [13, 37, 39–41]. However, previous studies have reported distinct contrasts between Saccharomyces cerevisiae chitinase and AcD1ChiA [26, 41, 42]. It has been suggested that the main role of ChBD was necessary to bind the chitin substrate efficiently and specifically, but ChBD did not enhance the catalytic activity of the enzyme molecule. The ChBD module might just play a role in maintaining the correct enzyme–substrate interactions, thereby assisting the enzyme catalysis. For example, a ChBD-truncation-induced conformation change of the catalytic center of Aeromonas family 19 chitinase was reported and proposed to demonstrate the participation of its two contiguous yet different ChBDs in this enzyme catalysis [41]. Thus, ChBD could have multiple functions in various chitinases. This domain could be required to maintain intact enzyme properties, perhaps through maintaining an optimal distance and correct orientation between the catalytic domain and the substrate binding in the enzyme catalysis.
The sequence alignments among the SmChiCG330, BcCatDChiA1, and SmChiA show that the exposed aromatic residues (e.g., Trp, Tyr, and Phe) in the SmChiA are not present in the corresponding SmChiCG330 [43, 44]. These aromatic residues located around the active site cleft and in the N-terminal domain and were aligned linearly toward the catalytic center of the SmChiA enzyme molecule. Previous research proved that these aromatic residues were essential for the chitin binding and hydrolysis in A. caviae CB101 Chi1 [38]. But, other evidence was also obvious from this study. Since the SmChiCG330 did not cause a complete loss of the insoluble substrate chitin-binding and hydrolyzing ability (Figs. 3 and 4), and other reports that the chitinases were still active on the insoluble chitin substrates without their C-terminal putative ChBD and FnIII motifs [45–47], the catalytic domain of the SmChiCG330 could interact with the substrate through contributions from neighboring amino acid residues without totally depending on the putative ChBD and FnIII motifs for the insoluble chitin substrate binding. These findings indicate that numerous bacterial chitinases contain structures of discrete substrate-binding domains as a general feature, other motifs, or amino acid residues might play cooperative roles in the enzyme catalysis [48].
The fluorescence and CD spectra results also imply that the C-terminal truncation of the putative ChBD and FnIII-like domains of the SmChiC might not have a critical or significant effect on its molecular integrity. The global foldings of these enzymes might not be seriously damaged by these C-terminal truncations (Fig. 5). Similar findings were found in the chitinases from Vibrio parahaemolyticus and Bacillus licheniformis by the same laboratory [27, 28]. However, a CD-monitored thermounfolding study could monitor thermostability changes in SmChiCG330. To my knowledge, this is the first fluorescence and CD spectroscopy analyses of SmChiC and its truncated mutants, which could make a valuably supplement information of the molecular characteristics of SmChiC.
In conclusion, SmChiC and its two truncated chitinases, SmChiCG426 and SmChiCG330, were characterized to determine the effects of the C-terminal ChBD and FnIII-like domain truncation on the enzyme properties. The putative C-terminal ChBD and FnIII-like domains do contribute cooperation with the N-terminus of this enzyme to attach the chitin. The necessity of the C-terminal region of the SmChiC for the insoluble chitin hydrolysis is, however, not absolutely required for the full catalytic power of the SmChiC.
References
Leah, R., Tommerup, H., Svendsen, I., & Mundy, J. (1991). Journal of Biology Chemistry, 266, 1564–1573.
Gohel, V., Singh, A., Vimal, M., Ashwini, P., & Chhatpar, H. S. (2006). African Journal of Biotechnology, 5, 54–72.
Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V., & Henrissat, B. (2009). Nucleic Acids Research, 37, D233–D238.
Henrissat, B., & Bairoch, A. (1993). Biochemistry Journal, 293, 781–788.
Robertus, J. D., & Monzingo, A. F. (1999). EXS, 87, 125–135.
Fukamizo, T. (2000). Current Protein & Peptide Science, 1, 105–124.
Koga, D., Mitsutomi, M., Kono, M., & Matsumiya, M. (1999). EXS, 87, 111–123.
Matsumoto, T., Nonaka, T., Hashimoto, M., Watanabe, T., & Mitsui, Y. (1999). Proceedings of the Japan Academy Series B, Physical and Biological Sciences, 75, 269–274.
Perrakis, A., Tews, I., Dauter, Z., Oppenheim, A. B., Chet, I., Wilson, K. S., & Vorgias, C. E. (1994). Structure, 2, 1169–1180.
van Aalten, D. M. F., Synstad, B., Brurberg, M. B., Hough, E., Riise, B. W., Eijsink, V. G. H., & Wierenga, R. K. (2000). Proceedings of the National Academy of Science, 97, 5842–5847.
Hashimoto, M., Ikegami, T., Seino, S., Ohuchi, N., Fukada, H., Sugiyama, J., Shirakawa, M., & Watanabe, T. (2000). Journal of Bacteriology, 182, 3045–3054.
Ikegami, T., Okada, T., Hashimoto, M., Seino, S., Watanabe, T., & Shirakawa, M. (2000). Journal of Biological Chemistry, 275, 13654–13661.
Watanabe, T., Ito, Y., Yamada, T., Hashimoto, M., Sekine, S., & Tanaka, H. (1994). Journal of Bacteriology, 176, 4465–4472.
Horn, S. J., Sorlie, M., Vaaje-Kolstad, G., Norberg, A. L., Synstad, B., Varum, K. M., & Eijsink, V. G. H. (2006). Biocatalysis and Biotransformation, 24, 39–53.
Horn, S. J., Sorbotten, A., Synstad, B., Sikorski, P., Sorlie, M., Varum, K. M., & Eijsink, V. G. H. (2006). FEBS Journal, 273, 491–503.
Seidl, V., Huemer, B., Seiboth, B., & Kubicek, C. P. (2005). FEBS Journal, 272, 5923–5939.
Papanikolau, Y., Tavlas, G., Vorgias, C. E., & Petratos, K. (2003). Acta Crystallographica Section D: Biological Crystallography, 59, 400–403.
van Aalten, D. M. F., Komander, D., Synstad, B., Gaseidnes, S., Peter, M. G., & Eijsink, V. G. H. (2001). Proceedings of the National Academy of Science, 98, 8979–8984.
Hirose, T., Maita, N., Gouda, H., Koseki, J., Yamamoto, T., Sugawara, A., Nakano, H., Hirono, S., Shiomi, K., & Watanabe, T. (2013). Proceedings of the National Academy of Sciences of the United States of America, 110, 15892–15897.
van Scheltinga, A. C. T., Armand, S., Kalk, K. H., Isogai, A., Henrissat, B., & Dijkstra, B. W. (1995). Biochemistry, 34, 15619–15623.
Suzuki, K., Taiyoji, M., Sugawara, N., Nikaidou, N., Henrissat, B., & Watanabe, T. (1999). Biochemistry Journal, 343(Pt 3), 587–596.
Synstad, B., Vaaje-Kolstad, G., Cederkvist, H., Saua, S. F., Horn, S. J., Eijsink, V. G. H., & Sorlie, M. (2008). Bioscience Biotechnology and Biochemistry, 72, 715–723.
van Scheltinga, A. C. T., Hennig, M., & Dijkstra, B. W. (1996). Journal of Molecular Biology, 262, 243–257.
Suzuki, K., Sugawara, N., Suzuki, M., Uchiyama, T., Katouno, F., Nikaidou, N., & Watanabe, T. (2002). Bioscience Biotechnology and Biochemistry, 66, 1075–1083.
Gal, S. W., Choi, J. Y., Kim, C. Y., Cheong, Y. H., Choi, Y. J., Lee, S. Y., Bahk, J. D., & Cho, M. J. (1998). FEMS Microbiology Letters, 160, 151–158.
Lin, F. P., Juang, W. Y., Chang, K. H., & Chen, H. C. (2001). Archives of Microbiology, 175, 220–225.
Chuang, H. H., & Lin, F. P. (2007). Applied Microbiology and Biotechnology, 76, 123–133.
Chuang, H. H., Lin, H. Y., & Lin, F. P. (2008). FEBS Journal, 275, 2240–2254.
Sampei, Z., Nagao, Y., Fukazawa, T., Fukagawa, S., Matsuo, T., Endo, K., Yatsunami, R., & Nakamura, S. (2004). Nucleic Acids Symposium Series (Oxford), 48, 167–168.
Lin, C. S., Chen, H. C., & Lin, F. P. (1997). Enzyme and Microbial Technology, 21, 472–478.
Marmur, J. (1961). Journal of Molecular Biology, 3, 208–218.
Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., & Struhl, K. (Eds.). (1993). Current Protocols in Molecular Biology. New York: Greene Publishing & Wiley-Interscience.
Fuminori, K., Masashi, T., Kengo, S., Taku, U., Naoki, N., Takamasa, N., Junji, S., & Takeshi, W. (2004). Journal of Biochemistry, 136, 163–168.
Laemmli, U. K. (1970). Nature, 227, 680–685.
Bradford, M. (1976). Analytical Biochemistry, 72, 248–254.
Krokeide, I.-M., Synstad, B., Gaseidnes, S., Horn, S. J., Eijsink, V. G. H., & Sorlie, M. (2007). Analytical Biochemistry, 363, 128–134.
Wang, F. P., Li, Q., Zhou, Y., Li, M. G., & Xiao, X. (2003). Proteins, 53, 908–916.
Li, Q., Wang, F. P., Zhou, Y., Li, M. G., & Xiao, X. (2005). Applied and Environmental Microbiology, 71, 7559–7561.
Blaak, H., & Schrempf, H. (1995). European Journal of Biochemistry, 229, 132–139.
Chang, M. C., Lai, P. L., & Wu, M. L. (2004). FEMS Microbiology Letters, 232, 61–66.
Kojima, M., Yoshikawa, T., Ueda, M., Nonomura, T., Matsuda, Y., Toyoda, H., Miyatake, K., Arai, M., & Fukamizo, T. (2005). Journal of Biochemistry, 137, 235–242.
Lin, F. P., Chuang, H. H., Liu, Y. H., Hsieh, C. Y., Lin, P. W., & Lin, H. Y. (2009). Archives of Microbiology, 191, 265–273.
Uchiyama, T., Katouno, F., Nikaidou, N., Nonaka, T., Sugiyama, J., & Watanabe, T. (2001). Journal of Biological Chemistry, 276, 41343–41349.
Watanabe, T., Ariga, Y., Sato, U., Toratani, T., Hashimoto, M., Nikaido, N., Kezuka, Y., Nonaka, T., & Sugiyama, J. (2003). Biochemistry Journal, 376, 237–244.
Kamei, K., Yamamura, Y., Hara, S., & Ikenaka, T. (1989). Journal of Biochemistry, 105, 979–985.
Tsujibo, H., Endo, H., Minora, K., Miyamoto, K., & Inamori, Y. (1993). Gene, 134, 113–117.
Tsujibo, H., Hatano, N., Endo, H., Miyamoto, K., & Inamori, Y. (2000). Bioscience Biotechnology and Biochemistry, 64, 96–102.
Vaaje-Kolstad, G., Horn, S. J., van Aalten, D. M. F., Synstad, B., & Eijsink, V. G. H. (2005). Journal of Biological Chemistry, 280, 28492–28497.
Acknowledgments
We thank Dr. Chin-Pan Chen of the Institute of Biomedical Science, Academia Sinica, for his considerable help in the CD spectroscopy analyses. This work was supported by the Ministry of Science and Technology, Taiwan and the Center of Excellence for the Oceans, National Taiwan Ocean University.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lin, FP., Wu, CY., Chen, HN. et al. Effects of C-Terminal Domain Truncation on Enzyme Properties of Serratia marcescens Chitinase C. Appl Biochem Biotechnol 175, 3617–3627 (2015). https://doi.org/10.1007/s12010-015-1530-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-015-1530-5