Abstract
Yarrowia lipolytica is an unconventional yeast, and is generally recognized as safe (GRAS). It provides a versatile fermentation platform that is used commercially to produce many added-value products. Here we report a multiple fragment assembly method that allows one-step integration of an entire β-carotene biosynthesis pathway (~11 kb, consisting of four genes) via in vivo homologous recombination into the rDNA locus of the Y. lipolytica chromosome. The highest efficiency was 21 %, and the highest production of β-carotene was 2.2 ± 0.3 mg per g dry cell weight. The total procedure was completed in less than one week, as compared to a previously reported sequential gene integration method that required n weeks for n genes. This time-saving method will facilitate synthetic biology, metabolic engineering and functional genomics studies of Y. lipolytica.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Yarrowia lipolytica is one of the most studied unconventional yeasts and is considered to be a generally recognized as safe (GRAS) organism, which has already been used for some biotechnological applications, including citric acid production and heterologous protein expression (Bankar et al. 2009; Beckerich et al.1998) and eicosapentaenoic acid production (Xue et al. 2013). Y. lipolytica can accumulate large amount of lipids either by de novo lipid synthesis or via the uptake of lipids and alkanes, and is considered to be a suitable host to produce valuable, lipid-derived compounds (Nicaud 2012). Among the oleaginous yeasts, Y. lipolytica is the only one for which genomic and efficient molecular genetic tools are currently available (Beopoulos et al. 2009). However, metabolic engineering of this strain is still time-consuming for sequential gene integrations (Celinska and Grajek 2013; Chuang et al. 2010; Xue et al. 2013). The assembly of large, recombinant DNAs encoding entire biochemical pathways represents a significant challenge. A DNA assembler, which allows the assembly of multiple fragments in a single step via in vivo homologous recombination, has been successfully applied in Saccharomyces cerevisiae (Gibson 2009; Shao et al. 2009), Xanthophyllomyces dendrorhous (Contreras et al. 2013) and Kluyveromyces marxianus (Heo et al. 2013). However, in vivo homologous recombination has not been used to assemble multiple-gene biochemical pathways in a one-step fashion in a chromosome of Y. lipolytica.
As a proof of concept, we report, for the first time, the use of a DNA assembler method to rapidly assemble β-carotene biosynthesis pathways, with sizes of approx. 11 kb, as well as their integration into a chromosome in Y. lipolytica.
Materials and methods
Strains, plasmids, reagents and medium
Strains and plasmids used in this study are listed in Supplementary Table 1. Yarrowia lipolytica ATCC 201249 was cultivated on yeast/peptone/dextrose (YPD) medium (10 g yeast extract/l, 20 g tryptone/l, and 20 g glucose/l). Synthetic complete minus uracil (SC-ura) medium (20 g glucose/l, 1.7 g yeast nitrogen base without amino acids/l, 5 g (NH4)2SO4/l, supplemented with 50 mg leucine and 50 mg lysine/l) was used to select integrants containing the assembled biochemical pathways. For plates, agar (2 %) was added. Growth was at 30 °C.
DNA manipulation
One set of DNAs, comprising the carB and carRP genes from Mucor circinelloides (Velayos et al. 2000a,b), and the GGS1 gene (Matthäus et al. 2013) and the selective marker ura3 (accession number: AJ306421.1) from the Y. lipolytica NRRL Y-1095 genome, was synthesized de novo (Crolla and Kennedy 2001; Wang et al. 2013). The other set comprised the crtE, crtI and crtYB genes from X. dendrorhous ATCC24202 (Verwaal et al. 2007), and also the selective marker ura3. Y. lipolytica promoters (TEF1p, EXP1p, FBAp and GPDp), terminators (xpr2t, mig1t and lip2t) and homologous fragments for integration into target regions (rDNAu and rDNAd) were obtained individually from the genomic DNA of Y. lipolytica strain NRRL Y-1095, and the terminator cyc1t was obtained from S. cerevisiae strain BY4742 (Blazeck et al. 2011). All primers used in this study are listed in Supplementary Table 3. The fragments were assembled by overlap extension PCR (OE-PCR) into the individual gene expression cassettes, and DNA mixtures of the individual cassettes were prepared for transformation using a previously described procedure (Shao et al. 2009). The PCRs were performed in 50 μl, as described in the instructions for the KOD-Plus-Neo kit (Toyobo, Japan).
Yeast transformation and genomic DNA extraction
Transformation of Y. lipolytica was performed using the Zymogen Frozen-EZ yeast transformation kit II (Zymo Research Corporation) (Blazeck et al. 2011). Genomic DNA (gDNA) was extracted from Y. lipolytica using the Axygen genomic DNA purification kit (Axygen, China).
Genotype confirmation
PCR analysis, restriction digestion of amplified fragments and Real–time qPCR were used to do genotype confirmation.
Confirmation of the correct DNA assembly was assessed by PCR amplification of each gene cassette from the genomic DNAs. The PCR products obtained were separated in agarose gels and purified. They were then subjected to digestion by the corresponding restriction endonucleases, and the correct restriction digest pattern was assessed via electrophoresis on 1 % agarose gels.
Real-time qPCR amplification and analysis were performed using a MyiQ2 Two Colour Real-Time PCR Detection System with iQ5 optical system software version 2.1 (Bio-Rad, USA). The host strain ATCC201249 was selected as the negative control. The GGS1, promoter EXP1 and GPD1 driven heterologous carB and carRP, and the residual ura3 fragment in the genome were chosen as reference copy (primers listed in Supplementary Table 3).
Functional analysis of the assembled pathways
Colonies of yeast harboring correctly assembled DNAs were picked into 4 ml YPD liquid medium in 20 ml test tubes and incubated overnight at 30 °C, with shaking at 230 rpm. These were used to inoculate 20 ml fresh media in 150 ml flasks. Cells were grown at 30 °C, with shaking at 230 rpm, for 96 h. Cells that were harvested and dried at 80 °C for 16 h were used to calculate dry cell weight (DCW). Cells were harvested by centrifugation at 4,000×g for 4 min, resuspended in 0.5 ml dimethyl sulfoxide, and incubated at 55 °C for 10 min and then at 45 °C for 15 min, after which an equal volume of acetone was added. Samples were then centrifuged at 13,000×g for 5 min and the supernatants containing β-carotene were transferred to a new tube. Cell extracts were analyzed by HPLC, as described by Shao et al. (2009). A sample (5 μl) was loaded onto an Agilent Diamonsil SB-C18 column and analyzed at 472 nm after elution with acetonitrile (A) and tetrahydrofuran (B). The elution program is described in Supplementary Table 3.
Results and discussion
Design of the Y. lipolytica DNA assembler
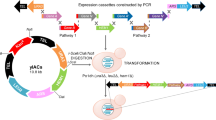
As proof of concept, we designed and constructed two sets of multiple-genes: β-carotene biosynthetic pathways, named as MC or XD, as shown in Fig. 1a and Supplementary Fig. 1. The accumulation of β-carotene resulting from the successful assembly and integration of the biosynthetic pathways turns transformants orange or red, which enables their identification. An rDNA sequence was previously used as the target locus for integration in the chromosome of Y. lipolytica (Nicaud 2012), and there are more than 200 rDNA repeat units in various chromosomes of Y. lipolytica, as well as δ sites in S. cerevisiae, that are available for integration (Heo et al. 2013; Verbeke et al. 2013). Compared to S. cerevisiae, in which the efficiency of homologous integration of exogenous DNA is very high, even with short (50 bp) homologous DNA flanking sequences, homologous recombination in Y. lipolytica only occurs at acceptable rates (80 %) with 0.5–1 kb 5′- and 3′-flanking regions of the target sequence (Verbeke et al. 2013). An rDNA site was selected as the target locus for integration into the chromosome, and ~0.6 kb 5′- and 3′-flanking regions were used as homologous flanking sequences. The two sets of β-carotene biosynthetic pathways (~11 kb) were divided into four cassettes, namely rDNAu-TEF1p-GGS1-xpr2t, EXP1p-carB-mig1t, GPDp-carRP-lip2t, and URA3-rDNAd, MC pathway or rDNAu-TEF1p-crtE-xpr2t, GPDp-crtI-lip2t, FBAp-crtYB-lip2t, and URA3-rDNAd, XD pathway, with overlaps between two successive cassettes of ~65 bp in MC or ~40 bp in XD.
Construction of a heterologous β-carotene biosynthetic pathway from Mucor circinelloides in Y. lipolytica, namely MC a The assembled β-carotene biosynthesis pathway with its corresponding verification primers b transformants on the selection plate c PCR analysis of the assembled, chromosomal β-carotene biosynthesis pathway using primers C1-F/C1-R, C2-F/C2-R, C3-F/C3-R, and C4-F/C4-Ralso demonstrated in Table S3 d Physical characterization of PCR analysis through restriction digestion. C1 was digested with PstI and XbaI; the correct clones should exhibit six bands, with sizes of 195, 242, 300, 330, 624 and 767 bp. C2 was digested with HindIII/NdeI; the correct clones should exhibit three bands, with sizes of 660, 1093 and 1896 bp. C3 was digested with HindIII/SacI; the correct clones should exhibit five bands, with sizes of 443, 500, 630, 837 and 1105 bp. C4 was digested with SmaI/SalI/PstI; the correct clones should exhibit four bands, with sizes of 347, 551, 708 and 1159 bp e HPLC analysis of the cell extracts from CIBTS1176 carrying the combined pathway. The host strain ATCC201249 was used as a negative control
Confirmation of the assembled β-carotene biosynthetic pathways
Prepared DNA mixtures were transformed into Y. lipolytica strain ATCC 201249. Around 92 colonies or 240 colonies appeared after 60 h on SC-ura medium for the MC or XD pathways, respectively. One-third of the MC and one-sixth of the XD transformants were orange/red (Table 1). Ten red colonies and 10 white colonies were randomly picked for amplification using four set of primers outside or inside the assembled fragment using junction PCRs as indicated in Fig. 1a and Supplementary Table 3, and the resulting 4 bands have overlaps of the assembling overlap region. the correct assemblies exhibited all four expected bands obtained following PCR (Fig. 1c and Supplementary Fig. 1c), and these four fragments were further confirmed by PstI/XbaI, HindIII/NdeI, HindIII/SacI, and SmaI/SalI/PstI digestion, as indicated in Fig. 1d. Of the MC or XD transformants, 60 or 70 %, respectively, were correctly assembled, while none of the white colonies were correct (Table 1). The total efficiency of one-step assembly of these four-gene pathways was 20.9 % for MC and 11.7 % for XD (Table 1). Higher efficiency can be further improved by extending the length of the overlap region between two cassettes as previously reported in S. cerevisiae (Shao et al. 2009), or deletion of KU70 in Y. lipolytica host (Verbeke et al. 2013). However, results of qPCR (supplementary information) indicate that CIBTS1176 assembled gene from MC which showed the deepest color underwent unexpected additional integrations of partial cassettes, including GGS1, carRP, and ura3 into the genome (supplementary information). This was caused by the high rate of non-homologous end-joining (NHEJ) in Y. lipolitica (Kretzschmar et al. 2013).
From the HPLC results, a peak appeared at 11.2 min from the correctly assembled strains, consistent with the elution time of authentic β-carotene, whereas no such peak was observed for the cell extracts from the wild-type strain ATCC 201209 (Supplementary Figs. 1e, d). Among all the engineered strains, the highest producing strain, CIBTS1176, yielded 2.22 ± 0.34 mg of β-carotene per g DCW of MC, while all the XD transformants produced less than 1 mg of β-carotene/g DCW. As an example, strain CIBTS1187, shown in Supplementary Fig. 1d, produced 0.84 ± 0.05 mg β-carotene/g DCW. Additionally, to our knowledge, this was the first report of β-carotene production in engineered strains of Y. lipolytica, and the production can be further optimized with additional metabolic engineering tools or our DNA assembler.
While sequential gene integration in Y. lipolytica requires n weeks for n genes, as described previously (Celinska and Grajek 2013; Matthäus et al. 2013), our finding enables the integration of multiple genes (four genes of ~11 kb) within one week. Furthermore, the method demonstrated in this study was also used to transform multiple genes into other Y. lipolytica strains, including ATCC 76861 and ATCC MYA2613 (data not shown). Thus, it seems that simultaneous gene integration is an efficient method for multiple-gene transformations in Y. lipolytica. Thus, this work contributes to the development of genetic tools that allow efficient genomic modifications in Yarrowia.
References
Bankar A, Kumar A, Zinjarde S (2009) Environmental and industrial applications of Yarrowia lipolytica. Appl Microbiol Biotechnol 84:847–865
Beckerich JM, Boisrame-Baudevin A, Gaillardin C (1998) Yarrowia lipolytica: a model organism for protein secretion studies. Intern Microbiol 1:123–130
Beopoulos A, Cescut J, Haddouche R, Uribelarrea J-L, Molina-Jouve C, Nicaud J-M (2009) Yarrowia lipolytica as a model for bio-oil production. Prog Lipid Res 48:375–387
Blazeck J, Liu L, Redden H, Alper H (2011) Tuning gene expression in Yarrowia lipolytica by a hybrid promoter approach. Appl Environ Microbiol 77:7905–7914
Celinska E, Grajek W (2013) A novel multigene expression construct for modification of glycerol metabolism in Yarrowia lipolytica. Microb Cell Factor 12:102
Chuang L-T, Chen D-C, Nicaud J-M, Madzak C, Chen Y-H, Huang Y-S (2010) Co-expression of heterologous desaturase genes in Yarrowia lipolytica. New Biotechnol 27:277–282
Contreras G, Barahona S, Rojas M, Baeza M, Cifuentes V, Alcaino J (2013) Increase in the astaxanthin synthase gene (crtS) dose by in vivo DNA fragment assembly in Xanthophyllomyces dendrorhous. BMC Biotechnol 13:84
Crolla A, Kennedy KJ (2001) Optimization of citric acid production from Candida lipolytica Y-1095 using n-paraffin. J Biotechnol 89:27–40
Gibson DG (2009) Synthesis of DNA fragments in yeast by one-step assembly of overlapping oligonucleotides. Nucleic Acid Res 37:6984–6990
Heo P et al (2013) Simultaneous integration of multiple genes into the Kluyveromyces marxianus chromosome. J Biotechnol 167:323–325
Kretzschmar A, Otto C, Holz M, Werner S, Hübner L, Barth G (2013) Increased homologous integration frequency in Yarrowia lipolytica strains defective in non-homologous end-joining. Curr Genet 59:63–72
Matthäus F, Ketelhot M, Gatter M, Barth G (2013) Production of lycopene in the non-carotenoid producing yeast Yarrowia lipolytica. Appl Environ Microbiol. doi:10.1128/aem.03167-13
Nicaud J-M (2012) Yarrowia lipolytica. Yeast 29:409–418
Shao Z, Zhao H, Zhao H (2009) DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acid Res 37:e16
Velayos A, Blasco JL, Alvarez MI, Iturriaga EA, Eslava AP (2000a) Blue-light regulation of phytoene dehydrogenase (carB) gene expression in Mucor circinelloides. Planta 210:938–946
Velayos A, Eslava AP, Iturriaga EA (2000b) A bifunctional enzyme with lycopene cyclase and phytoene synthase activities is encoded by the carRP gene of Mucor circinelloides. Eur J Biochem 267:5509–5519
Verbeke J, Beopoulos A, Nicaud J-M (2013) Efficient homologous recombination with short length flanking fragments in Ku70 deficient Yarrowia lipolytica strains. Biotechnol Lett 35:571–576
Verwaal R, Wang J, Meijnen J, Visser H, Sandmann G, van den Berg J, van Ooyen A (2007) High-level production of beta-carotene in Saccharomyces cerevisiae by successive transformation with carotenogenic genes from Xanthophyllomyces dendrorhous. Appl Environ Microbiol 73:4342–4350
Wang Z-P, Xu H-M, Wang G-Y, Chi Z, Chi Z-M (2013) Disruption of the MIG1 gene enhances lipid biosynthesis in the oleaginous yeast Yarrowia lipolytica ACA-DC 50109. Biochim Biophys Acta—Molec Cell Biol Lipids 1831:675–682
Xue Z, Sharpe PL, Hong SP, Yadav NS et al (2013) Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica. Nat Biotech 31:734–740
Acknowledgments
This work was supported by the National Basic Research Program of China (973: 2012CB721105 and 2014CB745101), National High-Tech Research and Development Program of China (863: 2012AA02A704) and the Knowledge Innovation Program of the Chinese Academy of Sciences (Y31M5A211). We thank Prof. Catherine Madzak for offering transformation protocols of Y. lipolytica.
Supporting information
Supplementary Table 1—Strains and plasmids used in this work
Supplementary Table 2—Primers used in this work
Supplementary Table 3—Procedure of elution for HPLC analysis
Supplementary Figure 1—Construction of engineering β-carotene biosynthesis pathway from X. dendrorhous in Y. lipolytica, namely XD
Supplementary Figure 2—A histogram representing the copy number of each cassette in mutant strain CIBTS 1176 by realtime qPCR
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supporting information
Supplementary Table 1—Strains and plasmids used in this work.
Supplementary Table 2—Primers used in this work.
Supplementary Table 3—Procedure of elution for HPLC analysis.
Supplementary Fig. 1—Construction of engineering β-carotene biosynthesis pathway from X. dendrorhous in Y. lipolytica, namely XD.
Supplementary Fig. 2—A histogram representing the copy number of each cassette in mutant strain CIBTS 1176 by realtime Qpcr.
Rights and permissions
About this article
Cite this article
Gao, S., Han, L., Zhu, L. et al. One-step integration of multiple genes into the oleaginous yeast Yarrowia lipolytica . Biotechnol Lett 36, 2523–2528 (2014). https://doi.org/10.1007/s10529-014-1634-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-014-1634-y