Abstract
Ankyrin repeat domain 52 (ANKRD52) is a regulatory component of the protein phosphatase 6 (PP6) holoenzyme. Evidence has emerged to suggest involvement of ANKRD52 in tumor metastases and cancer cell escape from T cell-mediated elimination and immunotherapy but there has been no research across different cancer types. The current study explored the biological functions of ANKRD52 by combining data from many databases. The aim was to expose new diagnostic or treatment biomarkers for malignant tumors. The roles of ANKRD52 with respect to immunotherapy in 33 human cancer types were analyzed by combining data from The Cancer Genome Atlas (TCGA), Genotype-Tissue Expression (GTEx), Cancer Cell Line Encyclopedia (CCLE), UCSC Xena, the Tumor Immune Estimation Resource (TIMER), TISIDB and Cellminer. Bioinformatics methods were used to analyze the association between ANKRD52 expression and prognosis, immunological indicators (immune cell infiltration, ESTIMATE scores and tumor microenvironment (TME) signatures), tumor mutational burden (TMB), microsatellite instability (MSI) and drug sensitivity. ANKRD52 expression was generally higher in 24 tumor tissues than in normal tissues and was associated with poor prognosis, especially in kidney chromophobe (KICH). Lower expression was observed in advanced cancer. ANKRD52 expression was strongly linked to major immunological indicators, such as immune cell infiltration, ESTIMATE scores, TME signatures, as well as expression of immune and tumor-related genes. Expression was also associated with indicators of immunotherapy efficacy and outcome, such as TMB in 7 cancer types and MSI in 12. In addition, ANKRD52 expression was linked to sensitivity to a number of anticancer drugs. ANKRD52 had a distinct immune function in breast invasive carcinoma (BRCA) that correlated negatively with most immune indicators. Expression was enriched in proliferation-, differentiation- and metabolism-related pathways and linked to other immune cells and TME signatures. A nomogram to predict 3- or 5-year overall survival (OS) of patients with BRCA was constructed. ANKRD52 may have utility as an oncological and immunological biomarker. New insights into oncogenesis are presented and the development of ANKRD52-targeting to increase the therapeutic efficacy of immunotherapy combined with chemotherapy explored.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cancer is the leading cause of worldwide morbidity and mortality (Sung et al. 2021; Siegel et al. 2022) and is the first or second leading cause of death before the age of 70 in 112 out of 183 countries, ranking third or fourth in a further 23 countries, according to estimates from the World Health Organization (WHO) for 2019 (World Health Organization (WHO) 2020). Moreover, the COVID-19 pandemic has threatened to impair cancer detection and treatment (Yabroff et al. 2022). Substantial diversity in the global demarcation of leading cancer types (Sung et al. 2021) indicates the necessity of pan-cancer research in examining the extraordinary variety of cancer types. Current cancer treatment strategies include surgery, chemotherapy, radiotherapy, targeted therapy and immunotherapy, of which immune checkpoint blockade (ICB) has become prominent (Ribas and Wolchok 2018; Sung et al. 2021). However, drug resistance, side effects and other issues contribute to prognosis and survival rates remaining poor (Sung et al. 2021). More sensitive tumor biomarkers and additional therapeutic targets are required to improve the detection and treatment of cancer.
Ankyrin repeat proteins are ubiquitous and common mediators of protein–protein interactions (Sedgwick and Smerdon 1999). Ankyrin repeat domain 52 (ANKRD52) comprises 28 copies of the ankyrin motif and is a regulatory subunit of the protein phosphatase 6 (PP6) holoenzyme (Stefansson et al. 2008), a eukaryotic serine-threonine phosphatase conserved from yeast to humans. The PP6 holoenzyme features catalytic (PP6c), first regulatory (PP6R1, PP6R2 or PP6R3) and second regulatory subunits that contain one of the three ankyrin repeat proteins (ANKRD28, ANKRD44 or ANKRD52) (Stefansson et al. 2008).
A previous CRISPR-Cas 9 screening study has indicated that ANKRD52 is a biomarker for tumor metastasis and that expression was related to late-stage in lung cancer (Chen et al. 2015). Further study of lung cancer has demonstrated suppression of tumor progression by ANKRD52-PP6c-mediated PAK1 dephosphorylation and suppression of ANKRD52 transcription by the transcriptional coactivator with PDZ-binding motif, TAZ (Lee et al. 2021), a Hippo signaling effector known to be elevated in the epithelial-mesenchymal transition and tumor invasion (Chan et al. 2008; Lei et al. 2008). In addition, ANKRD52 has been shown to have a role in cancer cell escape from T cell-mediated elimination and immunotherapy (Song et al. 2021). A combination of immune selection and CRISPR screen validation in mice, identified ANKRD52 as a modulator of JAK-STAT-interferon-γ signaling and antigen presentation in cancer cells by abolishing miR-155-targeted silencing of suppressor of cytokine signaling 1 (SOCS1). Thus, ANKRD52 is a novel tumor-associated gene and a promising target for immunotherapy. However, most previous studies have been restricted to a single cancer-type and pan-cancer research on ANKRD52 is lacking.
The current study combined data from TCGA, GTEX, CCLE, UCSC Xena, TIMER, TISIDB and Cellminer. ANKRD52 expression and prognostic value was investigated in 33 commonly occurring human cancers. Links between ANKRD52 expression and immunological indicators, such as immune cell infiltration, ESTIMATE scores, TME signatures, expression of immune and tumor-related genes were found. TMB and MSI were also analyzed to expose immunotherapy effectiveness and outcome in relation to ANKRD52 expression. In addition, ANKRD52 effects on drug sensitivity were evaluated through semi-inhibitory concentration (IC50) values of regularly used anticancer medications. Apart from conducting comprehensive studies on various types of cancers, our research focused specifically on the significance of ANKRD52 in predicting breast invasive carcinoma (BRCA) prognosis. Considering the alarmingly high prevalence and devastating impact of BRCA on women worldwide, the effectiveness of immunotherapy in combating this disease has not yet reached desirable levels. Hence, our investigation aimed to investigate ANKRD52 as a promising therapeutic target for enhancing patient outcomes. Our findings revealed that ANKRD52 exhibits distinct immune functions in BRCA. To provide accurate prognostic predictions, we developed nomograms capable of estimating the 3- or 5-year overall survival (OS) rates for affected individuals. In conclusion, this study found that ANKRD52 has potential as an oncological and immunological biomarker. Insights into oncogenesis are offered and the development of therapeutic ANKRD52-targeting strategies to improve the therapeutic efficacy of immunotherapy combined with chemotherapy explored.
Materials and Methods
Data Processing and Differential Expression Analysis
RNA sequencing and related clinical data of 33 kinds of different cancer and normal tissues were investigated by merging data from Genotype-Tissue Expression (GTEx) (https://commonfund.nih.gov/GTEx) (Consortium 2015) and The Cancer Genome Atlas (TCGA) (Cancer Genome Atlas Research 2013) downloaded via NCI’s Genomic Data Commons (GDC) portal (https://portal.gdc.cancer.gov/) (Grossman et al. 2016). Data on single nucleotide polymorphisms were also obtained from TCGA. Gene expression data from 30 different types of tumor cell lines were acquired from the Cancer Cell Line Encyclopedia (CCLE) database (https://portals.broadinstitute.org/ccle/) (Barretina et al. 2012). The mutation landscapes were drawn with R package “ComplexHeatmap” using SNP-related data from TCGA database.
Survival Analysis of Relationships Between ANKRD52 and Prognosis
Data downloaded from UCSC Xena (http://xena.ucsc.edu/) (Goldman et al. 2020) were divided into high- (Hexp) and low-expression (Lexp) groups, according to the median level of ANKRD52 expression. Overall survival (OS) and progression-free interval (PFI) were evaluated by Kaplan–Meier analysis (P < 0.05) using R packages, “survival” and “survminer”. Cox analysis was performed to ascertain the pan-cancer association between ANKRD52 expression and survival using the R packages “survival” and “forestplot”.
Correlation of the ANKRD52 Expression with Tumor Immune Microenvironment
Relative scores for 22 immune cells in 33 cancers were calculated by the metagene algorithm CIBERSORT to indicate immunocyte phenotypes (Chen et al. 2018a). The relationship between the ANKRD52 expression profile and the immune system was investigated using the Tumor Immune Estimation Resource (TIMER) database (https://cistrome.shinyapps.io/timer/) (Li et al. 2017, 2020). Interactions between ANKRD52 and immune-related genes, such as the major histocompatibility complex (MHC), immune checkpoint, immune activation, immunosuppressive, chemokine and chemokine receptor proteins, were assessed via TISIDB (http://cis.hku.hk/TISIDB) (Ru et al. 2019). Gene-expression signature scores reported to correlate with tumor microenvironment (TME), such as TMEscoreA and TMEscoreB, were also included (Mariathasan et al. 2018; Zeng et al. 2021). All results were evaluated and visualized using the R-packages, “ggplot2”, “ggpubr”, “cowplot”, “patchwork” and “showtext”, with a value of P < 0.05 considered significant.
Tumor Mutational Burden, Tumor Microsatellite Instability and ANKRD52 Expression
Tumor mutational burden (TMB) was defined as the total number of coding somatic mutations, insertions and deletions (indels) detected per million bases. The number of variants in exon length and the total number of nonsynonymous mutation sites for each tumor sample was calculated and divided by the total length of the protein-coding region. The microsatellite instability (MSI) score of each patient in the TCGA was derived from a previously published study (Bonneville et al. 2017).
Pan-Cancer Analysis of the Biological Significance of ANKRD52
Gene set enrichment analysis (GSEA) and gene set variation analysis (GSVA) were used to assess the biological roles of ANKRD52 in tumors. R-package, “limma”, was utilized to analyze differentially expressed genes in Hexp and Lexp groups. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was conducted to investigate biological functions and pathways correlated to ANKRD52 expression in the gene set, c2.cp.kegg.v7.4, and GSVA was conducted on the hallmark gene set, c2.all.v7.4, from the MSigDB database (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp) (Hanzelmann et al. 2013) using R-packages, “org.Hs.eg.db”, “clusterProfiler” and “enrichplot”.
Drug Sensitivity Associated with ANKRD52 Expression
The Cellminer database (http://discover.nci.nih.gov/cellminer/) (Reinhold et al. 2012) is a set of web-based genomic and pharmaceutical tools for investigating drug and transcript patterns in the NCI-60 cell line. Data on NCI-60 drug sensitivity and RNA-seq was downloaded to correlate ANKRD52 expression and sensitivity to commonly used anti-cancer drugs. A value of P < 0.05 was considered to indicate statistical significance.
Data from the largest pharmacogenomics database, Genomics Database for Drug Sensitivity in Cancer (GDSC) (https://www.cancerrxgene.org/), was analyzed by the R package, “pRRophetic” (Geeleher et al. 2014), to predict the chemotherapy sensitivity of BRCA samples. Semi-inhibitory concentration (IC50) values of chemotherapeutic drugs were obtained by regression and accuracy of regression and prediction tested by cross-validation with the GDSC training set.
Construction of a Predictive Nomogram
A nomogram was constructed from ANKRD52 expression levels and other clinicopathological parameters to enable clinical predictions, such as 3- and 5-year OS, for BRCA. Calibration curve analysis was performed to confirm the nomogram’s clinical reliability by using “rms” package. ROC curves are plotted using the “survivalROC” R package. The dataset was randomly divided into training and test sets in a ratio of 1:1.
Statistical Analysis
Statistical analyses were conducted using R language (version 4.0). A two-sided t-test was performed to compare differences among groups. Hazard ratios (HRs) and 95% confidence intervals were calculated using univariate survival analysis. Kaplan–Meier (K–M) analysis was conducted to estimate patient survival time according to ANKRD52 expression. Cox regression analysis, nomogram model and calibration curve analysis were used to evaluate the prognostic value of the ANKRD52 expression signature in BRCA with P < 0.05 set as the significance threshold for all statistical analyses.
Results
ANKRD52 Expression and Clinical Phenotype in Pan-Cancer
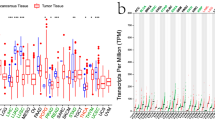
RNA-seq data of 33 different kinds of cancer (Table 1) and normal tissue types from TCGA and GTEX were analyzed. ANKRD52 expression was significantly higher in 24 cancer types, ACC, BLCA, BRCA, CESC, CHOL, COAD, ESCA, GBM, HNSC, KICH, LAML, LGG, LIHC, LUAD, LUSC, OV, PAAD, PRAD, SKCM, STAD, TGCT, THCA, UCEC and UCS (Fig. 1A). Similar results were obtained when the analysis was restricted to TCGA data and the expression of ANKRD52 shown statistical differences in 14 different tumor-normal tissue groups (Supplementary Fig. 1). ANKRD52 expression among normal tissues was generally lower. ANKRD52 expression in cancer cell lines from CCLE data is shown in Fig. 1B.
Differential expression of ANKRD52 and clinical correlation. A Differential expression of ANKRD52 in 33 normal and human tumor tissues; B ANKRD52 expression in 30 human cancer cells; C Significant clinical correlation between ANKRD52 and cancer stage in 6 cancer types, KICH, KIRP, LIHC, PAAD, TGCT and THCA. *P < 0.05, **P < 0.01 and ***P < 0.001
ANKRD52 expression was also measured in patients with stage I, II, III and IV cancers. Expression correlated with stage in KICH, KIRP, LIHC, PAAD, TGCT and THCA (Fig. 1C). There was greater variation in ANKRD52 expression when comparing stage I cancer with stages II or III but non-significant differences among higher stage III and IV tumors (Fig. 1C). LIHC patients showed the most statistically significant changes, especially between stages I and III (P < 0.001; Fig. 1C).
Prognostic Value of ANKRD52
The survival metrics, OS and PFI, were assessed in the Hexp and Lexp groups. Cox regression analysis of the 33 cancer types showed that ANKRD52 expression affected OS in 8 types of cancer, BRCA (P = 0.006), KICH (P = 0.005), LGG (P = 0.002), LIHC (P = 0.001), MESO (P < 0.001), PAAD (P = 0.004), SKCM (P < 0.001) and STAD (P < 0.001; Fig. 2A). Kaplan–Meier survival curves also linked high ANKRD52 expression with poor OS in patients with BRCA, KICH, LGG, LIHC, MESO and SKCM, although no relationship was seen for PAAD and STAD (P < 0.05; Fig. 2B). Cox regression analysis showed correlation of expression with PFI in 11 of the 33 cancers examined, BLCA (P = 0.043), BRCA (P = 0.006), KICH (P = 0.017), KIRP (P = 0.001), LGG (P < 0.001), LIHC (P < 0.004), MESO (P < 0.001), PAAD (P = 0.014), PRAD (P = 0.035), SKCM (P = 0.039) and UVM (P = 0.020; Fig. 2C). The association of ANKRD52 upregulation with poor PFI was particularly pronounced in BRCA, LGG, LIHC, MESO and UVM (P < 0.05; Fig. 2D).
Prognostic association between ANKRD52 expression and overall survival (OS) and progression-free interval (PFI). A Forest plot of hazard ratios of ANKRD52 related to OS; B OS curves for BRCA, KICH, LGG, LIHC, MESO and SKCM; C Forest plot of hazard ratios of ANKRD52 related to PFI; D PFI survival curves for BRCA, LGG, LIHC, MESO and UVM. A hazard ratio > 1 suggests that ANKRD52 expression is a risk factor impacting survival. *P < 0.05, **P < 0.01 and ***P < 0.001
ANKRD52 Expression, Immune Cell Infiltration and the Tumor Microenvironment
The TME is composed of the extracellular matrix, immune cells, growth factors, inflammatory mediators and cancer cells with the tumor immune microenvironment (TIME) being acknowledged to have an impact on immunotherapy. ANKRD52 has been reported to affect the efficacy of tumor immunotherapy (Song et al. 2021) and the relationships among ANKRD52 expression, immune cell infiltration and the tumor microenvironment were assessed. Infiltration by 22 immune-related cells in the Hexp and Lexp groups was evaluated using the CIBERSORT algorithm. Despite the presence of some variation, ANKRD2 expression showed the greatest correlation with infiltration by neutrophils in 11 types of cancers, M0 macrophages in 10 types, resting dendritic cells in 12 types, CD8+ T cells in 12 types and activated NK cells in 10 types (Fig. 3A). The greatest diversity of infiltrating immune cells was found for PRAD with 12 kinds, LUAD with 12 kinds, UCEC with 9 kinds, LIHC with 8 kinds, LUSC with 8 kinds and BRCA with 8 kinds (Fig. 3A). The link between immune cell infiltration and ANKRD52 expression was analyzed by 6 algorithms from the TIMER database (Fig. 3B).
ANKRD52 expression and immunocyte infiltration and tumor microenvironment signature. A Estimation of the association between ANKRD52 expression and immune infiltration by CIBERSORT algorithm; B Infiltration by different immune cell types and the association with ANKRD52 expression from TIMER database; C Correlation between ANKRD52 expression and stromal score, immune score, ESTIMATE score and tumor purity; D Association of ANKRD52 with different TME signatures. *P < 0.05, **P < 0.01 and ***P < 0.001
The ESTIMATE algorithm from the TIMER database was used to calculate immune scores, ESTEMATE scores, stromal scores and tumor purity (Meng et al. 2020). A negative correlation emerged between ANKRD52 expression and immune, ESTEMATE and stromal scores and a positive correlation with tumor purity for BRCA, KIRP, LAML, LUAD, LUSC, PCPG, SARC, SKCM and UCEC (Fig. 3C). Similar correlations were found for CESC, PRAD and THCA with the exception of a statistically insignificant relationship with stromal score (Fig. 3C). ANKRD52 also showed a strong positive correlation with some gene expression signatures reported to reflect TME, such as nucleotide excision repair, mismatch repair, DNA replication, DNA damage response and base excision repair and a negative correlation was found with TME score for most cancer types (Fig. 3D). A positive correlation was found between ANKRD52 expression and most gene-expression signature scores for LIHC, LGG, KIRP, HNSC, MESO and DLBC, but negative for SKCM and BRCA (Fig. 3D).
Co-expression of ANKRD52 with Key Regulatory Genes
Co-expression analyses of the ANKRD52 gene with those involved in immune checkpoints, immunostimulation, immunoinhibition, major histocompatibility complex (MHC), chemokines and chemokine receptors were performed. The heatmap shows a negative correlation of most immune-related genes with ANKRD52 expression (Fig. 4A).
Co-expression analyses of ANKRD52 expression with key immune- and tumor-related genes. A Correlation between ANKRD52 and immune-related genes encoding immune checkpoints, immunostimulatory, immunoinhibitory, major histocompatibility complex (MHC), chemokine and chemokine receptors; B Correlation between ANKRD52 and tumor-related genes implicated in TGF-beta signaling, TNFA signaling via NF-κB signaling, hypoxia, pyroptosis, DNA repair, autophagy and ferroptosis. *P < 0.05, **P < 0.01 and ***P < 0.001
Many cancer-related genes implicated in TGF-beta signaling, TNFA signaling via NF-κB signaling, hypoxia, pyroptosis, DNA repair, autophagy and ferroptosis also showed a negative correlation for co-expression with ANKRD52 in almost all samples (Fig. 4B).
ANKRD52 Expression, Tumor Mutational Burden and Tumor Microsatellite Instability
Patients with high TMB or high MSI are the most likely to benefit from immune checkpoint inhibitor therapy (Choucair et al. 2020; Velzen et al. 2020). ANKRD52 expression correlated positively with TMB for UCEC, READ, CESC, LUAD, COAD and ESCA and negatively for THCA (Fig. 5A). Similarly, a positive correlation was found between ANKRD52 expression and MSI for CESC, SARC, LUSC, COAD, ESCA, READ, LGG, LUAD, LIHC and UCEC and a negative correlation for DLBC and HNSC (Fig. 5A). A general conclusion may be drawn that upregulation of ANKRD52 tends to suggest higher TMB and MSI in the majority of cancers.
ANKRD52 as a Predictor of Drug Sensitivity
Chemotherapy combined with surgery is recommended for many early-stage cancers and the Cellminer database was used to assess the association of ANKRD52 expression with drug sensitivity. Tolerance to the following drugs showed the highest positive association with high ANKRD52 expression: Everolimus, Erlotinib, Ibrutinib, Irofulven, Rapamycin, Vandetanib, Lapatinib, Bleomycin, Gefitinib, Afatinib, Temsirolimus and 5-fluoro deoxy uridine whereas Paclitaxel, Oxaliplatin and Lapachone showed the highest negative associations (Table 2; Fig. 6A).
Clinical and Immunological Features of ANKRD52 in BRCA
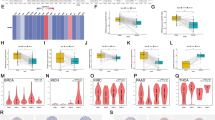
ANKRD52 expression was significantly higher on both transcriptional and translational levels in cancerous tissue samples from BRCA patients, in whom expression correlated negatively with immunological markers, but no clear relationship with cancer stage could be seen (Fig. 7A, B; Supplementary Fig. 2A). Cox regression analysis and Kaplan–Meier survival curves indicated ANKRD52 expression as a risk factor resulting in poor OS and PFI and as having prognostic utility for BRCA (Fig. 7C, D). Genes positively and negatively correlated with ANKRD52 and ANKRD52 mutation patterns in the Hexp and Lexp groups were also investigated (Supplementary Fig. 2B, C). GSEA analysis of the c2.cp.kegg.v7.4 dataset and GSVA analysis of the c2.all.v7.4 datasets were also used to identify enrichment patterns in Hexp and Lexp. The 5 pathways most clearly associated with ANKRD52 expression were Alzheimer’s disease, cytokine-cytokine receptor interaction, endocytosis, Huntington’s disease and ubiquitin-mediated proteolysis in the GSEA dataset. Endocytosis and ubiquitin-mediated proteolysis were related to high ANKRD52 expression (Fig. 7E). 19 out of 50 hallmark pathways were enriched in the GSVA dataset, including proliferation- and differentiation-related pathways (regulation of mitotic spindle, G2/M checkpoint, E2F transcription factors, cell polarity and hedgehog signaling), metabolic pathways (regulation of heme metabolism, protein secretion and bile acid metabolism) and sex hormone-related pathways (regulation of estrogen early and late response and androgen response; Fig. 7F).
Biological and immunological involvement of ANKRD52 in BRCA. A Transcription of ANKRD52 in normal and BRCA tumor tissues; B ANKRD52 protein in normal and BRCA tumor tissues; C, D OS and PFI curves with ANKRD52 expression; E Top 5 ANKRD52-related pathways enriched in KEGG analysis of GSEA; F Hallmark pathway analysis of ANKRD52 from GSVA; G Immune cell infiltration in Hexp and Lexp; H TME signatures in Hexp and Lexp; I ANKRD52 expression and chemosensitivity. *P < 0.05, **P < 0.01 and ***P < 0.001
ANKRD52 was shown to be negatively related to multiple TME indicators in BRCA and evaluation of immunological significance in the Hexp and Lexp groups produced similar results. Lexp showed greater enrichment of plasma cells, CD8+ T cells, T follicular helper cells, regulatory T cells (Treg), Gamma Delta T cells, activated NK cells and activated dendritic cells than Hexp. Resting memory CD4+ T cells, M2 macrophages and resting mast cells were more highly enriched in Hexp than Lexp. Most signature scores, including CD8+ T-effector, immunological checkpoint, antigen processing machinery, TMEscoreA, Pan-F-TBRs, EMT3 and TMEscoreB, were greater for Lexp than Hexp, except DNA damage response (Fig. 7H). ANKRD52 expression correlated negatively with that of most immune- and cancer-associated genes (Supplementary Fig. 3).
The R package, “pRRophetic”, was used to predict the effect of ANKRD52 expression on chemosensitivity to common chemotherapeutic agents. Comparison of IC50 values revealed a significant relationship between ANKRD52 expression and sensitivity to Axitinib, AZD8055, Lapatinib, Camptothecin, Gemcitabine and Erlotinib (Fig. 7I). In addition, the mutation landscapes of the top 30 mutation frequency genes in ANKRD52 high- and low-expression patients were drawn with the R package “ComplexHeatmap” (Supplementary Fig. 5). The results showed that GATA3 mutation frequency was high and HUWE1 mutation frequency was low in the ANKRD52 high-expression group of BRCA. (Supplementary Fig. 2C).
Prognostic Nomogram for BRCA Patients
A nomogram to estimate 3- and 5-year OS of BRCA patients was constructed from the results of logistic regression analysis. ANKRD52 expression was found to contribute to the prediction efficacy for BRCA samples (Fig. 8A). The calibration curves of the nomogram for 3- and 5-year OS prediction showed good correspondence with actual observations (Fig. 8B). The area under the ROC curve (AUC) was 0.547 and 0.535 for 3 and 5 years, respectively (Fig. 8C, D).
Discussion
Breast, lung and colon tumors are the three most common cancers contributing to high worldwide cancer morbidity and mortality rates (Sung et al. 2021; Siegel et al. 2022). Pan-cancer research is designed to overcome the diversity of prevalent cancer-types and to identify cancer signatures and contribute to cancer prevention. The current report identified higher levels of ANKRD52 expression among 33 types of cancer than in normal tissues using data from TCGA and GTEX. The potential for ANKRD52 to predict efficacy of immunotherapy was assessed by examination of differences between Hexp and Lexp. On this basis, the specific immunotherapeutic role of this gene in BRCA was further explored. Further study allowed the construction of a nomogram for 3- and 5-year OS prediction in BRCA.
Data from CCLE confirmed the tendency towards higher ANKRD52 expression at the cancer cell level. ANKRD52 has previously been described as a novel cancer suppressor due to its inhibitory effect on metastasis (Chen et al. 2015; Lee et al. 2021). By contrast, the current study focused on the differential in expression of this gene between tumor and normal tissues. The contrasting results obtained via these two approaches suggest a complex relationship between ANKRD52 expression and tumorigenesis. ANKRD52 expression was found to vary between low (stages I and II) and high (stages III and IV) stage cancer. The decrease in ANKRD52 level at later stages is consistent with similar findings in lung adenocarcinoma based on three datasets from the Oncomine database (Rhodes et al. 2004) and representative immunohistochemistry analysis (Lee et al. 2021).
Cox and K-M analyses were performed to evaluate any prognostic value of ANKRD52 level and found this to be a risk factor for poor OS and PFI. Low expression was associated with better prognosis in 8–11 cancers, especially KICH. Ting-Fang Lee et al. found high ANKRD52 expression to be non-significantly associated with a trend towards longer survival using data from GEPIA (Tang et al. 2017) and PROGgene (Goswami and Nakshatri 2013; Lee et al. 2021). The current investigation of LUAD produced a similar non-statistically significant result, indicating that the role of ANKRD52 in LUAD requires further validation.
Immunotherapy has been shown to be effective against various types of cancers (Ribas and Wolchok 2018; Sung et al. 2021). However, many patients receiving PD-1/PD-L1 ICB therapy fail to respond or develop resistance, often due to poor antigen presentation and signaling (Zaretsky et al. 2016; Sade-Feldman et al. 2017; Sharma et al. 2017; Sucker et al. 2017). A recent study combining immune selection and CRISPR screen validation in mice suggested that ANKRD52 was a key modulator of cancer immunity (Song et al. 2021). ANKRD52 knockout downregulated JAK-STAT-interferon-γ signaling and antigen presentation by controlling miR-155-targeted silencing of SOCS1. This gave a selective advantage for tumor cells against PD-1 independent T cell-mediated immunity and may explain the decrease of ANKRD52 in most high stage cancers. The proportions, although not the activation or cytotoxicity, of CD4+, CD8+ T cells and exhausted T cells (LAG3+ PD1+ or TIM3+ PD1+) were significantly decreased in ANKRD52-null mouse tumors. The current study found ANKRD52 gene expression to be highly associated with immune infiltration, positively correlated with neutrophil and M0 macrophage abundance and negatively correlated with resting dendritic cells, CD8+ T cells and activated NK cells in most of the cancers. TME has been used as a marker of tumor sensitivity to immunotherapy (Wu and Dai 2017). ESTIMATE scores showed a negative correlation of ANKRD52 expression with TME immune and stromal cell content and a positive correlation with tumor purity in 12 cancers, indicating prognostic value for ANKRD52 levels. ANKRD52 was found to be negatively co-expressed with most immune-related genes encoding immunological checkpoint, immunostimulatory, immunoinhibitory, MHC, chemokine and chemokine receptor proteins and cancer-related genes involved in TGF-beta signaling, TNFA signaling via NF-κB signaling, hypoxia, pyroptosis, DNA repair, autophagy and ferroptosis. Thus, ANKRD52 expression is related to immune infiltration, TME content, tumor purity and key regulatory genes and has potential as a new target for immunotherapy across a broad range of tumors.
Two immunotherapeutic biomarkers, TMB and MSI, were shown to be associated with ANKRD52 in some cancers. TMB is defined as the total number of somatic mutations per region of tumor genome, providing a useful estimation of tumor-neoantigen load. In general, a high TMB is linked to production of more neoantigens and better immunotherapy response. TMB has been suggested as a predictor of immunotherapy efficacy and patient prognosis after immunotherapy (Snyder et al. 2014; Rizvi et al. 2015; Schumacher and Schreiber 2015; Chalmers et al. 2017; Yarchoan et al. 2017; Hellmann et al. 2018a, b, c; Hellmann and Paz-Ares 2018; Rizvi et al. 2018; Offin et al. 2019). MSI is described as a robust mutator phenotype induced by defective DNA mismatch repair and is a potential predictor of immune-checkpoint inhibitor (ICI) efficacy and prognosis (Gryfe et al. 2000; Boland and Goel 2010; Lee et al. 2019; Yamamoto and Imai 2019). ANKRD52 expression was usually positively correlated with TMB in 7 cancer types and with MSI in 12 cancer types, suggesting an impact of ANKRD52 expression on a patient's response to immune checkpoint suppression therapy. ANKRD52 was also correlated with the TMB-linked processes of nucleotide excision repair, mismatch repair, DNA replication, DNA damage response and base excision repair in most cancers (Mariathasan et al. 2018). Thus, ANKRD52 may affect TMB and MSI through a series of DNA damage response and repair pathways, enabling levels to indicate likely immunotherapy outcomes.
Chemotherapy resistance may cause treatment failure, metastasis and cancer recurrence (Gottesman et al. 2002; Borovski et al. 2013; Holohan et al. 2013; Mathijssen et al. 2014). Examination of drug IC50 values revealed the involvement of ANKRD52 in drug tolerance. Thus, the gene may be a target for reversal of drug resistance, widening treatment choice when gene expression is changed in conjunction with chemotherapy. Future in vitro and in vivo experiments to measure ANKRD52 expression in drug-sensitive and drug-resistant clinical samples would allow changes in tumor cell sensitivity to chemotherapeutic drugs to be evaluated.
In light of the above comprehensive studies on pan-cancer, we have delved into the clinical significance of ANKRD52 in BRCA, which is also of interest to our department. Given its status as the predominant cancer among women and the limited efficacy of current immunotherapy approaches, there is a pressing need to delve into novel targets for enhancing BRCA immunotherapy. And in our analysis, it was found that ANKRD52 showed a distinct immunotherapeutic value in BRCA, where the expression of the gene was found to be negatively associated with most immunological indicators, including immunocytes, TME-related signature scores, immune- and cancer-related genes and MSI, which is in contrast to many other cancers. Mutation analysis showed a higher frequency of mutations in GATA3 and a lower frequency in HUWE1 in the ANKRD52 high-expression group of BRCA (Supplementary Fig. 2C). GATA3 is a valuable marker for confirming the diagnosis of many epithelial or mesenchymal neoplasms, both diagnostically and prognostically (Khazaeli Najafabadi et al. 2021). And HUWE1 is a key regulator of DNA damage response, transcription, autophagy, apoptosis and metabolism in a variety of cancers (Gong et al. 2020). Therefore, we will subsequently focus on how GATA3 and HUWE1 mutations regulate ANKRD52 expression by constructing wild-type or mutant cells of these two genes in breast invasive carcinoma cells. In the GSEA dataset, high ANKRD52 expression was associated with endocytosis and ubiquitin-mediated proteolysis and low expression with neurodegenerative disorders, such as Alzheimer’s disease and Huntington’s disease, as well as cytokine-cytokine receptor interactions. High ANKRD52 expression was enriched in proliferation- and differentiation-related pathways (e.g., regulation of mitotic spindle, G2/M checkpoint, E2F transcription factors, cell polarity, hedgehog signaling), metabolic- related pathways (e.g., regulation of heme metabolism, protein secretion and bile acid metabolism) and sex hormone-related pathways (e.g., regulation of estrogen early, late response and androgen response from GSVA analysis). ANKRD52 may affect pathways of cell proliferation, differentiation and metabolism through the JAK (janus tyrosine kinase)-STAT (signal transducer and activator of transcription) pathway in BRCA (Winthrop 2017). Examination of the TME in Hexp and Lexp did not reveal significant differences in neutrophils, M0 macrophages and resting dendritic cells associated with ANKR52 but 10 other immune cell types, plasma cells, CD8+ T cells, T follicular helper cells, regulatory T cells (Tregs), Gamma Delta T cells, activated NK cells and activated dendritic cells were more enriched in the Lexp group than in the Hexp, with resting memory CD4+ T cells, M2 macrophages and resting mast cells showing the opposite trend. ANKRD52 expression was more weakly associated with nucleotide excision repair, mismatch repair, DNA replication, base excision repair and TME score in BRCA than other cancers. However, the negative correlation with CD8+ T-effector, immunological checkpoint, antigen processing machinery, Pan-F-TBRs, EMT3, TMEscoreA and TMEscoreB was stronger, perhaps indicting a weaker ANKRD52-mediated DNA repair capacity in BRCA. ANKRD52 expression was also adversely associated with the expression of immune- and cancer-associated genes (Supplementary Fig. 3). Notably, in association with TNFA signaling via NF-κB signaling, ANKRD52 was found to be co-expressed with complement 1q binding protein (C1QBP) (Fig. 4B2), which is overexpressed in breast cancer and promotes cancer metastasis and paclitaxel resistance (Wu et al. 2020, 2021). In summary, the immunological significance of ANKRD52 in BRCA appears quite different from that in other cancers, suggesting an immunotherapeutic prognostic value for ANKRD52 in BRCA associated with different immune cell infiltration and TME signatures. In addition, sensitivities to Axitinib, AZD8055, Lapatinib, Camptothecin, Gemcitabine and Erlotinib were related to ANKRD52. The efficacy of Gemcitabine and Erlotinib on breast and pancreatic cancer cells has been reported to depend on JAK-STAT signaling (Thoennissen et al. 2009; Uluer et al. 2012; Chen et al. 2018b). JAK-STAT cross-talk may enable cancer cells to switch pathways and evade sensitivity to targeted drugs. This signaling pathway is thus highlighted for further studies of ANKRD52 and drug tolerance.
Given the considerable clinical significance attributed to ANKRD52 in BRCA, we incorporated this gene into risk factors to develop prognostic nomograms for predicting the 3- and 5-year outcomes of BRCA patients. To ensure the reliability and robustness of our model, we randomly divided the datasets into train and test sets at a 1:1 ratio (Supplementary Fig. 4). The findings revealed that the expression of ANKRD52 contributed to the predictive accuracy of prognosis in BRCA patients, but the AUC results were not satisfactory, measuring 0.547 and 0.535 for the 3- and 5-year predictions, respectively. A previous study encompassed five cuproptosis/ferroptosis-related genes (ANKRD52, HOXC10, KNOP1, SGPP1, TRIM45) to establish a risk score model for predicting patient survival, yielding more promising results with respect to predictive efficacy and ROC curves (Li et al. 2023). It is prudent to consider the inclusion of additional candidate genes to construct a proportional hazards regression model, thereby enhancing the AUC and augmenting the overall predictive value of our study.
To the best of our knowledge, the current is the first study to examine the significance of ANKRD52 in pan-cancer. Bioinformatics analysis demonstrated that ANKRD52 affected patient prognosis and may modulate immunological indicators, such as immune cell infiltration, TME signatures, expression of immune and tumor-related genes, TMB, MSI and drug sensitivity across a variety of cancers. Further studies involving ANKRD52 overexpression or knockdown and in vitro and in vivo experiments are required to establish, as well as whether ANKRD52 may be confirmed as a prognostic and immunotherapy biomarker.
References
Barretina J et al (2012) The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 492(7428):290–290
Boland CR, Goel A (2010) Microsatellite instability in colorectal cancer. Gastroenterology 138(6):2073–2087
Bonneville R et al (2017) Landscape of microsatellite instability across 39 cancer types. JCO Precis Oncol 1:1–5
Borovski T et al (2013) Therapy-resistant tumor microvascular endothelial cells contribute to treatment failure in glioblastoma multiforme. Oncogene 32(12):1539–1548
Cancer Genome Atlas Research N et al (2013) The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 45(10):1113–1120
Chalmers ZR et al (2017) Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 9(1):34
Chan SW et al (2008) A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res 68(8):2592–2598
Chen S et al (2015) Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 160(6):1246–1260
Chen B et al (2018a) Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol 1711:243–259
Chen L et al (2018b) Combination of gemcitabine and erlotinib inhibits recurrent pancreatic cancer growth in mice via the JAK-STAT pathway. Oncol Rep 39(3):1081–1089
Choucair K et al (2020) TMB: a promising immune-response biomarker, and potential spearhead in advancing targeted therapy trials. Cancer Gene Ther 27(Suppl 5):1–13
Consortium G. T. (2015) Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348(6235):648–660
Geeleher P et al (2014) pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS ONE 9(9):e107468
Goldman MJ et al (2020) Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol 38(6):675–678
Gong X et al (2020) The structure and regulation of the E3 ubiquitin ligase HUWE1 and its biological functions in cancer. Invest New Drugs 38(2):515–524
Goswami CP, Nakshatri H (2013) PROGgene: gene expression based survival analysis web application for multiple cancers. J Clin Bioinforma 3(1):22
Gottesman MM et al (2002) Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer 2(1):48–58
Grossman RL et al (2016) Toward a shared vision for cancer genomic data. N Engl J Med 375(12):1109–1112
Gryfe R et al (2000) Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med 342(2):69–77
Hanzelmann S et al (2013) GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform 14:7
Hellmann MD, Paz-Ares L (2018) Lung cancer with a high tumor mutational burden. N Engl J Med 379(11):1093–1094
Hellmann MD et al (2018a) Tumor mutational burden and efficacy of nivolumab monotherapy and in combination with ipilimumab in small-cell lung cancer. Cancer Cell 33(5):853–861
Hellmann MD et al (2018b) Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med 378(22):2093–2104
Hellmann MD et al (2018c) Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell 33(5):843–852
Holohan C et al (2013) Cancer drug resistance: an evolving paradigm. Nat Rev Cancer 13(10):714–726
Khazaeli Najafabadi M et al (2021) Role of GATA3 in tumor diagnosis: a review. Pathol Res Pract 226:153611
Lee DW et al (2019) Tumor mutation burden and prognosis in patients with colorectal cancer treated with adjuvant fluoropyrimidine and oxaliplatin. Clin Cancer Res 25(20):6141–6147
Lee TF et al (2021) TAZ negatively regulates the novel tumor suppressor ANKRD52 and promotes PAK1 dephosphorylation in lung adenocarcinomas. Biochim Biophys Acta Mol Cell Res 1868(2):118891
Lei QY et al (2008) TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol 28(7):2426–2436
Li T et al (2017) TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res 77(21):e108–e110
Li T et al (2020) TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 48(W1):W509–W514
Li J et al (2023) Cuproptosis/ferroptosis-related gene signature is correlated with immune infiltration and predict the prognosis for patients with breast cancer. Front Pharmacol 14:1192434
Mariathasan S et al (2018) TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554(7693):544–548
Mathijssen RH et al (2014) Determining the optimal dose in the development of anticancer agents. Nat Rev Clin Oncol 11(5):272–281
Meng Z et al (2020) Using ESTIMATE algorithm to establish an 8-mRNA signature prognosis prediction system and identify immunocyte infiltration-related genes in Pancreatic adenocarcinoma. Aging 12(6):5048
Offin M et al (2019) Tumor mutation burden and efficacy of EGFR-tyrosine kinase inhibitors in patients with EGFR-mutant lung cancers. Clin Cancer Res 25(3):1063–1069
Reinhold WC et al (2012) CellMiner: a web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res 72(14):3499–3511
Rhodes DR et al (2004) ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6(1):1–6
Ribas A, Wolchok JD (2018) Cancer immunotherapy using checkpoint blockade. Science 359(6382):1350–1355
Rizvi NA et al (2015) Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348(6230):124–128
Rizvi H et al (2018) Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J Clin Oncol 36(7):633–641
Ru B et al (2019) TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics 35(20):4200–4202
Sade-Feldman M et al (2017) Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat Commun 8(1):1136
Schumacher TN, Schreiber RD (2015) [Special Issue Review] Neoantigens in cancer immunotherapy. Science 348(6230):69–74
Sedgwick SG, Smerdon SJ (1999) The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem Sci 24(8):311–316
Sharma P et al (2017) Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168(4):707–723
Siegel RL et al (2022) Cancer statistics, 2022. CA Cancer J Clin 72(1):7–33
Snyder A et al (2014) Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 371(23):2189–2199
Song TY et al (2021) Tumor evolution selectively inactivates the core microRNA machinery for immune evasion. Nat Commun 12(1):7003
Stefansson B et al (2008) Protein phosphatase 6 regulatory subunits composed of ankyrin repeat domains. Biochemistry 47(5):1442–1451
Sucker A et al (2017) Acquired IFNgamma resistance impairs anti-tumor immunity and gives rise to T-cell-resistant melanoma lesions. Nat Commun 8:15440
Sung H et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71(3):209–249
Tang Z et al (2017) GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 45(W1):W98–W102
Thoennissen NH et al (2009) Cucurbitacin B induces apoptosis by inhibition of the JAK/STAT pathway and potentiates antiproliferative effects of gemcitabine on pancreatic cancer cells. Cancer Res 69(14):5876–5884
Uluer ET et al (2012) Effects of 5-fluorouracil and gemcitabine on a breast cancer cell line (MCF-7) via the JAK/STAT pathway. Acta Histochem 114(7):641–646
Velzen M et al (2020) MSI as a predictive factor for treatment outcome of gastroesophageal adenocarcinoma. Cancer Treat Rev 86:102024
Winthrop KL (2017) The emerging safety profile of JAK inhibitors in rheumatic disease. Nat Rev Rheumatol 13(5):320
World Health Organization (WHO) (2020) Global health estimates 2020: deaths by cause, age, sex, by country and by region, 2000–2019. http://who.int/data/gho/data/themes/mortality-and-global-health-estimates/ghe-leading-causes-of-death. Accessed 11 Dec 2020
Wu T, Dai Y (2017) Tumor microenvironment and therapeutic response. Cancer Lett 387:61–68
Wu H et al (2020) Isobaric tags for relative and absolute quantitation in proteomic analysis of potential biomarkers in invasive cancer, ductal carcinoma in situ, and mammary fibroadenoma. Front Oncol 10:574552
Wu H et al (2021) Hypoxia-mediated complement 1q binding protein regulates metastasis and chemoresistance in triple-negative breast cancer and modulates the PKC-NF-kappaB-VCAM-1 signaling pathway. Front Cell Dev Biol 9:607142
Yabroff KR et al (2022) Association of the COVID-19 pandemic with patterns of statewide cancer services. J Natl Cancer Inst 114(6):907–909
Yamamoto H, Imai K (2019) An updated review of microsatellite instability in the era of next-generation sequencing and precision medicine. Semin Oncol 46(3):261–270
Yarchoan M et al (2017) Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med 377(25):2500–2501
Zaretsky JM et al (2016) Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med 375(9):819–829
Zeng D et al (2021) Tumor microenvironment evaluation promotes precise checkpoint immunotherapy of advanced gastric cancer. J Immunother Cancer 9(8):e002467
Funding
Funding was provided by Haiyan Fund Project of Harbin Medical University Cancer Hospital (Grant nos. JJQN 2019-07, JJMS 2022-06), National Natural Science Foundation of China (Grant no. 82202996), The Fundamental Research Funds for the Provincial Universities (Grant no. 2022-KYYWF-0288), Heilongjiang Postdoctoral Financial Assistance (Grant no. LBH-Z22219), Heilongjiang Provincial Natural Science Foundation Outstanding Youth Project (Grant no. YQ2023H022), China Postdoctoral Science Foundation (Grant no. 2023MD 734172).
Author information
Authors and Affiliations
Contributions
H-ZY and M-CZ wrote the main manuscript text. H-ZY and HW prepared all figures. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10528_2023_10645_MOESM1_ESM.jpg
Supplementary file1 Supplementary Fig. 1: Expression of ANKRD52 in 14 types of tumour tissues and corresponding normal tissues from TCGA database. (JPG 1392 KB)
10528_2023_10645_MOESM2_ESM.jpg
Supplementary file2 Supplementary Fig. 2: Clinical and immunological features of ANKRD52 in BRCA. (A) Relationship of ANKRD52 expression and cancer stage. (B) Genes positively (B1) and negatively (B2) correlated with ANKRD52. (C) The mutation landscapes of top 30 genes with the highest mutation frequency in the ANKRD52 Hexp and Lexp groups. (JPG 4388 KB)
10528_2023_10645_MOESM3_ESM.jpg
Supplementary file3 Supplementary Fig. 3: The relationship of the expression of ANKRD52 and immune- and cancer-associated genes in BRCA. (JPG 4340 KB)
10528_2023_10645_MOESM4_ESM.jpg
Supplementary file4 Supplementary Fig. 4: Construction and validation of a nomogram for ANKRD52 in the train and test sets. (A) Test set of calibration curves for predicting 3- and 5-year OS in BRCA. (B) Test set of ROC curve for predicting 3-year OS in BRCA. (C) Test set of ROC curve for predicting 5-year OS in BRCA. (D) Train set of calibration curves for predicting 3- and 5-year OS in BRCA. (E) Train set of ROC curve for predicting 3-year OS in BRCA. (F) Train set of ROC curve for predicting 5-year OS in BRCA. (JPG 2820 KB)
10528_2023_10645_MOESM5_ESM.jpg
Supplementary file5 Supplementary Fig. 5: Mutation landscapes of top 30 genes with the highest mutation frequency between patients with high- and low-expression of ANKRD52. (A) ACC (B) BLCA (C) CESE (D) CHOL (E) COAD (F) DLBC (G) ESCA (H) GBM (I) HNSC (J) KICH (K) KIRC (L) KIRP (M) LAML (N) LGG (O) LIHC (P) LUAD (Q) LUSC (R) MESO (S) OV (T) PAAD (U) PCPG (V) PRAD (W) READ (X) SARC (Y) SKCM (Z) STAD (AA) TGCT (BB) THCA (CC) THYM (DD) UCEC (EE) UCS (FF) UVM. (JPG 8522 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yin, HZ., Zhang, MC. & Wu, H. Clinical and Immunological Significance of ANKRD52 in Pan-Cancer. Biochem Genet (2024). https://doi.org/10.1007/s10528-023-10645-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10528-023-10645-w