Abstract
A 14.5-kDa ribonuclease, with an optimum pH of 6 and a temperature optimum at 70°C, was isolated from fresh fruiting bodies of the edible mushroom Lyophyllum shimeiji. It was purified by ion exchange chromatography on DEAE cellulose, Q Sepharose, and SP Sepharose, followed by FPLC gel filtration on Superdex 75, and was adsorbed on all three ion exchangers. It showed the highest ribonucleolytic potency toward poly (U), 25% as much activity toward poly (C), and undetectable activity toward poly (A) and poly (G). Its ribonucleolytic activity at 100°C was similar to that at 20°C. It suppressed proliferation of hepatoma HepG2 cells and breast cancer MCF7 cells with an IC50 of 10 and 6.2 μM, respectively. It inhibited the activity of HIV-1 reverse transcriptase with an IC50 of 7.2 μM.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ribonucleases (RNases) are present in a diversity of organisms comprising bacteria (Hartley 1988; Zilhao et al. 1993), mushrooms (Kobayashi et al. 1992; Ng 2004; Ng and Wang 2004; Ngai and Ng 2004; Nomura et al. 1994; Wang and Ng 1999, 2001, 2003a, b, c, 2004a, b; Ye and Ng 2002a, b), flowering plants (Green 1994; Lam and Ng 2001b; Ng and Wang 2001; Wang and Ng 2000), and vertebrate animals (Adinolfi et al. 1995; Hofsteenge et al. 1989; Irie et al. 1988; Liao 1992; Sasso et al. 1991). In vertebrate animals, RNases have been reported from the brain (Sasso et al. 1991), kidney (Irie et al. 1988), liver (Hofsteenge et al. 1989), semen, and milk (Adinolfi et al. 1995; Matousek et al. 1995; Shapiro and Vallee 1987). Some of the ribonucleases demonstrate immunosuppressive antiviral (Silverman 1997), antiproliferative, and antitumor (Adinolfi et al. 1995) activities.
Mushrooms are a rich source of bioactive proteins, including lectins, antifungal proteins, ribosome inactivating proteins, and RNase (Adinolfi et al. 1995; Lam and Ng 2001a; Ng and Lam 2002; Ye and Ng 2002a, b). Different mushroom species produce RNases with different N-terminal sequences, pH, temperature optima, and polyhomoribonucleotide specificities (Kobayashi et al. 1992; Ng and Wang 2004; Ngai and Ng 2004; Nomura et al. 1994; Wang and Ng 1999, 2001, 2003a, b, c, 2004a, b; Ye and Ng 2002a, b). Pleurotes sajor-caju ribonuclease has a variety of activities, including antimitogenic activity toward mouse splenocytes, antiproliferative activity toward tumor cells, and growth-inhibitory activity toward fungi and bacteria (Ngai and Ng 2004). Thus, it would be worthwhile to isolate RNases from mushrooms that have not been investigated to see if they have unique characteristics. From the fruiting bodies of Lyophyllum shimeiji, a lectin, an antifungal protein and a ribosome inactivating protein have been isolated (Lam and Ng 2001a; Ng and Lam 2002). The purpose of the present study was to isolate and characterize an RNase from L. shimeiji.
Materials and Methods
Isolation of RNase
All chemicals used in this study were from Sigma Chemical Company, St. Louis, MO, USA, unless otherwise stated. Fresh edible mushroom Lyophyllum shimeiji (1 kg) was obtained from a local supplier of Yunnan Province, China. The fruiting bodies were homogenized in distilled water (3 ml/g). Following centrifugation of the homogenate, Tris–HCl buffer (pH 7.4, 1 M) was added to the resulting supernatant until its concentration reached 10 mM. The supernatant was fractionated by ion exchange chromatography on a 5 × 20 cm column of DEAE cellulose. After elution of unadsorbed proteins (fraction D1) with 10 mM Tris–HCl buffer (pH 7.4), adsorbed proteins were desorbed sequentially with 0.2 M NaCl and 1 M NaCl in the 10 mM Tris–HCl buffer (pH 7.4) to form fractions D2 and D3, respectively. Fraction D2 with ribonuclease activity was chromatographed on a 2.5 × 20 cm column of Q Sepharose (GE Healthcare). Unadsorbed proteins were eluted as fraction Q1, and adsorbed proteins were eluted as fractions Q2 and Q3 with a linear 0–1 M NaCl concentration gradient. Fraction Q2 with ribonuclease activity was dialyzed and subsequently applied on a 2.5 × 20 cm column of SP Sepharose (GE Healthcare). Unadsorbed proteins were eluted as fraction SP1, and adsorbed proteins were eluted as fractions SP2, SP3, and SP4 with a linear 0–1 M NaCl concentration gradient. Fraction SP3 with ribonuclease activity was dialyzed and subsequently further purified on a Superdex 75 h 10/30 column (GE Healthcare) in 0.2 M NH4HCO3 buffer (pH 8.5). The second resulting peak (SU2) represented purified RNase.
Molecular Mass Determination
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was carried out following the procedure of Laemmli and Favre (1973), using a 12% resolving gel and a 5% stacking gel. At the end of electrophoresis, the gel was stained with Coomassie Brilliant Blue. FPLC gel filtration was conducted in a Superdex 75 h 10/30 column calibrated with molecular-mass markers (GE Healthcare) using an AKTA Purifier (GE Healthcare).
Analysis of N-Terminal Amino Acid Sequence
Amino acid sequence analysis was performed using an HP G1000A Edman degradation unit and an HP1000 HPLC system (Lam et al. 1998).
Assay for Activity of Ribonuclease
The activity of the purified RNase toward yeast tRNA was determined by measuring the formation of acid-soluble, UV-absorbing species with the method of Wang and Ng (1999). The RNase was incubated with 200 μg tRNA in 150 μg of 100 mM MES buffer (pH 6) at 37°C for 1 h. The reaction was stopped by addition of 350 μl ice-cold 3.4% perchloric acid. After leaving on ice for 15 min, the sample was centrifuged (15,000g, 15 min) at 4°C. The OD260 of the supernatant was measured after appropriate dilution. One unit of enzymatic activity is defined as the amount of enzyme that brings about an increase in OD260 of one per minute in the acid-soluble fraction per milliliter of reaction mixture under the specified conditions.
Activity of RNase Toward Polyhomoribonucleotides
The ribonucleolytic activity of the purified RNase toward polyhomoribonucleotides was assayed with a modification of the method of Wang and Ng (2001). Incubation of RNase with 100 μg poly (A), poly (C), poly (G), or poly (U) in 250 μl of 100 mM MES buffer (pH 6.0) was performed at 37°C for 1 h, before introduction of 250 μl ice-cold 1.2 N perchloric acid containing 20 mM lanthanum nitrate to stop the reaction. After leaving on ice for 15 min, the sample was centrifuged at 15,000g for 15 min at 4°C. The absorbance of the supernatant, after appropriate dilution, was read at 260 nm, in the case of poly (A), poly (G), and poly (U), or at 280 nm, in the case of poly (C).
Assay of Antifungal Activity
Antifungal activity toward Mycosphaerella arachidicola and Physalospora piricola was assayed on 100 mm × 15 mm petri plates containing 10 ml potato dextrose agar. After the mycelial colony had formed, sterile blank paper disks (0.625 cm in diameter) were placed at a distance of 0.5 cm from the periphery of the mycelial colony. An aliquot (15 μl) of the mushroom RNase was added to a disk. The plates were incubated at 23°C for 72 h until mycelial growth had surrounded the disks containing the control and had formed crescents of inhibition around disks containing samples with antifungal activity (Ng and Wang 2001).
Assay of Hemagglutinating Activity
The assay was conducted using rabbit erythrocytes (Wang and Ng 2005).
Assay of Antiproliferative Activity on Tumor Cell Lines
HepG2 and MCF 7 cells were cultured in RPMI medium supplemented with 10% (v/v) fetal bovine serum (Invitrogen), 100 mg/l streptomycin, and 100 IU/ml penicillin, at 37°C in a humidified atmosphere of 5% (v/v) CO2. Cells were subsequently seeded into 96-well plates with a concentration of 2 × 103 cells/well and incubated for 24 h. Different concentrations of L. shimeiji RNase in 100 μl complete RPMI medium were then added to the wells and incubated for 72 h (Ngai and Ng 2004). After that MTT quantification assays were carried out to measure the cells’ viability. Briefly, 20 μl of a 5 mg/ml solution of 3-(4,5-dimethythiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) in phosphate-buffered saline was spiked into each well and the plates were incubated for 4 h. The plates were then centrifuged at 2500 rpm for 5 min. The supernatant was carefully removed, and 150-μl dimethyl sulfoxide was added in each well to dissolve the MTT formazan at the bottom of the wells. After 10 min, the absorbance at 590 nm was measured with a microplate reader. PBS was added into wells instead of protease as control.
Assay for HIV Reverse Transcriptase Inhibitory (HIV RT) Activity
The assay for HIV RT inhibitory activity was performed, as described by Zhao et al. (2009), using a nonradioactive enzyme-linked immunosorbent assay (ELISA) kit from Boehringer-Mannheim (Germany). The assay was carried out as stated in the protocol that came with the kit, except that each well contained 2 ng recombinant HIV-1 reverse transcriptase in a total reaction volume of 60 μl. It made use of the ability of reverse transcriptase to synthesize DNA, starting from the template/primer hybrid poly (dA)-oligo (dT) 15. In this assay, nucleotides labeled with digoxigenin and biotin in an optimized proportion were incorporated into the DNA, which was freshly synthesized by the reverse transcriptase (RT). The detection and quantification of synthesized DNA as a parameter for RT activity follows a sandwich ELISA protocol. The surface of the microtiter plate modules, which had been coated with streptavidin, allowed the binding of biotin-labeled DNA. An antibody, which had been conjugated to peroxidase, to digoxigenin (anti-DIG-POD) was then used to bind to the digoxigenin-labeled DNA. This was followed by the addition of peroxidase substrate. The peroxidase enzyme, conjugated to the antibody, then catalyzed the cleavage of the substrate to produce a colored product. The absorbance was measured at 405 nm with a microtiter (ELISA) reader and correlated to the level of RT activity. A fixed amount (4–6 ng) of recombinant HIV-1 reverse transcriptase was used. The activity of inhibition exhibited by the L. shimeiji RNase was determined as compared to a control without RNase.
Results
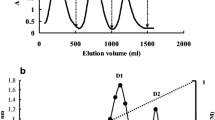
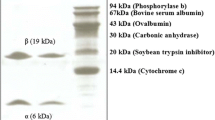
Chromatography of the fruiting body extract on DEAE cellulose gave rise to an unadsorbed fraction D1 and an adsorbed fraction D2 containing slightly less protein than D1. Ribonuclease activity was concentrated in fraction D2 (Table 1). Fraction D2 was resolved on Q Sepharose to produce a small unadsorbed fraction Q1, with negligible RNase activity, and two larger adjacent adsorbed fractions, Q2 and Q3, of approximately the same size. Ribonuclease activity was enriched in fraction Q2 (Table 1). Fraction Q2 was separated on SP Sepharose into an unadsorbed fraction SP1 and several adsorbed fractions, SP1-SP4 (Fig. 1). The bulk of ribonuclease activity resided in fraction SP3 (Table 1). Fraction SP3 was resolved on Superdex 75 into two fractions, SU1 and SU2, of about the same size (Fig. 2). Ribonuclease activity was confined to fraction SU2 (Table 1). Fraction SU2 displayed a single band with a molecular mass of 14.5 kDa in SDS–PAGE (Fig. 3).
The ribonucleolytic activity of the purified ribonuclease increased steadily from pH 3 until it reached its maximum at pH 6 and then decreased steadily from pH 6 until it reached its residual level at pH 9 (Fig. 4). The activity fell precipitously when the temperature was raised from 80 to 90°C. The activity observed at 100°C was similar to that at 20°C (Fig. 5). The ribonuclease exerted a specific ribonucleolytic activity of 0.1 U/mg toward poly (A) and poly (G), 3.5 U/mg toward poly (C), and 14.2 U/mg toward poly (U). The N-terminal sequence of the purified ribonuclease exhibited little similarity to counterparts from other mushrooms (Table 2). The protein was devoid of antifungal activity and hemagglutinating activity when tested at 300 μg.
At the concentrations of 2.5, 5, 10, and 20 μM, the purified RNase inhibited the proliferation of HepG2 cells by 3.4, 20.1, 50.4, and 75.3% and the proliferation of MCF7 cells by 14.6, 41.7, 68.5, and 84.9%, respectively. The IC50 values toward HepG2 cells and MCF7 cells were 10.0 and 6.2 μM, respectively.
At the concentrations of 0.8, 4, and 20 μM, the purified RNase inhibited HIV-1 reverse transcriptase by 5.2, 37.0, and 93.4%, respectively. The IC50 value was 7.2 μM.
Discussion
Like most other mushroom RNases (Ye and Ng 2002a, b), Lyophyllum shimeiji RNase is adsorbed on a cation exchanger. It resembles RNases from Pleurotus tuber-regium (Wang and Ng 2001) and Thelephora ganbajun (Wang and Ng 2004b), in that it is adsorbed on DEAE cellulose and Q Sepharose.
A comparison of the N-terminal ribonucleases of mushroom RNase (Table 2) reveals that mushroom RNases, with the exception of RNase from Irpex lacteus and Lentinus edodes, do not exhibit significant sequence homology to each other or to RNase from other organisms, such as mammals, plants, and bacteria. A similar picture is seen in the case of mushroom antifungal proteins (Ng 2004).
The molecular mass of L. shimeiji mushroom RNase (14.5 kDa) is larger than that of Pleurotus ostreatus RNase (11 kDa; Nomura et al. 1994). It is similar to those of P. sajor-caju RNase (12 kDa; Ngai and Ng 2004), Pleurotus pulmonarius (Ye and Ng 2002a), Agaricus bisporus RNase (14 kDa; Wang and Ng 2006), and Pleurotus eryngii RNase (16 kDa; Ng and Wang 2004). It is smaller than those of Russulus virescens RNase (28 kDa; Wang and Ng 2003c), P. tuber-regium RNase (29 kDa; Wang and Ng 2001), T. ganbajun RNase (30 kDa; Wang and Ng 2004b), and straw mushroom RNase (42 kDa; Wang and Ng 1999).
Pleurotus pulmonarius RNase is poly (C)-specific (Ye and Ng 2002a). RNases from R. virescens (Wang and Ng 2003c) and Termitomyces globulus (Wang and Ng 2003a) manifest co-specificity for poly (A) and poly (C). Portabella mushroom (Agaricus bisporus) RNase exhibits ribonucleolytic activity toward poly (A), poly (C), and poly (U) (Wang and Ng 2006). Lyophyllum shimeiji RNase is specific for poly (U) and poly (C), with much higher activity toward poly (U). Pleurotus sajor-caju RNase is poly (U)-specific (Ngai and Ng 2004). On the other hand, P. tuber-regium (Wang and Ng 2001) and P. ostreatus (Nomura et al. 1994) RNases are specific for poly (G).
Lyophyllum shimeiji RNase is markedly more thermostable than Thelephora ganbajun RNase. In the latter, activity is much attenuated at 80°C compared with 20°C and is indiscernible at 100°C (Wang and Ng 2004b). On the other hand, the activity of L. shimeiji RNase at 80 and 100°C is higher than that observed at 20°C, but activity at 100°C is more or less the same as that observed at 20°C. Thus, L. shimeiji RNase would lose its RNase activity when the mushroom is eaten after it has been boiled, but retain some activity after treatment at 80°C.
The optimum pH for L. shimeiji RNase (pH 6.0) is similar to that for P. tuber-regium (Wang and Ng 2001) and P. eryngii (Ng and Wang 2004) RNases (pH 6.5). It is different from the pH 4.5 for portabella mushroom RNase (Wang and Ng 2006) and R. virescens RNase (Wang and Ng 2003c), and pH 8.0 for P. pulmonarius RNase (Ye and Ng 2002a) and P. ostreatus RNase (Nomura et al. 1994).
Ribonucleases from several types of ginseng (Lam and Ng 2001b; Ng and Wang 2001; Wang and Ng 2000) show antifungal activity, but L. shimeiji ribonuclease lacks similar activity. Many other mushroom ribonucleases (Wang and Ng 1999, 2003a, b, c, 2004a, b; Ye and Ng 2002a), except P. sajor-caju RNase (Ngai and Ng 2004), are also without antifungal activity. Bullfrog oocyte RNase (Liao 1992), but not L. shimeiji RNase, has lectin activity.
Lyophyllum shimeiji RNase is a new protein as suggested by its novel N-terminal sequence. It has some novel characteristics, including activity at high temperatures, a pH optimum and base specificity different from some of the isolated mushroom RNases, antiproliferative activity toward tumor cells and anti-HIV-1 reverse transcriptase activity, which have not been demonstrated for the majority of the isolated mushroom RNases.
References
Adinolfi BS, Cafaro V, Dalessio G, Didonato A (1995) Full antitumor action of recombinant seminal ribonuclease depends on the removal of its N-terminal methionine. Biochem Biophys Res Commun 213:525–532
Green PJ (1994) The ribonucleases of higher plants. Annu Rev Plant Biol 45:421–445
Hartley RW (1988) Barnase and barstar. Expression of its cloned inhibitor permits expression of a cloned ribonuclease. J Mol Biol 202:913–915
Hofsteenge J, Matthies R, Stone SR (1989) Primary structure of a ribonuclease from porcine liver, a new member of the ribonuclease superfamily. Biochemistry 28:9806–9813
Irie M, Nitta R, Ohgi K, Niwata Y, Watanabe H, Iwama M, Beintema JJ, Sanda A, Takizawa Y (1988) Primary structure of a non-secretory ribonuclease from bovine kidney. J Biochem 104:289–296
Kobayashi H, Inokuchi N, Koyama T, Watanabe H, Iwama M, Ohgi K, Irie M (1992) Primary structure of a base non-specific and adenylic acid preferential ribonuclease from the fruit bodies of Lentinus edodes. Biosci Biotechnol Biochem 56:2003–2010
Laemmli UK, Favre M (1973) Maturation of the head of bacteriophage T4. I. DNA packaging events. J Mol Biol 80:575–599
Lam SK, Ng TB (2001a) First simultaneous isolation of a ribosome inactivating protein and an antifungal protein from a mushroom (Lyophyllum shimeji) together with evidence for synergism of their antifungal effects. Arch Biochem Biophys 393:271–280
Lam SK, Ng TB (2001b) Isolation of a novel thermolabile heterodimeric ribonuclease with antifungal and antiproliferative activities from roots of the sanchi ginseng Panax notoginseng. Biochem Biophys Res Commun 285:419–423
Lam SSL, Wang HX, Ng TB (1998) Purification and characterization of novel ribosome inactivating proteins, alpha-and beta-pisavins, from seeds of the garden pea Pisum sativum. Biochem Biophys Res Commun 253:135–142
Liao YD (1992) A pyrimidine-guanine sequence-specific ribonuclease from Rana catesbeiana (bullfrog) oocytes. Nucl Acids Res 20:1371–1377
Matousek J, Soucek J, Riha J, Zankel TR, Benner SA (1995) Immunosuppressive activity of angiogenin in comparison with bovine seminal ribonuclease and pancreatic ribonuclease. Comp Biochem Physiol B 112:235–241
Ng TB (2004) Peptides and proteins from fungi. Peptides 25:1055–1073
Ng TB, Lam YW (2002) Isolation of a novel agglutinin with complex carbohydrate binding specificity from fresh fruiting bodies of the edible mushroom Lyophyllum shimeiji. Biochem Biophys Res Commun 290:563–568
Ng TB, Wang HX (2001) Panaxagin, a new protein from Chinese ginseng possesses anti-fungal, anti-viral, translation-inhibiting and ribonuclease activities. Life Sci 68:739–749
Ng TB, Wang HX (2004) A novel ribonuclease from fruiting bodies of the common edible mushroom Pleurotus eryngii. Peptides 25:1365–1368
Ngai PHK, Ng TB (2004) A ribonuclease with antimicrobial, antimitogenic and antiproliferative activities from the edible mushroom Pleurotus sajor-caju. Peptides 25:11–17
Nomura H, Inokuchi N, Kobayashi H, Koyama T, Iwama M, Ohgi K, Irie M (1994) Purification and primary structure of a new guanylic acid specific ribonuclease from Pleurotus ostreatus. J Biochem 116:26–33
Sasso MP, Carsana A, Confalone E, Cosi C, Sorrentino S, Viola M, Palmier M, Russo E, Furia A (1991) Molecular cloning of the gene encoding the bovine brain ribonuclease and its expression in different regions of the brain. Nucl Acids Res 19:6469–6474
Shapiro R, Vallee BL (1987) Human placental ribonuclease inhibitor abolishes both angiogenic and ribonucleolytic activities of angiogenin. Proc Natl Acad Sci USA 84:2238–2241
Silverman RH (1997) 2-5A-Department RNase L: a regulated endoribonuclease in the interferon system. In: D’Alessio G, Riordan J (eds) Ribonucleases: structures and functions. Academic Press, New York, pp 515–551
Wang HX, Ng TB (1999) Isolation of a new ribonuclease from fresh fruiting bodies of the straw mushroom. Biochem Biophys Res Commun 264:714–718
Wang HX, Ng TB (2000) Quinqueginsin, a novel protein with anti-human immunodeficiency virus, antifungal, ribonuclease and cell-free translation-inhibitory activities from American ginseng roots. Biochem Biophys Res Commun 269:203–208
Wang HX, Ng TB (2001) Purification and characterization of a potent homodimeric guanine-specific ribonuclease from fresh mushroom (Pleurotus tuber-regium) sclerotia. Int J Biochem Cell Biol 33:483–490
Wang HX, Ng TB (2003a) Isolation of a ribonuclease from fruiting bodies of the wild mushroom Termitomyces globulus. Peptides 24:973–977
Wang HX, Ng TB (2003b) A novel ribonuclease from the veiled lady mushroom Dictyophora indusiata. Biochem Cell Biol 81:373–377
Wang HX, Ng TB (2003c) A ribonuclease with distinctive features from the wild green-headed mushroom Russulus virescens. Biochem Biophys Res Commun 312:965–968
Wang HX, Ng TB (2004a) Isolation of a new ribonuclease from fruiting bodies of the silver plate mushroom Clitocybe maxima. Peptides 25:935–939
Wang HX, Ng TB (2004b) Purification of a novel ribonuclease from dried fruiting bodies of the edible wild mushroom Thelephora ganbajun. Biochem Biophys Res Commun 324:855–859
Wang HX, Ng TB (2005) First report of an arabinose-specific fungal lectin. Biochem Biophys Res Commun 337:621–625
Wang HX, Ng TB (2006) A novel ribonuclease from fresh fruiting bodies of the portabella mushroom Agaricus bisporus. Biochem Cell Biol 84:178–183
Wang HX, Ng TB, Chiu SW (2004) A distinctive ribonuclease from fresh fruiting bodies of the medicinal mushroom Ganoderma lucidum. Biochem Biophys Res Commun 314:519–522
Watanabe H, Hamid F, Iwami M, Onda T, Ohgi K, Irie M (1995) Primary structure of RNase from Irpex lacteus. Biosci Biotechnol Biochem 59:2092–2103
Ye XY, Ng TB (2002a) A novel and potent ribonuclease from fruiting bodies of the mushroom Pleurotus pulmonarius. Biochem Biophys Res Commun 293:857–861
Ye XY, Ng TB (2002b) A novel peptide with ribonuclease and translation-inhibitory activities from fruiting bodies of the oyster mushroom Pleurotus ostreatus. J Pept Sci 8:235–240
Zhao JK, Wang HX, Ng TB (2009) Purification and characterization of a novel lectin from the toxic wild mushroom Inocybe umbrinella. Toxicon 53:360–366
Zilhao R, Camelo L, Arraiano CM (1993) DNA sequencing and expression of the gene rnb encoding Escherichia coli ribonuclease II. Mol Microbiol 8:43–51
Acknowledgments
This work was financially supported by National Grants of China (nyhyzx07-008 and 2007BAD89B00).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Zhang, R.Y., Zhang, G.Q., Hu, D.D. et al. A Novel Ribonuclease with Antiproliferative Activity from Fresh Fruiting Bodies of the Edible Mushroom Lyophyllum shimeiji . Biochem Genet 48, 658–668 (2010). https://doi.org/10.1007/s10528-010-9347-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-010-9347-y