Abstract
Tunicamycin (TN), one of the endoplasmic reticulum stress inducers, has been reported to inhibit tumor cell growth and exhibit anticarcinogenic activity. However, the mechanism by which TN initiates apoptosis remains poorly understood. In the present study, we investigated the effect of TN on the apoptotic pathway in U937 cells. We show that TN induces apoptosis in association with caspase-3 activation, generation of reactive oxygen species (ROS), and downregulation of survivin expression. P38 MAPK (mitogen-activated protein kinase) and the generation of ROS signaling pathway play crucial roles in TN-induced apoptosis in U937 cells. We hypothesized that TN-induced activation of p38 MAPK signaling pathway is responsible for cell death. To test this hypothesis, we selectively inhibited MAPK during treatment with TN. Our data demonstrated that inhibitor of p38 (SB), but not ERK (PD) or JNK (SP), partially maintained apoptosis during treatment with TN. Pre-treatment with NAC and GSH markedly prevented cell death, suggesting a role for ROS in this process. Ectopic expression of survivin in U937 cells attenuated TN-induced apoptosis by suppression of caspase-3 cleavage, mitochondrial membrane potential, and cytochrome c release in U937 cells. Taken together, our results show that TN modulates multiple components of the apoptotic response of human leukemia cells and raise the possibility of a novel therapeutic strategy for hematological malignancies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
TN, a naturally occurring antibiotic, blocks the first step in the biosynthesis of N-linked oligosaccharides in cells [1]. TN is a prototype of substances that exert potent inhibitory effects on protein maturation [2, 3]. TN has been applied in vitro, primarily to discern the functional significance of N-glycosylation in living systems including its role in cell proliferation and survival [4–7], sensitivity and resistance of tumor cells to antineoplastic drugs [8–10], and programmed cell death or apoptosis [11–13].
ER plays a central role in the synthesis, maturation, and assembly of secretary and structural proteins [14], and under some conditions ER stress can disrupt ER function [15]. Recent studies have identified an intrinsic apoptotic pathway that is regulated by the ER [16, 17]. In addition to controlling Ca2+ homeostasis and protein trafficking, the ER senses stress stimuli that lead to the accumulation of unfolded proteins and triggers a signaling cascade termed the unfolded protein response (UPR) [18]. The UPR is mediated by the activation of three ER transmembrane receptors, pancreatic ER kinase-like kinase (PERK), activating transcription factor 6 (ATF6), and inositol-requiring enzyme-1 (IRE-1), which under physiologic conditions are maintained in an inactive state by the ER chaperone binding immunoglobulin protein/glucose-regulated protein78 (BiP/GRP78). Identification of stress-associated ER protein expression gains importance in potential therapeutic targeting for ER stress-provoked disease. Furthermore, experimental ER stress responses have been induced using pharmacological agents such as thapsigargin, A23187 and brefeldin A, with upregulated levels of GRP78 [19]. One typical agent, TN, induces ER stress by interfering with N-linked protein glycosylation in ER [20]. The accurate signaling pathway of ER stress-mediated apoptosis was not still fully understood in leukemia cells.
Although the mechanisms responsible for survivin up-regulation during leukemia cells have not been clearly remained, epigenetic, genetic and post-translational mechanisms for survivin gene regulation have been described in other cell types. The survivin promoter was found to be methylated in ovarian cancers and unmethylated in normal ovarian tissues [21], although such differences in promoter methylation were not found in earlier comparisons of normal and malignant tissues [22]. While p53 was shown to repress survivin transcription by direct binding to the survivin promoter in a lung adenocarcinoma line [23], another study in an ovarian carcinoma cell line found that negative regulation of survivin by p53 was not affected by mutation or deletion of the putative p53-binding site in the survivin promoter [24]. It has been little know that the expression of survivin is affected by TN treatment. We showed a direct correlation between TN-induced apoptosis and the survivin expression.
MAPKs can be activated and exert their roles in response to stressors, such as DNA damage. One of the potential stressors that induces DNA damage and activation of the MAPK pathway is oxidative stress due to ROS. Many anticancer agents induce ROS generation and trigger cancer cell apoptosis via the ROS–MAPK pathway. In particular ROS can act as a second messenger by activating a diverse redox-sensitive signaling transduction cascade, including p38 MAPK and its subsequent target, the stress-related transcription factor activating transcription factor-2 (ATF-2) [25, 26].
In this study, we hypothesized that ER stress results in the accumulation of ROS generation and inhibition of survivin are required to significantly enhance ER stress-induced apoptosis. This study therefore demonstrates the importance of targeting survivin to enhance sensitivity to TN-induced apoptosis, and suggests that tumor cell survival may be inhibited by specific targeted approaches, including targeting of survivin.
Materials and methods
Materials
RPMI 1640 medium, fetal bovine serum (FBS), penicillin, streptomycin, and all other tissue culture reagents were obtained from GIBCO/BRL Life Technologies (Grand Island, NY). Tunicamycin (TN) was acquired from Aldrich (Milwaukee, WI, USA). Antibodies against PARP-1, Bcl-2, Bcl-xL, Mcl-1, and actin were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA), anti-survivin from R&D systems (Minneapolis, MN, USA), and anti-phospho-JNK, anti-phospho-p38, and anti-phospho-ERK antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA). Benzyl carbonyl-Val-Ala-Asp-fluoromethyl ketone (z-VAD-fmk) was purchased from Biomol (Plymouth Meeting, PA, USA); 2′7′-dichlorodihydrofluorescein diacetate (H2DCFDA) was from Molecular Probes (Eugene, OR, USA); and N-acetylcysteine (NAC) and all other chemicals used in this study were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Cell culture and chemical treatments
Human leukemia cells (U937 cells) were obtained from the American Type Culture Collection (Rockville, MD, USA). The U937 cells were cultured in RPMI 1640 medium supplemented with 10 % heat-inactivated fetal bovine serum (FBS), penicillin (100 units/mL) and streptomycin (100 units/mL) at 37 °C in a humidified incubator with 5 % CO2 and 95 % air. When the cells were subconfluent, the medium was replaced with fresh medium and 25–100 ng/mL of TN was added for 24 or 48 h.
Cellular viability assay
For morphological evaluation of cell death, approximately 5 × 105 U937 cells were plated into 60-mm cell culture dishes and incubated overnight. For the trypan blue exclusion assay, trypsinized cells were pelleted and resuspended in 0.2 mL of medium, 0.5 mL of 0.4 % trypan blue solution, and 0.3 mL of phosphate-buffered saline solution (PBS). The samples were mixed thoroughly, incubated at room temperature for 15 min, and examined under a light microscope. At least 300 cells were counted for each survival determination.
Flow cytometric assay of DNA content
After treatment with the indicated agent, the cells were harvested by trypsinization, fixed with 70 % (v/v) alcohol at 4 °C for 30 min and washed with PBS. After centrifugation, cells were incubated in 0.1 mL of phosphate-citric acid buffer (0.2 M NaHPO4, 0.1 M citric acid, pH 7.8) for 30 min at room temperature and then centrifuged and resuspended in 0.5 mL propidium iodide (PI) solution containing phosphate-citric acid buffer and 50 μg/mL PI. DNA content was analyzed with a FACScan flow cytometer (Beckman Coulter, Inc., Hialeah, FL, USA).
Analysis of cytochrome c release
A total of 2 × 106 cells were harvested, washed once with ice-cold PBS, and gently lysed for 2 min in 100 μL ice-cold lysis buffer. Lysates were centrifuged at 13,000×g at 4 °C for 20 min to obtain supernatants (cytosolic extracts free of mitochondria) and pellets (fraction containing mitochondria). The resulting cytosolic fractions were used for Western blot analysis with an anti-cytochrome c antibody.
Western blot analysis
For Western blot analyses, U937 cells were lysed with 1× Laemmli lysis buffer (2.4 M glycerol, 0.14 M Tris, pH 6.8, 0.21 M SDS, 0.3 mM bromophenol blue) and boiled for 10 min. Protein content was measured with the BCA Protein Assay Reagent (Pierce, Rockford, IL, USA). The samples were diluted with 1× Laemmli lysis buffer containing 1.28 M β-mercaptoethanol, and equal amounts of protein were loaded on 8–12 % SDS–polyacrylamide gels. Proteins were separated by SDS-PAGE and electrophoretically transferred to a nitrocellulose membrane. The membrane was blocked with 5 % nonfat dry milk in PBS-Tween-20 (0.1 %, v/v) for 1 h and then incubated with primary antibody (diluted according to the manufacturer’s instructions) at room temperature for 1.5 h. Horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG was used as the secondary antibody. Immunoreactive protein was visualized by the chemiluminescence protocol (ECL, Amersham, Arlington Heights, IL, USA). To ensure equal protein loading, each nitrocellulose membrane was stripped and reprobed with an anti-actin antibody.
DNA fragmentation and DAPI staining assay
To assess DNA fragmentation in U937 cells after treatment with TN for 24 h, approximately 1 × 106 cells were lysed for 30 min on ice in buffer containing 10 mM Tris (pH 7.4), 150 mM NaCl, 5 mM EDTA, and 0.5 % Triton X-100. Lysates were vortexed and cleared by centrifugation at 13,000×g for 30 min. Fragmented DNA in the supernatant was extracted with an equal volume of a mixture of neutral phenol:chloroform:isoamyl alcohol (25:24:1) and analyzed electrophoretically on 1.5 % agarose gels containing 0.1 g/mL EtBr. The cells were fixed on slide glass by the application of 4 % paraformaldehyde for 30 min at room temperature. After washing with PBS, 300 nM 4′, 6′-diamidino-2-phenylindole (DAPI) was added to the fixed cells for 10 min, and the cells were examined by fluorescence microscopy. Apoptotic cells were identified by condensation and fragmentation of nuclei. All DAPI staining experiments were performed in duplicate.
Measurement of reactive oxygen species
The generation of ROS was measured by staining with 2′,7′-dichlorofluorescein diacetate (DCF-DA). Briefly, U937 cells were seeded in 6-well plates (1 × 105 cells per well), allowed to attach overnight, and treated with TN. The cells were stained with 20 μM DCF-DA for 30 min at 37 °C and fluorescence was detected using a fluorescence microscope.
JC-1 mitochondrial membrane potential assay
To monitor mitochondrial membrane potential (MMP), JC-1 dye was used as previously described [27]. In cells with undamaged mitochondria the aggregated dye appears as red fluorescence, whereas in cells with altered MMP that are undergoing apoptosis the dye remains as monomers in the cytoplasm and emits diffuse green fluorescence. The red/green fluorescence ratio is dependent on the MMP. After treatment with TN, U937 cells were stained with a JC-1 MMP detection kit for 10 min and analyzed by flow cytometry. The fluorescence intensity was measured with a FACScan flow cytometer.
Data analysis
All experiments were repeated at least three times. The results are represented as mean ± standard deviation (SD). The difference between two mean values was analyzed using Student’s t test and was considered statistically significant when p < 0.05.
Results
Tunicamycin has anti-proliferation activity and induces cell death in U937 cells
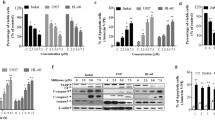
To investigate whether TN exerts antiproliferation activity and induces apoptosis, U937 cells were stimulated with the indicated concentrations of TN for 24 and 48 h and cell growth and viability was determined by a trypan blue assay. As shown in Fig. 1a, treatment with tunicamycin for 24 and 48 h inhibited the growth of U937 cells in a dose-dependent manner. Flow cytometric analysis of DNA staining showed that TN induced a time- and dose-dependent increase in hypodiploid (sub-G1) DNA content (Fig. 1b). Programmed cell death was also demonstrated by microscopic examination of DAPI staining (Fig. 1c) and by a DNA fragmentation assay (Fig. 1d). These results suggest that the growth inhibitory effect of TN in U937 cells was the result of induction of apoptosis.
Effect of tunicamycin on the growth and viability of U937 cells. Cells were treated with various concentrations of TN for 24 and 48 h. a Effect of TN on cell growth. U937 cells were seeded in 96-well plates and cultured in triplicate in the presence or absence of various concentrations of TN (25–100 ng/mL) for 24 and 48 h. Viable cells were quantified by the trypan blue assay. b Effect of TN on the viability of U937 cells. Cells were treated for 24 and 48 h with the indicated concentrations of TN, and DNA content was evaluated by propidium iodide staining. The fraction of apoptotic cells is indicated. Data are mean values obtained from three independent experiments. *p < 0.05 compared with the control cells. c U937 cells were treated in the absence or presence of TN at the indicated concentrations for 24 h. Microscopic examination was performed to detect apoptosis by nuclear staining with DAPI. The data are expressed as means of three independent experiments. d DNA fragmentation was analyzed by extraction of DNA and electrophoresis on a 1.5 % agarose gel (Color figure online)
Tunicamycin induces activation of the caspase cascade in U937 cells
To address the significance of caspase activation in TN-induced apoptosis, we examined caspase activity after treatment with TN. Treatment of U937 cells with various concentrations of TN resulted in a dramatic increase in cleavage of the caspase substrate PARP, and cleavage of procaspase-8, procaspase-9, and procaspase-3 (Fig. 2a) in a time-dependent manner (Fig. 2b). To confirm that the activation of caspases is a dependent manner in TN-induced apoptosis, U937 cells were pretreated with z-VAD-fmk, a cell-permeable caspase inhibitor, followed by TN treatment for 24 h. As shown in Fig. 2c, TN-induced apoptosis was prevented by pretreatment with z-VAD-fmk, as indicated by reduced PARP cleavage and procaspase-3 cleavage. We also found that z-VAD-fmk prevented the dose-dependent increase in the accumulation of apoptotic DNA after treatment with TN (Fig. 2d). These results suggest that tunicamycin-induced cell death is associated with caspase activation.
Tunicamycin treatment induced caspase activation in U937 cells. a Protein expression of caspase-3 (cas-3), caspase-8 (cas-8), caspase-9 (cas-9), and PARP after treatment with different doses of TN for 24 h. Total cell lysates were subjected to Western blotting with specific antibodies. Actin was used as a loading control. Proteolytic cleavage of PARP, cas-3, cas-8, and cas-9 is indicated by arrows. b U937 cells were treated with 100 ng/mL tunicamycin for 1.5, 3, 6, 12, and 24 h. PARP, cas-3, and actin were detected by Western blotting. Proteolytic cleavage of PARP and cas-3 is indicated by arrows. c U937 cells were incubated with 50 μM z-VAD-fmk for 1 h before treatment with 100 ng/mL tunicamycin for 24 h. Equal amounts of cell lysates (40 μg) were subjected to electrophoresis and analyzed by Western blotting for PARP and cas-3. Proteolytic cleavage of PARP and cas-3 is indicated by arrows. d DNA fragmentation was analyzed by extraction of DNA and electrophoresis on a 1.5 % agarose gel
Tunicamycin induces activation of the p38 pathway in U937 cells
Next, we investigated the effect of TN treatment on the expression and activities of MAPKs to determine whether this signaling pathway is involved in mediating the observed apoptotic response. As shown in Fig. 3a, TN treatment induced a strong increase in phosphorylated p38 MAPK level, which peaked at 1.5 h and slowly declined thereafter. However, phosphorylation of ERK and JNK was not increased after TN treatment. We then evaluated the possible roles of MAPKs in TN-induced apoptosis. As shown in Fig. 3b, pretreatment with PD (a specific inhibitor of ERK MAPK) and SP (a specific inhibitor of JNK) did not affect TN-induced proteolytic cleavage of PARP, whereas treatment with SB (a specific inhibitor of p38 MAPK) significantly decreased TN-induced proteolytic cleavage of PARP. To further evaluate the effect of p38 signaling on TN-induced apoptosis, we investigated whether TN induces cell death in the presence of SB using the trypan blue assay. As shown in Fig. 3c, pretreatment of U937 cells with SB before exposure to TN markedly increased cell viability. Taken together, these results indicate that activation of the p38 pathway plays an important role in regulating tunicamycin-induced apoptosis in U937 cells.
The p38 pathway plays an important role in tunicamycin-induced apoptosis. a U937 cells were treated with 100 ng/mL TN for the indicated asterisk. Equal amounts of cell lysates (40 μg) were subjected to electrophoresis and analyzed by immunoblotting using phosphorylation-specific antibodies. b Effect of MAPK inhibitor on TN-induced apoptosis. U937 cells were incubated with ERK inhibitor (PD, 25 μM), p38 inhibitor (SB, 25 μM), JNK inhibitor (SP, 25 μM), or solvent as indicated for 30 min before treatment with 100 ng/mL tunicamycin for 24 h. Proteolytic cleavage of PARP is indicated by arrows. c Cells were pretreated for 30 min with specific inhibitors before 24 h treatment with TN and then evaluated for cell viability by the trypan blue assay. Data are mean values obtained from three independent experiments and error bars represent standard deviation. Statistics: Student’s t test for unpaired values. *p < 0.05 versus control cells
Generation of reactive oxygen species in tunicamycin-induced apoptosis
Many antineoplastic agents eliminate tumor cells by inducing apoptosis, and oxidative stress-mediated cellular changes are frequently observed in cells that are exposed to cytotoxic drugs [28, 29]. Therefore, we examined whether TN affects the cellular levels of ROS by measuring changes in the fluorescence of H2DCF-DA. The green fluorescence of DCF was markedly increased after treatment of U937 cells with 100 ng/mL TN for 0.5 h, indicating that TN slightly increase intracellular ROS levels (Fig. 4a). Co-treatment with 10 mM NAC for 24 h completely blocked the TN-induced ROS production (Fig. 4b), whereas NAC alone did not alter the fluorescence intensity of DCF (Fig. 4b). Exposure of the cells to NAC or GSH plus TN for 24 h resulted in remarkable inhibition of caspase-3 activation and reduction of PARP cleavage (Fig. 4c). Co-treatment with NAC reduced cell death of TN-treated cells by approximately 40 % (Fig. 4d). Taken together, these results indicate that tunicamycin-induced caspase activation is preceded by ROS generation in U937 cells.
Tunicamycin increases ROS levels in leukemia cancer cells. a U937 cells were treated with 100 ng/mL TN for 0.5 h. ROS levels were measured by incubation with 5 μM H2DCF-DA and analysis of fluorescence by flow cytometry. H2O2; positive control. b U937 cells were treated with or without 100 ng/mL TN and N-acetyl-cysteine (NAC) for 24 h. At the end of treatment, the cells were stained with H2DCF-DA and observed under a confocal microscope. Green color indicates the location of DCF fluorescence. c U937 cells were co-treated with or without 10 mM NAC or 10 mM glutathione (GSH) and 100 ng/mL TN for 24 h and apoptosis was determined by Western blotting. Total protein extracts were subjected to Western blot analysis using specific antibodies for PARP, cas-3, and actin. Representative Western blot data are shown for one of three separate experiments with similar findings. d U937 cells were co-treated with or without 10 mM NAC and 100 ng/mL TN for 24 h and apoptosis was determined by FACS. Their DNA content was measured after propidium iodide staining. Populations of total apoptotic cells were quantified using flow cytometric analysis. Data are shown as mean ± S.E. *p < 0.01 indicates a significant difference between untreated and TN-treated samples. **p < 0.01 indicates significant difference between TN alone and co-treatment with NAC (Color figure online)
Tunicamycin induces downregulation of survivin protein and mRNA expression
To investigate the underlying mechanisms involved in TN-induced apoptosis, we analyzed changes in the expression levels of various antiapoptotic proteins. As shown in Fig. 5a and b, protein levels of Bcl-2, Bcl-xL, Bax, and XIAP were not altered in response to TN. However, the protein level of survivin was remarkably reduced by TN in a dose- and time-dependent manner. Survivin was recently reported to modulate the balance between cell death and viability in cancer [30]. To investigate whether TN exposure triggered a decrease in survivin protein levels at the transcriptional level, we evaluated the effect of TN on survivin mRNA levels by RT-PCR. As shown in Fig. 5c and d, TN treatment resulted in a large reduction in the survivin mRNA level. Taken together, these results indicate that downregulation of survivin may be important for tunicamycin-induced apoptosis.
Tunicamycin exposure leads to downregulation of antiapoptotic protein and RNA expression. a Expression levels of apoptotic-related proteins after treatment with tunicamycin. U937 cells were treated with the indicated concentrations of TN for 24 h. Equal amounts of cell lysate were resolved by SDS-PAGE, transferred to nitrocellulose, and probed with specific antibodies against Bcl-2, Bcl-xL, Bax, survivin, and XIAP. A representative study is shown; two additional experiments yielded similar results. b U937 cells were treated with 100 ng/mL TN for the indicated asterisk. Equal amounts of cell lysate were resolved by SDS-PAGE, transferred to nitrocellulose, and probed with specific antibodies. c The expression of survivin mRNA was detected by RT-PCR after treatment with TN at the indicated concentrations. The level of total actin was used as a control for equal loading of mRNA in different lanes. d Quantification of survivin mRNA level in U937 cells treated with TN for different times in three separate experiments
Ectopic expression of survivin reduces tunicamycin-induced apoptosis
To determine whether survivin downregulation was responsible for TN-induced cytotoxicity, we examined the effect of TN in U937 cells that were stably transfected with a plasmid encoding myc-survivin or pcDNA. As shown in Fig. 6a, the cell viability of U937 cells treated with TN was increased in the survivin-transfected cells compared with the vector-transfected cells. We next investigated the role of survivin in TN-induced cell death. Western blotting confirmed that the survivin-transfected cells showed increased survivin expression and decreased the cleavage of caspase-3 proteins (Fig. 6b). As shown in Fig. 6a, absolutely reduced cytotoxicity was observed in cells transfected with the survivin overexpression plasmid compared with cells transfected with control vector. These results demonstrate that overexpression of survivin can counteract the cytotoxicity induced by TN in U937 cells. The induction of cell death by TN is therefore due to the downregulation of survivin. To further investigate the underlying mechanisms involved in TN-induced apoptosis, we examined the role of mitochondria by examining the effect of TN on the mitochondrial membrane potential (MMP). As shown in Fig. 6c, exposure of U937/vector cells to 100 ng/mL tunicamycin for 24 h led to a significant reduction in the MMP level, which was rescued in U937 cells that overexpressed survivin. In addition, accumulating evidence suggests that mitochondria play an essential role in apoptosis by releasing apoptogenic effectors such as cytochrome c. Western blot analysis of cytosolic fractions to measure the release of mitochondrial cytochrome c showed that TN treatment markedly induced the release of cytochrome c into the cytoplasm in U937/vector cells, but not in U937/survivin cells (Fig. 6d). Taken together, these results indicate that ectopic expression of survivin inhibits tunicamycin-induced apoptosis by suppressing the release of cytochrome c.
Survivin overexpression confers resistance to tunicamycin-mediated cell death. a U937 cells were stably transfected with pcDNA3 (vector) or pcDNA3myc-survivin (survivin) using Lipofectamine 2000 reagent. U937/vector and U937/survivin cells were seeded in 96-well plates and cultured in triplicate in the presence or absence of various concentrations of TN for 24 h and cell viability was determined by the MTT assay. Cell survival is shown as a percentage of control untreated cells. Statistics: Student’s t test for unpaired values. *p < 0.05 versus control cells. b U937/vector and U937/survivin cells were treated for 24 h with the indicated concentrations of TN and their DNA content was measured by FACS after propidium iodide staining. Cells treated as above were harvested in lysis buffer and equal amounts of cell lysate (40 μg) were subjected to electrophoresis and analyzed by Western blotting for PARP, cas-3 and survivin. Proteolytic cleavage of PARP and cas-3 is indicated by the arrow. Western blotting using an anti-survivin antibody was also performed to confirm overexpression of survivin in the transfected cells. c Effects of TN on the mitochondrial membrane potential (MMP). U937/vector and U937/survivin cells were treated with 100 ng/mL TN for 24 h and the MMP was measured using a flow cytometer. Quantification of the MMP in U937/vector and U937/survivin cells treated with TN in three separate experiments. Statistics: Student’s t test for unpaired values. *p < 0.05 versus U937/vector cells. d U937/vector and U937/survivin cells were treated for 24 h with TN and cytosolic extracts were prepared as described in Materials and Methods. Thirty micrograms of cytosolic proteins was resolved by 15 % SDS-PAGE, transferred to a membrane, and probed with specific anti-cytochrome c antibody or with anti-actin antibody as a control for protein loading
Discussion
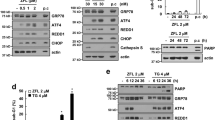
In this study, we investigate the apoptotic signaling pathways that are activated by TN as a result of activation of the UPR and induction of GRP78 and ATF6 (supplemental data). ER stress participates in the pathogenesis of various cancers and cardiovascular diseases and promotes disease progression [31]. A variety of extracellular stresses, such as nutrient deprivation, hypoxia, oxidative stress, and calcium imbalance, activate the UPR in the ER. GRP78 and GRP94 are upregulated in the early stages of ER stress, followed by overexpression of CCAAT/enhancer-binding protein-homologous protein (CHOP) in prolonged ER stress. In cardiomyocytes, CHOP is accumulated in the nucleus and promotes cell apoptosis by depleting glutathione and increasing the production of ROS, causing depolarization of the mitochondrial membrane potential and the release of cytochrome c [32–35]. There is a growing body of evidence that antioxidants such as NAC protect cancer cells by inhibiting ER stress. These studies suggested that antioxidants could reduce ER stress-mediated apoptosis by directly decreasing ROS generation [36]. In the current study we demonstrated that TN induced cell death in cultured U937 cells, as shown by propidium iodide staining and DNA fragmentation assay (Fig. 1). Moreover, NAC suppressed the TN-induced cell death and mitochondria dysfunction that are induced by ER stress. We found that treatment with TN at low concentrations downregulated survivin expression (Fig. 5), upregulated ROS generation (Fig. 4), potently inhibited cell growth, and promoted cell death through the mitochondrial pathway in leukemic cell lines. We also found that treatment with TN in other cells, THP-1 induced apoptosis through downregulation of survivin expression and upregulation of ROS generation (supplemental data).
Inhibition of caspases by the pan caspase inhibitor z-VAD-fmk blocked the induction of cell death by TN, suggesting that this process was caspase-dependent (Fig. 2). In addition, TN-induced cell death involved caspase-9 activation, indicating that activation of caspase-9 by the mitochondrial pathway and subsequent cleavage of the effector caspase-3 is critical for the action of TN. This model was further supported by the findings that TN decreased survivin levels, enhanced cytochrome c release, and induced changes in MMP. Moreover, overexpression of survivin suppressed the induction of cell death by TN. Survivin is a potent caspase-9 and caspase-3 inhibitor that blocks cell death both upstream and downstream of the apoptotic cell death cascade. The ability of TN to reduce the levels of survivin makes it a powerful inducer of apoptosis. Regarding the underlying mechanism, TN was shown to effectively inhibit survivin expression at both protein and RNA levels (Fig. 5). In addition, we found that the decrease in survivin protein levels caused by TN was mediated through both degradation of survivin protein and transcriptional inhibition of survivin mRNA. We also other ER stress inducer, thapsigargin was shown to successfully reduce survivin expression and induce apoptosis (supplemental data). The control of survivin protein expression may occur at several other levels in addition to transcription. Survivin is transcriptionally up-regulated by Dec1 [37], c-myc [38] and KLF5 [39], all of which show aberrant activation in tumors as compared with normal tissues. Various transcription factors including Stat3 [40], HIF-1a [41] and Rb-E2F1 [42] have been shown to interact directly with the survivin promoter in breast cancer and lung cancer cell lines. Sp1 [43], phosphatidylinositol-3 kinase (PI3 K) [44], mitogen-activated protein kinase/extracellular-signal regulated kinase (MEK/ERK) [45] have also been shown to regulate survivin expression. The fact that ER stress has been shown to inhibit Sp1 [46] further suggests that the transcriptional downregulation of survivin by TN is mediated, at least in part, through inhibition of Sp1. Most of the work on survivin gene regulation has been in malignant cells, and little is known regarding mechanisms of survivin repression in normal cells. After 24 h of TN treatment, control U937 cells (U937/vector) were more sensitive to TN than cells that overexpressed survivin (U937/survivin) (Fig. 6). The notion that TN induces cell death through inhibition of survivin is thus further supported by the finding that forced survivin overexpression attenuates TN-induced cell death.
In this study we show that TN possesses anti-leukemia activity through ROS generation and downregulation of survivin expression mediated by the ER stress pathway. This mechanism is supported by the following evidence: (i) TN dose-dependently decreases cell viability and induces apoptosis in leukemia cells; (ii) TN stimulates ROS accumulation, and blocking ROS generation strongly inhibits TN-induced apoptosis; (iii) Upregulation of survivin expression markedly inhibits TN-induced apoptosis and decreases TN-induced ROS generation in leukemia cancer cells; and (iv) TN triggers ER stress. The potent effects of tunicamycin in leukemia cells reported in this study warrant further investigation of this compound for the treatment of leukemia.
Abbreviations
- DAPI:
-
4′,6′-Diamidino-2-phenylindole
- DCFDA:
-
2′7′-Dichlorodihydrofluorescein diacetate
- NAC:
-
N-acetylcysteine
- PBS:
-
Phosphate-buffered saline
- PI:
-
Propidium iodide
- ROS:
-
Reactive oxygen species
- UPR:
-
Unfolded protein response
- z-VAD-fmk:
-
Benzyl carbonyl-Val-Ala-Asp-fluoromethyl ketone
References
Noda I, Fujieda S, Seki M, Tanaka N, Sunaga H, Ohtsubo T, Tsuzuki H, Fan GK, Saito H (1999) Inhibition of N-linked glycosylation by tunicamycin enhances sensitivity to cisplatin in human head-and-neck carcinoma cells. Int J Cancer 80:279–284
Dennis JW, Granovsky M, Warren CE (1999) Protein glycosylation in development and disease. BioEssays 21:412–421
Goss PE, Baker MA, Carver JP, Dennis JW (1995) Inhibitors of carbohydrate processing: a new class of anticancer agents. Clin Cancer Res 1:935–944
Bentley J, Quinn DM, Pitman RS, Warr JR, Kellett GL (1997) The human KB multidrug-resistant cell line KB-C1 is hypersensitive to inhibitors of glycosylation. Cancer Lett 115:221–227
Carlberg M, Larsson O (1993) Role of N-linked glycosylation in cell-cycle progression and initiation of DNA synthesis in tumor-transformed human fibroblasts. Anticancer Res 13:167–171
Martínez JA, Torres-Negrón I, Amigó LA, Banerjee DK (1999) Expression of Glc3Man9GlcNAc2-PP-Dol is a prerequisite for capillary endothelial cell proliferation. Cell Mol Biol (Noisy-le-grand) 45:137–152
Hiss D, Gabriels G, Jacobs P, Folb P (1996) Tunicamycin potentiates drug cytotoxicity and vincristine retention in multidrug resistant cell lines. Eur J Cancer 32:2164–2172
Kramer R, Weber TK, Arceci R, Ramchurren N, Kastrinakis WV, Steele G Jr, Summerhayes IC (1995) Inhibition of N-linked glycosylation of P-glycoprotein by tunicamycin results in a reduced multidrug resistance phenotype. Br J Cancer 71:670–675
Lou LX, Geng B, Yu F, Zhang J, Pan CS, Chen L, Qi YF, Ke Y, Wang X, Tang CS (2006) Endoplasmic reticulum stress response is involved in the pathogenesis of stress induced gastric lesions in rats. Life Sci 79:1856–1864
Rao RV, Poksay KS, Castro-Obregon S, Schilling B, Row RH, del Rio G, Gibson BW, Ellerby HM, Bredesen DE (2004) Molecular components of a cell death pathway activated by endoplasmic reticulum stress. J Biol Chem 279:177–187
Reimertz C, Kögel D, Rami A, Chittenden T, Prehn JH (2003) Gene expression during ER stress-induced apoptosis in neurons: induction of the BH3-only protein Bbc3/PUMA and activation of the mitochondrial apoptosis pathway. J Cell Biol 162:587–597
Walker BK, Lei H, Krag SS (1998) A functional link between N-linked glycosylation and apoptosis in Chinese hamster ovary cells. Biochem Biophys Res Commun 250:264–270
Kleizen B, Braakman I (2004) Protein folding and quality control in the endoplasmic reticulum. Curr Opin Cell Biol 16:343–349
Aridor M, Balch WE (1999) Integration of endoplasmic reticulum signaling in health and disease. Nat Med 5:745–751
Xu C, Bailly-Maitre B, Reed JC (2005) Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest 115:2656–2664
Kim I, Xu W, Reed JC (2008) Cell death and endoplasmic reticulum stress: disease relevance and herapeutic opportunities. Nat Rev Drug Discov 7:1013–1030
Rutkowski DT, Kaufman RJ (2004) A trip to the ER: coping with stress. Trends Cell Biol 14:20–28
Elbein AD (1987) Inhibitors of the biosynthesis and processing of N-linked oligosaccharide chains. Annu Rev Biochem 56:497–534
Natsume Y, Ito S, Satsu H, Shimizu M (2009) Protective effect of quercetin on ER stress caused by calcium dynamics dysregulation in intestinal epithelial cells. Toxicology 258:164–175
Xu C, Bailly-Maitre B, Reed JC (2005) Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest 115:2656–2664
Hattori M, Sakamoto H, Satoh K, Yamamoto T (2001) DNA demethylase is expressed in ovarian cancers and the expression correlates with demethylation of CpG sites in the promoter region of c-erbB-2 and survivin genes. Cancer Lett 169:155–164
Li F, Altieri DC (1999) Transcriptional analysis of human survivin gene expression. Biochem J 344Pt 2:305–311
Hoffman WH, Biade S, Zilfou JT, Chen J, Murphy M (2002) Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J Biol Chem 277:3247–3257
Mirza A, McGuirk M, Hockenberry TN, Wu Q, Ashar H, Black S, Wen SF, Wang L, Kirschmeier P, Bishop WR, Nielsen LL, Pickett CB, Liu S (2002) Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene 21:2613–2622
Simoncini S, Sapet C, Camoin-Jau L, Bardin N, Harlé JR, Sampol J, Dignat-George F, Anfosso F (2005) Role of reactive oxygen species and p38 MAPK in the induction of the pro-adhesive endothelial state mediated by IgG from patients with anti-phospholipid syndrome. Int Immunol 17:489–500
Kim BM, Lee KH, Hong IS, Hong SH (2012) p38 mitogen-activated protein kinase is a key regulator of 5-phenylselenyl- and 5-methylselenyl-methyl-2′-deoxyuridine-induced apoptosis in human HL-60 cells. Biochem Biophys Res Commun 417:237–244
Kim MJ, Lee TH, Kim SH, Choi YJ, Heo J, Kim YH (2010) Triptolide inactivates Akt and induces caspase-dependent death in cervical cancer cells via the mitochondrial pathway. Int J Oncol 37:1177–1185
Fleury C, Mignotte B, Vayssière JL (2002) Mitochondrial reactive oxygen species in cell death signaling. Biochimie 84:131–141
Wen J, You KR, Lee SY, Song CH, Kim DG (2002) Oxidative stress-mediated apoptosis. The anticancer effect of the sesquiterpene lactone parthenolide. J Biol Chem 277:38954–38964
Jin HO, Yoon SI, Seo SK, Lee HC, Woo SH, Yoo DH, Lee SJ, Choe TB, An S, Kwon TJ, Kim JI, Park MJ, Hong SI, Park IC, Rhee CH (2006) Synergistic induction of apoptosis by sulindac and arsenic trioxide in human lung cancer A549 cells via reactive oxygen species-dependent down-regulation of survivin. Biochem Pharmacol 72:1228–1236
Guan L, Han B, Li Z, Hua F, Huang F, Wei W, Yang Y, Xu C (2009) Sodium selenite induces apoptosis by ROS-mediated endoplasmic reticulum stress and mitochondrial dysfunction in human acute promyelocytic leukemia NB4 cells. Apoptosis 14:218–225
Ono Y, Shimazawa M, Ishisaka M, Oyagi A, Tsuruma K, Hara H (2012) Imipramine protects mouse hippocampus against tunicamycin-induced cell death. Eur J Pharmacol 696:83–88
Shore GC, Papa FR, Oakes SA (2011) Signaling cell death from the endoplasmic reticulum stress response. Curr Opin Cell Biol 23:143–149
Okada K, Minamino T, Tsukamoto Y, Liao Y, Tsukamoto O, Takashima S, Hirata A, Fujita M, Nagamachi Y, Nakatani T (2004) Prolonged endoplasmic reticulum stress in hypertrophic and failing heart after aortic constriction: possible contribution of endoplasmic reticulum stress to cardiac myocyte apoptosis. Circulation 110:705–712
Zhao H, Liao Y, Minamino T, Asano Y, Asakura M, Kim J, Asanuma H, Takashima S, Hori M, Kitakaze M (2008) Inhibition of cardiac remodeling by pravastatin is associated with amelioration of endoplasmic reticulum stress. Hypertens Res 31:1977–1987
Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, Ron D (2004) CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev 18:3066–3077
Li Y, Xie M, Yang J, Yang D, Deng R, Wan Y, Yan B (2006) The expression of antiapoptotic protein survivin is transcriptionally upregulated by DEC1 primarily through multiple sp1 binding sites in the proximal promoter. Oncogene 25:3296–3306
Cosgrave N, Hill AD, Young LS (2006) Growth factor-dependent regulation of survivin by c-myc in human breast cancer. J Mol Endocrinol 37:377–390
Zhu N, Gu L, Findley HW, Chen C, Dong JT, Yang L, Zhou M (2006) KLF5 Interacts with p53 in regulating survivin expression in acute lymphoblastic leukemia. J Biol Chem 281:14711–14718
Gritsko T, Williams A, Turkson J, Kaneko S, Bowman T, Huang M, Nam S, Eweis I, Diaz N, Sullivan D, Yoder S, Enkemann S, Eschrich S, Lee JH, Beam CA, Cheng J, Minton S, Muro-Cacho CA, Jove R (2006) Persistent activation of stat3 signaling induces survivin gene expression and confers resistance to apoptosis in human breast cancer cells. Clin Cancer Res 12:11–19
Peng XH, Karna P, Cao Z, Jiang BH, Zhou M, Yang L (2006) Cross-talk between epidermal growth factor receptor and hypoxia-inducible factor-1alpha signal pathways increases resistance to apoptosis by up-regulating survivin gene expression. J Biol Chem 281:25903–25914
Dasgupta P, Kinkade R, Joshi B, Decook C, Haura E, Chellappan S (2006) Nicotine inhibits apoptosis induced by chemotherapeutic drugs by up-regulating XIAP and survivin. Proc Natl Acad Sci USA 103:6332–6337
Kim YH, Lee DH, Jeong JH, Guo ZS, Lee YJ (2008) Quercetin augments TRAIL-induced apoptotic death: involvement of the ERK signal transduction pathway. Biochem Pharmacol 75:1946–1958
Ringshausen I, Schneller F, Bogner C, Hipp S, Duyster J, Peschel C, Decker T (2002) Constitutively activated phosphatidylinositol-3 kinase (PI-3 K) is involved in the defect of apoptosis in B-CLL: association with protein kinase Cdelta. Blood 100:3741–3748
Hu P, Han Z, Couvillon AD, Exton JH (2004) Critical role of endogenous Akt/IAPs and MEK1/ERK pathways in counteracting endoplasmic reticulum stress-induced cell death. J Biol Chem 279:49420–49429
Abdelrahim M, Liu S, Safe S (2005) Induction of endoplasmic reticulum-induced stress genes in Panc-1 pancreatic cancer cells is dependent on Sp proteins. J Biol Chem 280:16508–16513
Acknowledgments
This work was supported by Basic Science Research Program through the NRF funded by the Ministry of Education, Science and Technology (2010-0009064).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lim, E.J., Heo, J. & Kim, YH. Tunicamycin promotes apoptosis in leukemia cells through ROS generation and downregulation of survivin expression. Apoptosis 20, 1087–1098 (2015). https://doi.org/10.1007/s10495-015-1135-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-015-1135-z