Abstract
Introduction
In this study, we delineated the apoptotic signaling pathways activated by sodium selenite in NB4 cells.
Materials and methods
NB4 cells were treated with 20 μM sodium selenite for different times. The activation of caspases and ER stress markers, ROS levels, mitochondrial membrane potential and cell apoptosis induced by sodium selenite were analyzed by immunoblotting analysis, DCF fluorescence and flow cytometric respectively. siRNA was used to detect the effect of GADD153 on selenite-induced cell apoptosis.
Conclusions
Sodium selenite-induced reactive oxygen species generation is an early event that triggers endoplasmic reticulum stress mitochondrial apoptotic pathways in NB4 cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Apoptosis is a major mechanism to eliminate cancer cells. The activation of caspase family proteases is the key step in apoptosis, and several pathways lead to caspase activation [1]. Up to now, three predominant apoptotic pathways, namely death receptor-mediated extrinsic pathway, mitochondria-mediated intrinsic pathway, and endoplasmic reticulum (ER) stress-mediated apoptotic pathway have been elucidated.
The accumulation of unfolded or misfolded protein in ER can induce ER stress. As a protective mechanism, ER triggers the UPR to protect cells against ER stress [2, 3]. UPR is mediated by one ER molecular chaperon GRP78 and three ER-resident transmembrane proteins, PERK, IRE1 and ATF6 [4, 5]. Activated PERK blocks general protein synthesis by phosphorylating eIF2α. ATF6 is cleaved to produce an active ATF6 (50 kDa), which regulates the expression of ER chaperones and XBP-1. XBP-1 mRNA is spliced by IRE1 to create a potent transcription factor XBP-1S, which controls the transcription of chaperones, as well as genes involved in protein degradation. Collectively, these UPR signaling pathways initiate cell protective mechanisms to protect cells against ER stress-induced damage. However, if protein aggregation is persistent and the stress cannot be alleviated, cell apoptotic pathways will be initiated [6]. CHOP, also known as growth-arrest and DNA-damage inducible gene 153 (GADD153), is a key pro-apoptotic transcription factor that is closely related to ER stress [7, 8]. During ER stress, all the three arms of the UPR act together to induce transcription of GADD153. Of these three arms, the PERK-eIF2α-ATF4 branch plays an essential role in up regulating GADD153 protein [8].
Selenium is an essential nutrient element with a chemopreventive role against cancer at nutritional levels and anticancer potential at supranutritional doses [9, 10]. Apoptosis is one mechanism by which selenium expresses its anticancer effect. Sodium selenite, a common dietary form of selenium, can effectively induce several cancer cell lines to undergo apoptosis, raising a new idea for its clinical application [11–13]. The human acute promyelocytic leukemia-derived NB4 cell is derived from the marrow of a patient with APL in relapse [14]. All-trans retinoic acid (ATRA) and arsenic trioxide have been successful in treating APL, but both drugs have some limitations [15, 16]. In previous studies, we discovered that 20 μM sodium selenite markedly inhibited the proliferation and induced apoptosis of NB4 cells in a time-dependent manner [17, 18]. In the present study, we further investigated the molecular mechanisms by which 20 μM sodium selenite-induced apoptosis in NB4 cells.
Materials and methods
Reagents and antibodies
Sodium selenite (the purity is 98%) and Rh123 were purchased from Sigma-Aldrich. M-MLV was purchased from Promega. MnTMPyP was purchased from Calbiochem. The antibodies purchased from Cell Signaling Technology are cleaved caspases antibody sampler kit, anti-eIF2α, anti-phospho-eIF2α (Ser51), anti-AKT, anti-phospho-AKT (Ser308); antibodies purchased from Santa Cruz are anti-phospho-PERK (Thr981), anti-GADD153, anti-CREB2 (ATF4), anti-ATF6, anti-caspase-4. Anti-XBP-1 was purchased from Biolegend, anti-GRP78 from BD transudation laboratories, and anti-β-actin from Sigma-Aldrich. ROS detection kit was purchased from Beyotime Company (Jiangsu, China).
Cell culture
NB4 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37°C in a humidified atmosphere of 5% CO2.
Immunoblotting analysis
Approximately 1 × 107 cells were collected, washed twice with ice-cold PBS, and lysed in cell lysis RIPA buffer (20 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerolphosphate, 1 mM Na3VO4, 1 μg/ml leupeptin, 1 mM PMSF) for 5 min on ice and then subjected to sonication for 20 s. The lystates were centrifuged at 12,000g for 10 min at 4°C. The supernatant was collected, and protein concentration was determined by the Bradford assay. Equal amounts of protein were separated by 12% (or 15%, depending on the protein of interest) SDS-PAGE and transferred onto nitrocellulose membranes. The membranes were blocked with TBST containing 5% non-fat milk and incubated with primary antibodies overnight at 4°C. After washing with TBST, membranes were incubated with HRP-conjugated secondary antibodies for 60 min at room temperature. After a second round of washing with TBST, the blots were probed with the ECL system.
ROS measurement
Cells in 6-well culture plates were incubated with or without 10 μM MnTMPyP for 1.5 h before being subjected to sodium selenite treatment. Harvested cells were washed with serum-free culture medium and incubated with 10 μM DCFH-DA at 37°C for 20 min. The DCF fluorescence distribution of 4 × 105 cells was recorded every 10 min by a fluorospectrophotometer at an excitation wavelength of 488 nm and at an emission wavelength of 535 nm for up to 80 min.
Small interference RNA
The GADD153-specific siRNA (sense 5′-CCAGGAAACGGAAACAGA-3′; antisense 5′-UCUGUUUCCGUUUCCUGG-3′) [19], and non-silencing scrambled siRNA were synthesized and purchased from GenePharma (Shanghai Co. Ltd., China). For siRNA transfection, NB4 cells were plated in 6-well culture plates at density of 3.5 × 105 and transfected with siRNA by using Lipofectamine™ 2000 reagent (Invitrogen) according to the manufacturer’s protocol. Briefly, for each well, 5 μl Lipofectamine™ 2000 was diluted in 250 μl Opti-MEMI medium (Invitrogen). This mixture was carefully added to a solution containing 200 nM siRNA in 250 μl Opti-MEMI medium. The solution was incubated for 20 min at room temperature, and then gently dripped into the NB4 cells in 2 ml antibiotic free medium. Regular growth medium was added 6–12 h after transfection. 24 h after transfections, cells were treated with 20 μM sodium selenite.
RT-PCR
Total RNA was extracted by Trizol reagents according to the manufacturer’s instructions. Reverse transcription was carried out by M-MLV and the obtained cDNA was subjected to PCR. Primers were as follows: EDEM: F: -5′-ATATGGTGCCCTCCCTGAGAG-3′; R: 5′-GATTGCAGTTGGAGCTGGAGT-3′. GADD153: F: 5′-GCCTTTCTCTTCGGACACTG-3′; R: 5′-TCACCATTCGGTCAATCAGA-3′. GAPDH: F: 5′-CAACAGCCTCAAGATCATCAGC-3′; R: 5′-TTCTAGACGGCAGGTCAGGTC-3′.
Flow cytometric analysis of mitochondrial membrane potential (Δψm)
NB4 cells were pretreated with or without 10 μM MnTMPyP for 1.5 h and then treated with 20 μM sodium selenite for 24 h. Approximately 1 × 106 cells were collected, washed twice with ice-cold PBS, and incubated in 1 ml staining solution (PBS containing 10 μg/ml Rh123) for 30 min in the dark at 37°C. Then, after rinsing with PBS, cells were resuspended in 0.5 ml PBS. The fluorescent intensities of Rh123 were determined by an EPICS XL-MCL flow cytometry.
Flow cytometric analysis of cell apoptosis
Approximately 1 × 106 cells were collected, washed twice with ice-cold PBS, and fixed with 70% ethanol at 4°C overnight. The cells were then collected by centrifugation and resuspended in 0.5 ml PBS containing 50 μg/ml RNase A. The system was incubated at 37°C for 30 min, and then kept on ice to stop the reaction. PI solution was then added to achieve a final concentration of 50 μg/ml, and cells were stained for at least 30 min on ice in the dark. The resultant cell suspension was then subjected to flow cytometry analysis.
Statistical analysis
Data were analyzed by ANOVA analysis with the Student t test. A value of P < 0.05 was considered statistical significance.
Results
Sodium selenite activates caspase cascades in NB4 cells
We first assessed the effects of sodium selenite on caspases activation in NB4 cells by western blot. The results showed that the activated cleavage fragment of caspase-9 increased after 12 h exposure to sodium selenite, whereas the activated cleavage fragment of caspase-8 was not detected (data not shown). Caspase-4 was also activated by treatment with sodium selenite, as shown from the decrease of pro-caspase-4 and the increase of cleavage fragment. In parallel with the activation of caspase-9 and -4, there were also increases in the cleavage of effector caspases, namely caspase-3, -6, and -7 (Fig. 1). The activation of caspase-9 and -4 indicated that mitochondria and ER stress-mediated apoptotic pathways might be involved in sodium selenite-induced cell apoptosis in NB4 cells [1, 20]. Sodium selenite-induced mitochondrial apoptotic pathway has been demonstrated by our and some other studies [11, 17]. Next, we asked whether sodium selenite can induce ER stress-mediated apoptotic pathway.
Sodium selenite induces ER stress in NB4 cells
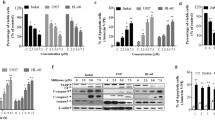
To determine whether sodium selenite-induced ER stress, we investigated several crucial ER stress markers, including phospho-PERK, phospho-eIF2α, ATF4, XBP-1S, ATF6 and molecular chaperon GRP78 (Fig. 2a). Phosphorylation of PERK was induced at 3 h, and then gradually decreased. The level of phosphorylated form of eIF2α began to increase at 6 h, which occurred just a slightly later than the phosphorylation of PERK, peaked at 9 h and declined at longer periods of incubation. Total eIF2α protein level did not change throughout the incubation periods examined. Combining these results, we infer that the increase of eIF2α phosphorylation is likely due to the activation of PERK. Phosphorylation of eIF2α by PERK inhibits its activity as an initiating factor, thus leading to suppression of protein synthesis. However, phosphorylated eIF2α selectively induces the translation of ATF4 mRNA [21, 22]. In line with this, the expression of ATF4 in NB4 cells was induced after sodium selenite treatment (Fig. 2).
Sodium selenite induces ER stress in NB4 cells. a Time-dependent effects of sodium selenite on activation of UPR effectors. Total cell lysates were prepared from cells at indicated times following sodium selenite exposure, and subjected to western blot. b Effects of sodium selenite on transcription of EDEM. Total RNAs were extracted from cells at indicated times following sodium selenite exposure, and EDEM gene transcription was examined by RT-PCR. GADPH was taken as an internal control for loading normalizations. c Time-dependent effect of sodium selenite on GRP78 expression. β-actin was used as a loading control. The results shown are representative of at least three independent experiments
Two other ER stress-related proteins IRE1 and ATF6 were also investigated. Activated product of ATF6, the 50 kDa fragment increased slightly first and then returned to its basal level or a bit below (Fig. 2a). Activated IRE1 results in XBP-1 mRNA splicing and the spliced-XBP-1 mRNA encodes a potent transcription factor XBP-1S [23]. Using IRE1-mediated XBP-1 splicing as a surrogate marker, we further assessed the effect of sodium selenite on IRE1 activation. 54 kDa XBP-1S protein was induced soon after exposure to sodium selenite, and then its level gradually decreased (Fig. 2a). As a transcription factor, XBP-1S protein induces EDEM gene transcription [24], which plays a role in ER-related protein degradation [25]. Using RT-PCR, we assessed EDEM mRNA production (Fig. 2b). Consistent with the induction of XBP-1S protein, the level of EDEM mRNA increased soon after sodium selenite exposure and then decreased. Based on these results, we conclude that sodium selenite induces all three UPR signaling pathways, including PERK-eIF2α-ATF4, IRE1-XBP-1S and ATF6 in NB4 cells, but the activation of ATF6 is less sensitive to sodium selenite than the other. Although activated UPR signaling pathways, sodium selenite treatment did not change ER molecular chaperone GRP78 protein level (Fig. 2c), which functions as part of the protective mechanisms induced by the UPR pathways to maintain protein folding-competent state [5, 26]. As a consequence, NB4 cells were deprived of one of the main pro-survival mechanisms during sodium selenite-induced ER stress.
GADD153 involves in ER stress-mediated cell apoptosis
Having confirmed the initiation of ER stress by sodium selenite treatment, we next investigated whether sodium selenite-induced ER stress actually contributed to NB4 cell apoptosis. GADD153 is induced during ER stress and regularly participates in ER stress-mediated apoptosis [8]. RT-PCR and western blot results showed that GADD153 mRNA and protein levels increased markedly at 1.5 h after exposure to sodium selenite and peaked at 9 h (Fig. 3a). To further establish a functional role of GADD153 in sodium selenite-induced NB4 cell apoptosis, GADD153-specific siRNA was used to inhibit GADD153 expression [19]. A scrambled RNA duplex was used as a negative control. GADD153-specific siRNA effectively down-regulated GADD153 mRNA and protein levels and reduced sodium selenite-induced cell apoptosis significantly compared with scrambled siRNA (Fig. 3b, c). These results indicate that GADD153 participates in ER stress-mediated apoptosis induced by sodium selenite in NB4 cells.
Sodium selenite induces GADD153-mediated cell apoptosis. a Time-dependent effect of sodium selenite on GADD153 expression. Total RNA and cell lysates were prepared from cells at indicated times following sodium selenite exposure, and subjected to RT-PCR and western blot, respectively. b The mRNA and protein levels of GADD153 were determined by RT-PCR and western blot, respectively. Cells were transfected with GADD153-specific siRNA and scrambled siRNA. 24 h after transfection, cells were treated with 20 μM sodium selenite for 24 h. c Apoptosis was examined by flow cytometry. *P < 0.05 compared with the scrambled siRNA (n = 3). β-actin was used as a loading control. The results shown are representative of at least three independent experiments
GADD153 inhibits the activity of AKT
What is the downstream signal of GADD153-mediated cell apoptosis? AKT plays a critical role in controlling cell survival by resisting ER stress-induced apoptotic signal [27]. Western blot result showed that phospho-Thr 308-AKT (the activated form of AKT) decreased in a time-dependent manner after sodium selenite treatment; while no apparent change of AKT level was observed (Fig. 4a). This probably points to an anti-apoptotic role that activated AKT plays in sodium selenite-induced apoptosis. Silencing of GADD153 gene expression prevented the reduction of phospho-Thr 308-AKT in response to sodium selenite (Fig. 4b). Thus GADD153-induced cell apoptosis is probably mediated by inhibition the activation of anti-apoptotic kinase AKT.
GADD153-siRNA inhibits the inactivation of AKT induced by sodium selenite. a Time-dependent effect of sodium selenite on the activity of AKT. b GADD153-specific siRNA prevented dephosphorylation of AKT in response to sodium selenite. β-actin was used as loading control. The results shown are representative of at least three independent experiments
Sodium selenite-induced ER stress requires ROS generation
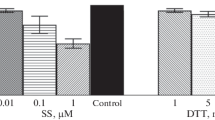
Next, we investigated the upstream regulatory mechanisms leading to sodium selenite-induced ER stress. Sodium selenite has been reported to induce the generation of ROS during apoptosis [28, 29]. As anticipated, DCFH-DA-dependent measurements showed that intracellular ROS levels were significant increase in cells treated with 20 μM sodium selenite (Fig. 5a). Pretreatment with antioxidant 10 μM Mn(III) tetrakis (1-methyl-4-pyridyl) porphyrin pentachloride MnTMPyp, a cell-permeable superoxide scavenger, abrogated the increase in ROS and simultaneity completely inhibited cell apoptosis induced by sodium selenite (Fig. 5a, b), suggesting that ROS plays an important role in sodium selenite-induced cell apoptosis. To test the relationship between sodium selenite-induced ROS production and ER stress, we incubated NB4 cells with MnTMPyP, prior to sodium selenite treatment. The results showed that MnTMPyp significantly attenuated sodium selenite-induced activation of UPR, as well as abrogated the expression of ATF-4 and GADD153 following sodium selenite stimulation (Fig. 5c). Next, we also investigate the effect of scavenging ROS by MnTMPyP on selenite-induced mitochondrial dysfunction. Pretreatment with MnTMPyp prevented sodium selente-induced mitochondrial membrane permeabilization and the activation of caspase-9, -3, and -6 (Fig. 5d, e). These results indicate that ROS plays a major role in sodium selenite-induced ER stress and mitochondrial dysfunction in NB4 apoptosis process.
Sodium selenite-induced ROS generation triggers ER stress and mitochondrial dysfunction. NB4 cells were pretreated with or without 10 μM MnTMPyP for 1.5 h and then treated with sodium selenite for indicated times. a The level of ROS in cells detected by DCFH-DA-dependent measurements. b The effects of MnTmPyP on sodium selenite-induced cells apoptosis by flow cytometry analysis. *P < 0.05 compared with sodium selenite-treated group (n = 3). c The effects of MnTmPyP on ER stress markers. Total cell lysates were prepared from cells at indicated times following sodium selenite exposure and subjected to western blot. d The effect of MnTmPyP on mitochondrial membrane potential by flow cytometry analysis. e The effects of MnTmPyP on activation of caspase-9, -3 and -6. β-actin was used as a loading control. The results shown are representative of at least three independent experiments
Discussion
In this study, we delineate the apoptotic signaling pathways activated by sodium selenite in NB4 cells. Sodium selenite induces ER stress by activating UPR and inducing GADD153, which contribute to NB4 cell apoptosis by inhibition of AKT activity. Antioxidant suppresses sodium selenite-induced ER stress and mitochondrial dysfunction. Taken together, these results indicate that sodium selenite-induced ROS generation is an early event that triggers ER stress and mitochondrial dysfunction in NB4 cells. The proposed apoptotic pathways induced by sodium selenite are depicted in Fig. 6.
Apoptotic signaling pathways activated by sodium selenite in NB4 cells. Sodium selenite induces the production of ROS which triggers ER stress and mitochondrial apoptotic pathways in NB4 cells. Sodium selenite induces the activation of UPR first and than activates GADD153, which contributes to cell apoptosis by inhibition of anti-apoptotic kinase AKT activity. Inactivated AKT may cause mitochondrial dysfunction by dephosphorylation of bad. ER stress and mitochondrial dysfunction induce activation of caspase-4 and -9, respectively and in concert mediate selenite-induced NB4 cell apoptosis
It has been reported that selenium can cause global thiol/disulfide redox modification of numerous proteins, which may result in accumulation of misfolded or unfolded proteins in the ER, thus triggering ER stress [2–4]. The present work showed that the three UPR transducer pathways, PERK-eIF2α, IRE1-XBP1S and ATF6 were activated at earlier times of sodium selenite exposure, although their activation receded at later stages of sodium selenite exposure. Activation of PERK-eIF2α signaling blocks general protein synthesis and reduces the burden caused by more unfolded proteins. Nevertheless phosphorylated eIF2α selectively activates the translation of the transcription factor ATF4, which induces GADD153 expression at later stages of sodium selenite exposure [21, 22]. The primary function of UPR is to protect cells against ER stress-induced damage. If the stress level is too severe to be repaired by cells, the apoptotic signals will be triggered [6]. It is thus not surprised that sodium selenite activates UPR pathways at early stages of treatment and induces apoptotic markers GADD153 later on.
The robust expression of GADD153 by the classic ER stress inducers has been well documented [7, 8]. The present study suggests that GADD153 is important in sodium selenite-induced NB4 cell apoptosis as GADD153-specific siRNA diminished cell apoptosis. However, the down stream effectors of GADD153 are poorly understood. GADD153 expression can result in the down-regulation of Bcl-2 expression, depletion of cellular glutathione, and exaggerated production of ROS [30]. Zu et al. [26] found that GADD153 might play a positive role in upregulating the expression of p21 in a p53-independent manner. In this study we found that silencing of GADD153 gene expression by siRNA prevented sodium selenite-induced the inactivation of anti-apoptotic kinase AKT. These data suggest that GADD153 acts as a critical negative regulator of AKT in sodium selenite-induced NB4 cell apoptosis. Activated AKT can induce the phosphorylation of bad and cause a decrease of cytochrome c release [31]. How GADD153 mediates dephosphorylation of AKT is unknown. TRB3 has recently been reported to be the target of GADD153/ATF4, and involved in GADD153-dependent cell apoptosis during ER stress [32]. It was also reported to bind to unphosphorylated AKT and prevent its activation [33]. So it is possible that GADD153 mediates the inactivation of AKT through induction of TRB3 expression. Although sodium selenite clearly activated UPR signaling, the primary ER chaperone GRP78 was not induced by sodium selenite, suggesting that the enhancement of protein folding capacity as a cell protective mechanism was probably not up-regulated by sodium selenite.
We demonstrated that sodium selenite-induced NB4 cell apoptosis was associated with the production of ROS and mitochondrial damage [17]. What are the relationships between these three signals? Sodium selenite-induced the activation of UPR at early times and than activated GADD153, which may initiate and amplify mitochondrial membrane permeabilization by dephosphorylation of AKT and Bad. Thus GADD153 may mediate the apoptotic signals from ER to mitochondria. The activation of UPR is probably earlier than mitochondrial dysfunction. It was reported that high levels of ROS can be an effective inducer of cell apoptosis, probably through activation of ER stress-mediated apoptotic pathway [34]. Removal ROS by antioxidant MnTMPyP could attenuate sodium selenite-induced the activation of UPR and at the same time abolish the expression of ATF4 and GADD153 induced by sodium selenite. MnTMPyP also completely prevented sodium selenite-induced mitochondrial membrane permeability, and the activation of caspase-9 and -3. In conclusion, these results show that the production of ROS is an early event that initiates mitochondria and ER stress-mediated apoptotic pathways in NB4 cells.
Abbreviations
- ER:
-
Endoplasmic reticulum
- MnTMPyp:
-
Mn(III) tetrakis(1-methyl-4-pyridyl) porphyrin pentachloride
- ROS:
-
Reactive oxygen species
- UPR:
-
Unfolded protein response
References
Lavrik IN, Golks A, Krammer PH (2005) Caspases: pharmacological manipulation of cell death. J Clin Invest 115:2665–2672. doi:10.1172/JCI26252
Rutkowski DT, Kaufman RJ (2004) A trip to the ER: coping with stress. Trends Cell Biol 14:20–28. doi:10.1016/j.tcb.2003.11.001
Faitova J, Krekac D, Hrstka R, Vojtesek B (2006) Endoplasmic reticulum stress and apoptosis. Cell Mol Biol Lett 11:488–505. doi:10.2478/s11658-006-0040-4
Szegezdi E, Logue SE, Gorman AM et al (2006) Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep 7:880–885. doi:10.1038/sj.embor.7400779
Zhang KZ, Kaufman RJ (2004) Signaling the unfolded protein response from the endoplasmic reticulum. J Biol Chem 279:25935–25938. doi:10.1074/jbc.R400008200
Kadowaki H, Nishitoh H, Ichijo H (2004) Survival and apoptosis signals in ER stress: the role of protein kinases. J Chem Neuroanat 28:93–100
Wang XZ, Lawson B, Brewer JW et al (1996) Signals from the stressed endoplasmic reticulum induce C/EBP-homologous protein CHOP/GADD153). Mol Cell Biol 16:4273–4280
Oyadomari S, Mori M (2004) Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ 11:381–389. doi:10.1038/sj.cdd.4401373
Patrick L (2004) Selenium biochemistry and cancer: a review of the literature. Altern Med Rev 9:239–258
Zeng H, Combs GF Jr (2007) Selenium as an anticancer nutrient: roles in cell proliferation and tumor cell invasion. J Nutr Biochem 19:1–7. doi:10.1016/j.jnutbio.2007.02.005
Xiang N, Zhao R, Zhong W (2009) Sodium selenite induces apoptosis by generation of superoxide via the mitochondrial-dependent pathway in human prostate cancer cells. Chemother Pharm 63:351–362
Gopee NV, Johnson VJ, Sharma RP (2004) Sodium selenite-induced apoptosis in murine B-lymphoma cells is associated with inhibition of protein kinase C-δ, nuclear factor-κB, and inhibitor of apoptosis protein. Toxicol Sci 78:204–214. doi:10.1093/toxsci/kfh072
Xiao R, Qiao JT, Zhao HF (2006) Sodium selenite induces apoptosis in cultured cortical neurons with special concomitant changes in expression of the apoptosis-related genes. Neurotoxicology 27:478–484. doi:10.1016/j.neuro.2006.01.008
Lanotte M, Martin-Thouvenin V, Najman S et al (1991) NB4, a maturation inducible cell line with t(15;17) marker isolated from a human acute promyelocytic leukemia (M3). Blood 77:1080–1086
Chen Z, Chen CG, Shen ZX et al (2001) Treatment of acute promyelocytic leukemia with arsenic compounds: in vitro and in vivo studies. Semin Hematol 38:26–36. doi:10.1053/shem.2001.20863
Shen ZX, Shi ZZ, Fang J et al (2004) All-trans retinoic acid/As2O3 combination yields a high quality remission and survival in newly diagnosed acute promyelocytic leukemia. Proc Natl Acad Sci USA 101:5328–5335. doi:10.1073/pnas.0400053101
Li J, Zuo L, Shen T, Xu CM et al (2003) Induction of apoptosis by sodium selenite in human acute promyelocytic leukemia NB4 cells: involvement of oxidative stress and mitochondria. J Trace Elem Med Biol 17:19–26. doi:10.1016/S0946-672X(03)80041-X
Han BS, Wei W, Hua FY et al (2007) Requirement for ERK activity in sodium selenite-induced apoptosis of acute promyelocytic leukemia-derived NB4 cells. J Biochem Mol Biol 40:196–204
Wu Y, Zhang HT, Dong Y et al (2005) Endoplasmic reticulum stress signal mediators are targets of selenium action. Cancer Res 65:9073–9079. doi:10.1158/0008-5472.CAN-05-2016
Hitomi J, Katayama T, Eguchi Y et al (2004) Involvement of caspase-4 in endoplasmic reticulum stress-induced apoptosis and Aβ-induced cell death. J Cell Biol 165:347–356. doi:10.1083/jcb.200310015
Fels DR, Koumenis C (2006) The PERK/eIF2α/ATF4 module of the UPR in hypoxia resistance and tumor growth. Cancer Biol Ther 5:723–728. doi:10.1158/1535-7163.MCT-05-0164
Jiang HY, Wek RC (2005) Phosphorylation of the α-subunit of the eukaryotic initiation factor-2α (eIF2α) reduces protein dynthesis and enhances apoptosis in response to proteasome inhibition. J Biol Chem 280:14189–14202. doi:10.1074/jbc.M413660200
Yoshida H, Matsui T, Yamamoto A et al (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107:881–891. doi:10.1016/S0092-8674(01)00611-0
Lee AH, Iwakoshi NN, Glimcher LH (2003) XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol 23:7448–7459. doi:10.1128/MCB.23.21.7448-7459.2003
Ruddock LW, Molinari M (2006) N-glycan processing in ER quality control. J Cell Sci 119:4373–4380. doi:10.1242/jcs.03225
Zu K, Bihani T, Lin A et al (2006) Enhanced selenium effect on growth arrest by BiP/GRP78 knockdown in p53-null human prostate cancer cells. Oncogene 25:546–554
Hu P, Han Z, Couvillon AD, Exton JH (2004) Critical role of endogenous AKT/IAPs and MEK1/ERK pathways in counteracting endoplasmic reticulum stress-induced cell death. J Biol Chem 279:49420–49429. doi:10.1074/jbc.M407700200
Shen HM, Yang CF, Ong CN (1999) Sodium selenite-induced oxidative stress and apoptosis in human hepatoma HepG2 cells. Int J Cancer 81:820–828. doi:10.1002/(SICI)1097-0215(19990531)81:5<820::AID-IJC25>3.0.CO;2-F
Li GX, Hu H, Jiang C, Schuster T (2007) Differential involvement of reactive oxygen species in apoptosis induced by two classes of selenium compounds in human prostate cancer cells. Int J Cancer 120:2034–2043. doi:10.1002/ijc.22480
Karen DM, Jennifer LM, Lars-oliver K et al (2001) Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating bcl2 and perturbing the cellular redox state. Mol Cell Biol 21:1249–1259. doi:10.1128/MCB.21.4.1249-1259.2001
Kuo CT, Hsu MJ, Chen BC et al (2008) Denbinobin induces apoptosis in human lung adenocarcinoma cells via Akt inactivation, bad activation, and mitochondrial dysfunction. Toxicol Lett 177:48–58
Nobumichi O, Satoshi Y, Takayuki H (2005) TRB3, a novel ER stress-inducible gene, is induced via ATF4–CHOP pathway and is involved in cell death. EMBO J 24:1243–1255. doi:10.1038/sj.emboj.7600596
Kato S, Du K (2007) TRB3 modulates C2C12 differentiation by interfering with AKT activation. Biochem Biophys Res Commun 353:933–938. doi:10.1016/j.bbrc.2006.12.161
Xue X, Piao JH, Nakajima A et al (2005) Tumor necrosis factor-α (TNF-α) induces the unfolded protein response (UPR) in a reactive oxygen species (ROS)-dependent fashion, and the UPR counteracts ROS accumulation by TNF-α. J Biol Chem 280:33917–33925. doi:10.1074/jbc.M505818200
Acknowledgments
This work was supported by grants from National Natural Sciences Foundation of China (no. 30370348 and no. 30770491), Doctoral Point Foundation of National Educational Committee (no. 20010023029), and Natural Sciences Foundation of Beijing (no. 7032034 and no. 5082015).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Guan, L., Han, B., Li, Z. et al. Sodium selenite induces apoptosis by ROS-mediated endoplasmic reticulum stress and mitochondrial dysfunction in human acute promyelocytic leukemia NB4 cells. Apoptosis 14, 218–225 (2009). https://doi.org/10.1007/s10495-008-0295-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-008-0295-5