Abstract
Knowledge graph completion(KGC) has attracted increasing attention in recent years, aiming at complementing missing relationships between entities in a Knowledge Graph(KG). While the existing KGC approaches utilizing the knowledge within KG could only complement a very limited number of missing relations, more and more approaches tend to study the completion of the multi-relationship knowledge graph. However, the existing completion methods of multi-relation knowledge graph regard knowledge graph as an undirected graph, which ignores the directionality of knowledge graph, so that the potential characteristics of multi-relation cannot be learned. Besides, most algorithms fail to explore the local information of knowledge because they ignore the different importance of entity adjacencies. In this paper, we propose to use local information fusion to join the entity and its adjacency relation, to acquiring the multi-relation representation. In addition, we try to specify distinct weights to model the direction of the relationship and apply the attention mechanism between entity nodes to obtain local information between entity nodes. Experiments conducted on three benchmark datasets and a medical domain knowledge graph dataset that we collect demonstrate the effectiveness of the proposed framework.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Knowledge Graphs(KG) is a knowledge base adopted by Google to enhance its search engine’s results with information gathered from an enormous variety of sources, and it represents the knowledge bases (KBs) in the form of a directed graph structure. In recent years, the knowledge graph’s construction and application have developed rapidly with the in-depth study of artificial intelligence. There are several KGs contain abundant and various facts of the real world, such as, DBPedia [14], Freebase [1], YAGO [22] and the Google knowledge vault [8]. These KGs play a significant role in a great deal of knowledge-driven applications, such as information search [6], question answering [3], and recommended systems [5]. The majority of the KGs mentioned above include a significant number of single relationships, however, a large number of multi-relational KGs have been constructed and applied in professional domains. Nonetheless, there are many unresolved tasks in multi-relationship KG, the hidden multiple relationships between a large number of entities and the local information of entity nodes have not been fully mined.
Compared with the knowledge graph include a lot of single relationships, the entities of the multi-relational knowledge graph have complicated relationships and local structure. As illustrated in Fig. 1, this is a part of the biomedical knowledge graph that depicts the relationship between calcium and other genes, proteins, etc. We can observe that there have multiple directed relationships between a pair of entities in the biomedical knowledge graph, such as the triple (Calium, affect reaction, caspase 3) and (Calium, affect cotreatment, caspase 3). Similarly, there is a relationship increase phosphorytation between a pair of entities (Calium, AKT serine kinase 1). And we need to detect another relation decrease reaction between the pair of entities,i.e, find the triple ((Calium, decrease reaction, AKT serine kinase 1)). Therefore, it is necessary to put forward a method of multi-relational knowledge graph completion (KGC).
In recent years, a family of the Trans model has been proposed for knowledge graph completion. These models employ a transitional characteristic to model relationships between entities in low dimensional vector space and obtain excellent performance. Specifically, they utilize a limited number of parameters and adopt some simple operations, such as inner products, matrix multiplications in an embedding space. In terms of the current deep neural network model, the majority of these models are based on convolutional neural networks (CNN), which explore expressive features in complex relationships using a multi-layer neural network. However, these models only learn the triple’s information but ignore the structure features of the knowledge graph. Therefore, some models based on graph convolutional networks (GCN) have been proposed that utilize graph convolution networks for mining the graph structure features of the knowledge graph. But they treat the knowledge graph as an undirected single relation graph and neglect the multiple relationship characteristics and local information of the knowledge graph. As shown in Fig. 1, to detect the triple ((Calium, decrease reaction, AKT serine kinase 1)), we need to learn the feature of the directed relation decrease reaction from the fact (Calium, decrease reaction, paraoxonase 1), and explore the local information from the correlation degree between the four adjacency relationships and the entity Calium. In addition, some models incorporate external information into an entity or relationship embedding, ignoring the graph structure feature and directional relations in KG.
What’s more, as demonstrated in Table 1, these techniques are incapable of learning local graph structure information or directed multi-relational embedding. Therefore, it is necessary to propose a framework that can learn different directed multiple relationship characteristics of knowledge graph and dig out the importance of adjacency to extract the local information for the multi-relational knowledge graph completion.
According to the above analysis, we propose a relation-based Joint Graph Attention Networks (JGAN) model that takes into account the multiple relation features of knowledge graphs and the importance of different local directional relationships. In our model, we joint entities and relationships of triples to obtain the features representation of directed relations in DKGs. Meanwhile, we focus on the most relevant adjacency of the input entity to make decisions and utilize the self-attention mechanism to weigh the importance of different relationships. Finally, we acquire the embedding of the entity and directional relation for the multi-relational knowledge graph completion.
The main contributions of our work are the following:
-
Using a joint operation from knowledge graph embedding approaches, we learn the direction multiple relation feature.
-
Our method extracts local information features of entity nodes by estimating the significance of distinct multiple adjacency relations of the entity and investigating the graph structure in a multi-relational knowledge graph using a graph convolutional network (GCN).
-
We collect and release a medical domain knowledge graph dataset Footnote 1 by working with companies, consisting of 27100 entities and 41 relations, to evaluate our proposed model.
The remainder of this paper is organized as follows: we review related work in Section 2, introduce our proposed model in Section 3, report experiments and results in Section 4, and finally we conclude our work with future work in Section 5.
2 Related work
In this section, we introduce some existing work relevant to our research. In general, our research is related to the knowledge graph embedding and graph convolutional network.
2.1 Knowledge graph embedding
Various kinds of knowledge graph embedding models have been proposed to learn continuous low-dimensional vector representations of entities and relations in the last few years.
Translating embedding models represent entities and relationships in low dimensional vector space. TransE [2] is the first and most representative knowledge graph embedding model. It supposes that the representation of the head entity plus the representation of relation should be close to the representation of the tail entity in an irrefutable fact. After that, more variant models of TransE [2] have been proposed. Such as TransH [32], TransR [15], TransD [10], TranSparse [11] and so on.
Semantic matching models exploit similarity-based scoring functions. RESCAL [19] is a classical semantic matching model, which associates each entity with a vector to capture the underlying semantics of the triple. DistMult [35] simplifies RESCAL by constraining the relation matrix to a diagonal matrix. ComplEx [26] extends DistMult [35] by introducing complex-valued embeddings so as to better model asymmetric relations. RotatE [23] defines each relation as a rotation from the head entity to the tail entity in the complex vector space.
Besides, several models adopt convolution neural networks to mine the potential features of triples for knowledge graph embedding. For example, ConvE [7] and ConKB [18] has been proposed for knowledge graph embedding. In ConvE [7], a one-dimensional input vector is reshaped to a two-dimensional vector and fed into the convolution layer for extracting features. ConKB [18] composed of a convolution layer, a projection layer to the embedding dimension, and an inner product layer.
Some researchers consider adding some external information for the KGC task. The external data, including entity attribute information, entity description information, and relation description information. The most representative model is DKRL [33] that adding entity description information to the model. Other than this, TEKE [31] also merges external context information to represent learning for knowledge graph embedding.
These methods and models mentioned above only deal with triples separately, ignoring the local graph structure information and the directional multiple relations of the knowledge graph. Therefore, we need to provide a technique for extracting directional properties of multiple relations as well as local graph structure features for multi-relational knowledge graph completion.
2.2 Graph convolutional network
GCNs [4] have been first proposed for image and audio recognition tasks, which consider possible generalizations of CNNs to signals defined on more general domains without the action of a translation group [4]. After that, GCNs were proposed for semi-supervised learning on graph-structured data based on an efficient variant of convolutional neural networks that operate directly on graphs, and they motivate the choice of our convolutional architecture via a localized first-order approximation of spectral graph convolutions [13]. In recent years, with the rise of graph convolutional network and its ability to deal graph structure data, more varients of GCNs have been proposed, such as GraphSAGE [9], GAT [29], ConfGCN [28], LCNs [34] and so on. GraphSAGE leverages node feature information to efficiently generate node embeddings for previously unseen data [9]. It learns a function that generates embeddings by sampling and aggregating features from a node’s local neighborhood. GAN presents graph attention networks (GATs), neural network architectures that operate on graph-structured data, leveraging masked self-attentional layers to learn relation embedding [29]. ConfGCN estimates label scores along with confidences jointly in a GCN-based setting [28]. LCNs use the embeddings of nodes that arise from Lovász’s orthogonal representations as an implicit regularizer [34].
What’s more, the methods based on GCNs for relational graphs have been proposed. The most representative model is R-GCN [20], it is related to a recent class of neural networks operating on graphs, and were developed specifically to handle the highly multi-relational data characteristic of realistic knowledge bases [20]. WGCN [21] has been proposed, it has learnable weights that adapt the amount of information from neighbors used in local aggregation, leading to more accurate embeddings of graph nodes [21]. However, these GCN-based models ignore the multiple relationships of the graph.
3 Proposed method
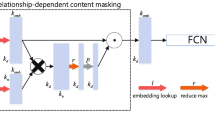
In this section, we introduce the related notations and problem definition we will use in this work, followed by a detailed description of our model, as shown in Fig. 2. We take into account the complexity of local graph structure and the multi-relational of knowledge graph in our method. In order to obtain more expressive entities and relationship embedding, our JGAN module based on GCNs extracts local graph structure features and the multiple relationship characteristic at the same time, so as to further complete the knowledge graph.
The overall architecture of our model. For a triple (h1,r1,h2), tail entity h2 and relation r1 are combined into entity representation that include feature of the relation r1 by a ϕ operation, then compute the importance α12, α14, α21 and α23. Last, the entity and relation embedding are inputted into the score function to get the score
As illustrated in Fig. 2, for a triplet (h1,r1,h2) in the given knowledge graph, the model combines tail entity h2 and its corresponding relationship r1 into vector representation with relation features by combining operations. At the same time, it combines tail entity with h4, and its corresponding relationship r4 is fused to get vector representation. The embedded representation of neighborhood information is then obtained by combining the vector representation of two different neighborhood relations of entity h1, and the importance of different neighbor nodes of entity h1 is calculated, namely:α12, α14, α21, α23. Next, combining the neighborhood relationship features of entities, the entity representation including neighborhood relations and multiple repetitive complex relations features is obtained. Finally, the vector representation of entities and relationships is input into the target function to calculate scores.
3.1 Notations and problem definition
In this part, we formally introduce some notations as below. For the given domain knowledge graph G = \(\{V, R, E, \mathcal {X}, \mathcal {Z}\}\), where V represents the set of all vertices, E represents the set of all entities and R represents the set of all relationships. What’s more, \(\mathcal {X} \in \mathbb {R}^{|V| \times d_{0}}\) denotes the initial entity embedding, \(\mathcal {Z} \in \mathbb {R}^{|R| \times d_{0}}\) denotes the initial relation embedding.
The triple of knowledge graph be denoted as (h, r, t), where h ∈ E is the head entities, t ∈ E is the tail entities, and r ∈ R is a relation corresponding to an edge in the KG. KGC task aims to find missing triple in the KG, it is to answer queries (h,r,?) or (?,r,t). In our model, the score for the triple is defined as f(h,r,t), and the score determines whether the triple is valid or not.
3.2 Attention-based relation conjunction
Our method is inspired by the GCNs that is a scalable approach for semi-supervised learning on graph-structured data, and it is based on an efficient variant of convolutional neural networks that operate directly on graphs [13]. Unlike other graph-structured data, edges in the knowledge graph represent relationships and have directions. Hence, we allow information to spread in the directed relation like Syntactic GCN [17]. We extend a reverse relationship and a self-loop relationship between the two entities, i.e., \(E^{\prime }=E \cup \{\left (j, i, r^{-1}\right ) \mid (i, j, r) \in \) E}∪{(i,i,L)∣i ∈ V )}, \(R^{\prime }=R \cup R_{i n v} \cup \{L\}\). Where \(R_{i n v}=\left \{r^{-1} \mid r \in R\right \}\) denotes the inverse edges and L denotes the self loop.
The same pair of entities may have different relationships in the DKG. Therefore, our method extract the relation characteristic \(\mathcal {Z}\) as the initial embedding to require the relation embedding \(\boldsymbol {g}_{r} \in \mathbb {R}^{d}, \forall r \in R\) and the entity representation \(\boldsymbol {g}_{v} \in \mathbb {R}^{d}, \forall v \in E\). In order for GCN to obtain the characteristics embedding of a relationship, we conjunct relationships with their corresponding entities by operations that utilizing in knowledge embedding methods [2] as shown in Fig. 3. The formula is as follows:
Where \(\phi : \mathbb {R}^{d} \times \mathbb {R}^{d} \rightarrow \mathbb {R}^{d}\) denotes the conjunction operate, \(e_{h} \in \mathbb {R}^{d}\) indicates head entity embedding, \(e_{t} \in \mathbb {R}^{d}\) indicates tail entity embedding, \(e_{r} \in \mathbb {R}^{d}\) indicates relation embedding.
However, the importance of different neighborhood nodes of each entity is different, motivated by the attention mechanism, we assign different scores to different adjacent nodes. Given a entity h, we denote the set of adjacency nodes of h as \(\mathbf {N}=\left \{h_{1}, h_{2}, \ldots , h_{j}, \ldots , h_{N}\right \},\) \( h_i \in \mathbb {R}^{d}\), then computing the attention coefficients through the formula:
Where \(e_r^{i j}\) represent the importance of node j to node i, the W is a weight matrix, a: \(\mathbb {R}^{d} \times \mathbb {R}^{d} \rightarrow \mathbb {R}\) denotes a single-layer feedforward neural network, ∥ denotes the concatenate operation. Therefore, we can compute the attention score by the following formula:
Thus, our entity embedding acquires the information of the corresponding relationship, and relation representation has a different attention score.
3.3 Joint graph attention network
The R-GCNs show that the GCN framework can be applied to modeling relational data [20], and its update formula like the following:
Where \(\mathcal {N}(i)\) is the set of adjacent nodes of node h. However, the formula can not acquire the relation feature of the DKGs. As shown in Fig. 3, our JGAN module combines the relationship with its corresponding adjacent nodes by jointing operations,i.e. \(\phi \left (e_{t_{1}}, e_{r_{1}}\right )\), and computes different importance score to the edges of each pair of adjacent nodes, i.e. α1 denotes the \(\alpha _{t_{1} h}\). Therefore, the update formula of our JGAN module is as follows:
Where xj denotes the initial embedding of entity j, zr is the initial embedding of relation r, eh represents the updated representation of entity i, and \(\boldsymbol {W}_{d_(r)}\) is a parameter rely on relation.
Because of the different relationship directions and the importance of varying adjacency relations, we define different weights \(\boldsymbol {W}_{d_(r)}\) for different relationship directions in our JGAN module, for example, r is the relationship between node i and node j, then \(\boldsymbol {W}_{d_(r)}\) are defined as follows:
After that, we can acquire the relation embedding update formula:
In this way, we can obtain the embedding of all entities and relationships.
3.4 Score computing
Through the JGAN module, could get a representation of an entity eh and its connection er. In our structure, we select the ConvE [7] as the score function:
Where \(\overline {\mathbf {e}_{h}}\) is the reshaped two-dimensional representation of eh, \(\overline {\mathbf {e}_{r}}\) is the reshaped two-dimensional representation of er, W denotes the parameter matrix.
4 Experiment
We start this section by introducing our Medical KG, the biomedical KG dataset, and two standard datasets, followed by the experimental settings, including evaluation protocol and baselines. At last, we present the experiment results in Medical KG and the biomedical KG dataset. What’s more, in order to study the Variability and Generality of our JGAN model, we present and analyze experiment results on two standard datasets.
4.1 Knowledge graph datasets
In order to evaluate our proposed model, we select four knowledge graph datasets as follows:
We collect and release a Medical knowledge graph dataset by working with companies, which describes the relationship between disease, drugs, and symptoms. It covers 27100 entities and 41 relation types.
Paper_reading dataset is a biomedical knowledge graph, which has been proposed by [30]. The dataset extracted from biomedical papers describes the relationship between disease, chemicals, and genes. It covers 14858 entities and 134 relation types.
WN18RR [7] is a benchmark dataset that removes the reversible relations of WN18, and it contains 40943 entities and 11 relation types. For WN18 [2] dataset, is an English dictionary that includes semantic information and extracted from wordnet, which covers 40943 entities and 18 relation types. Each entity corresponds to a different meaning, and there is a relationship between different senses.
FB15K-237 [25] also is a benchmark dataset of FB15K [2], and it includes 14541 entities and 237 relation types. With respect to FB15K dataset, it is a large collaborative knowledge base composed of metadata that preserved general knowledge about the world and sub-sampling from the FreeBase dataset, which is composed of 14951 entities and 1345 relation types.
Table 2 presents the statistics of four datasets.
4.2 Experimental setup
- Cross-validation :
-
The datasets are uniformly divided into training, validation, and test sets. We conducted a 10-fold cross-validation experiment for all datasets to ensure an unbiased evaluation.
- Evaluation protocol :
-
In the knowledge graph completion and link prediction task, the target is to identify the missing entity for a given entity and relation. i.e., for each fact (h, r, ?), predict the tail entity. Similarly, for each fact (?, r, t), predict the head entity. Following the evaluation criteria of existing models, we apply the popular evaluation criteria: Mean Rank(MR), Mean Reciprocal Ranking(MRR) and Hits@k, Mean Rank(MR) means the objective function score ranking of the entities that need to be predicted in test triples. As the name suggests, Mean Reciprocal Ranking (MRR) refers to that reciprocal of Mean Rank. Calculation of Hits@k demand to arrange the f function values according to MR, and then check whether the correct answer for each testing triple is ranked in the first k of the sequence, and if so, count plus one.
Therefore, lower Mean Rank(MR), higher Mean Reciprocal Ranking(MRR), or higher Hits@k show better performance.
- Baselines :
-
In the knowledge graph completion task, we compare our model with the following baseline models: TransE [2], TransD [10], TransH [32], SimplE [12], RotatE [24], ComplEx [26], DistMult [35], Analogy [16], ConvE [7] , CompGCN [27], and SACN [21].
4.3 Experimental results and analysis
In this part, we show the experimental results in Tables 3 and 4. In addition, we analyze the Variability and Generality of our JGAN model.
- Experimental Results :
-
In order to compare with baseline models, we experiment with the paper_reading dataset on these models. Table 3 reports
Hits@10, Hits@3, Hits@1, MR, and MRR results of eleven different baseline models and our models on our medical dataset and the biomedical knowledge graphs datasets. From the table, we can see that our JGAN model achieves the best performance on MR, Hits@10, Hits@3, and Hits@1. Specifically, our JGAN model improves upon the best SACN’s MR by a margin of 43, upon Rotate’s Hits@3 by a margin of 2.6 % for the test. What’s more, our model’s performance is similar to the best baseline in MRR, Hits@10, and Hits@1. At the same time, our model obtains a relatively good result and the best MR, Hits@k on our medical dataset. Concretely, the MR value improves 127 points compared to the best result 2508 in our medical dataset. Moreover, we get the best Hit@3 0.203 and MRR 0.188. From the above experimental results, our model has achieved excellent results in different datasets, especially in the domain knowledge graph paper_reading. In a word, we conclude that our JGAN model learns the multiple relation representation and information of different adjacency relations, so as to achieve better performance.
- Variability Analysis :
-
For the sake of verifying the variability and generality of our JGAN model, we evaluate our model in two classic datasets, i.e., WN18RR, FB15K237. Table 4 reports Hits@10, Hits@3, Hits@1, MR, and MRR results of eleven different baseline models and our models on WN18RR and FB15K237 datasets. From Table 4, we can learn that our model obtains the best MR, MRR, Hits@3, and Hits@1 results in the FB15K237 dataset. More concretely, our model acquires the MR value 160 in the FB15K237 dataset, and this result is more than 17 points higher than the best Rotate’s MR value, MRR, and the hits@k also receive the best work. In the WN18RR dataset, we obtain the best MRR value of 2415 compared with all the baseline methods. However, our JGAN doesn’t work well on other indicators. According to our analysis, WN18RR dataset is an English dictionary that includes semantic information, which covers only 18 relation types, therefore it includes less complicated relations. However, FB15K-237 dataset is a general knowledge graph, which includes 237 relation types and more complex relations. thus, the reason for this result may be that our JGAN model learns complicated relation but ignore more simple relationship.
In order to further analyze the experimental results in WN18RR, we analyzed the data again. Our proposed method models the multiple relationships in the knowledge graph. As shown in Table 5, we can see that the Medical, Paper_reading, and FB15K237 datasets contain a large proportion of multiple relationships, especially Paper_reading dataset contains 99% of the multiple relationships. Furthermore, based on Table 3, our model achieves the best performance on the Paper_reading dataset. As for WN18RR dataset, which includes less multiple relation data, so our model obtains a poor result.
5 Conclusion
In this paper, we introduce the JGAN model, a relation-based knowledge graph completion model for a multi-relations knowledge graph. The JGAN model fuses the entity with its corresponding relationship by a joint operation to learn the multiple relationship’s representations. What’s more, inspired by the attention mechanism, our model adopts its idea to measure the importance of different adjacency relationships of entities. Meanwhile, our experiments proved that the JGAN model outperforms other models on multi-relation knowledge graph datasets.
However, our model has low computational efficiency and high complexity due to the graph convolution network in the model. After preliminary analysis, We find that many methods based on graph convolution networks also have high complexity, and the complexity of the JGAN model is equivalent. In future work, we consider reducing the adjacency matrix by pooling the input model’s adjacency matrix in future work, to improve the computational efficiency and reduce the complexity of the model. In this way, our model can be extended to a larger scale knowledge graph.
References
Bollacker K, Evans C, Paritosh P, Sturge T, Taylor J (2008) Freebase: a collaboratively created graph database for structuring human knowledge. In: Proceedings of the 2008 ACM SIGMOD International conference on management of data, SIGMOD ’08. https://doi.org/10.1145/1376616.1376746. ACM, New York, pp 1247–1250
Bordes A, Usunier N, Garcia-Duran A, Weston J, Yakhnenko O (2013) Translating embeddings for modeling multi-relational data. In: Neural Information Processing Systems (NIPS), pp 2787–2795. https://hal.archives-ouvertes.fr/hal-00920777
Bordes A, Weston J, Usunier N (2014) Open question answering with weakly supervised embedding models. In: Joint european conference on machine learning and knowledge discovery in databases. Springer, Berlin, pp 165–180
Bruna J, Zaremba W, Szlam A, LeCun Y (2014) Spectral networks and locally connected networks on graphs. In: Bengio Y, LeCun Y (eds) 2nd International conference on learning representations, ICLR 2014, Banff, AB, Canada, April 14-16, 2014, conference track proceedings. arXiv:1312.6203
Catherine R, Cohen W (2016) Personalized Recommendations Using Knowledge Graphs: a probabilistic logic programming approach. Recsys ’16. Association for Computing Machinery, New York, pp pp 325–332. https://doi.org/10.1145/2959100.2959131
Clark P, Thompson JA, Holmback H, Duncan L (2000) Exploiting a thesaurus-based semantic net for knowledge-based search. In: Seventeenth national conference on artificial intelligence & twelfth conference on innovative applications of artificial intelligence
Dettmers T, Minervini P, Stenetorp P, Riedel S (2018) Convolutional 2d knowledge graph embeddings. In: Proceedings of the AAAI Conference on Artificial Intelligence, vol 32. https://ojs.aaai.org/index.php/AAAI/article/view/11573
Dong X, Gabrilovich E, Heitz G, Horn W, Lao N, Murphy K, Strohmann T, Sun S, Zhang W (2014) Knowledge vault: a web-scale approach to probabilistic knowledge fusion. In: Proceedings of the 20th ACM SIGKDD international conference on knowledge discovery and data mining, KDD ’14. Association for Computing Machinery, New York, pp 601–610, DOI https://doi.org/10.1145/2623330.2623623, (to appear in print)
Hamilton W, Ying Z, Leskovec J (2017) Inductive representation learning on large graphs. CoRR arXiv:abs/1706.02216, pp 1024–1034. 1706.02216
Ji G, He S, Xu L, Liu K, Zhao J (2015) Knowledge graph embedding via dynamic mapping matrix. In: Proceedings of the 53rd annual meeting of the association for computational linguistics and the 7th international joint conference on natural language processing (volume 1: long papers). https://doi.org/10.3115/v1/P15-1067. Association for Computational Linguistics, Beijing, pp 687–696
Ji G, Liu K, He S, Zhao J (2016) Knowledge graph completion with adaptive sparse transfer matrix. In: Proceedings of the AAAI Conference on Artificial Intelligence 30(1). https://ojs.aaai.org/index.php/AAAI/article/view/10089
Kazemi SM, Poole D (2018) Simple embedding for link prediction in knowledge graphs. In: Bengio S, Wallach HM, Larochelle H, Grauman K, Cesa-Bianchi N, Garnett R (eds) Advances in neural information processing systems 31: annual conference on neural information processing systems 2018, NeurIPS 2018, 3-8 December 2018, Montréal, Canada, pp 4289–4300. http://papers.nips.cc/paper/7682-simple-embedding-for-link-prediction-in-knowledge-graphshttp://papers.nips.cc/paper/7682-simple-embedding-for-link-prediction-in-knowledge-graphshttp://papers.nips.cc/paper/7682-simple-embedding-for-link-prediction-in-knowledge-graphs
Kipf TN, Welling M (2017) Semi-supervised classification with graph convolutional networks. In: 5th International Conference on Learning Representations, ICLR 2017, Toulon, France, April 24-26, 2017, Conference Track Proceedings. OpenReview.net. https://openreview.net/forum?id=SJU4ayYgl
Lehmann J, Isele R, Jakob M, Jentzsch A, Kontokostas D, Mendes PN, Hellmann S, Morsey M, Van Kleef P, Auer S et al (2015) Dbpedia–a large-scale, multilingual knowledge base extracted from wikipedia. Semantic Web 6(2):167–195
Lin Y, Liu Z, Sun M, Liu Y, Zhu X (2015) Learning entity and relation embeddings for knowledge graph completion. https://ojs.aaai.org/index.php/AAAI/article/view/9491
Liu H, Wu Y, Yang Y (2017) Analogical inference for multi-relational embeddings. In: Precup D, Teh YW (eds) Proceedings of the 34th international conference on machine learning, ICML 2017, Sydney, NSW, Australia, 6-11 August 2017, proceedings of machine learning research. http://proceedings.mlr.press/v70/liu17d.html, vol 70. PMLR, pp 2168–2178
Marcheggiani D, Titov I (2017) Encoding sentences with graph convolutional networks for semantic role labeling. In: Proceedings of the 2017 conference on empirical methods in natural language processing. 1703.04826
Nguyen DQ, Nguyen TD, Nguyen DQ, Phung DQ (2018) A novel embedding model for knowledge base completion based on convolutional neural network. In: Walker MA, Ji H, Stent A (eds) Proceedings of the 2018 conference of the north american chapter of the association for computational linguistics: Human Language Technologies, NAACL-HLT, New Orleans, Louisiana, USA, June 1-6, 2018, Volume 2 (Short Papers), Association for Computational Linguistics, pp 327–333. https://doi.org/10.18653/v1/n18-2053
Nickel M, Tresp V, Kriegel HP (2011) A three-way model for collective learning on multi-relational data. ICML 11:809–816. https://icml.cc/2011/papers/438_icmlpaper.pdf
Schlichtkrull M, Kipf TN, Bloem P, Van Den Berg R, Titov I, Welling M (2018) Modeling relational data with graph convolutional networks. In: European semantic web conference, Springer, pp 593–607
Shang C, Tang Y, Huang J, Bi J, He X, Zhou B (2019) End-to-end structure-aware convolutional networks for knowledge base completion. Proc AAAI Conf Art Intell 33(01):3060–3067. https://doi.org/10.1609/aaai.v33i01.33013060. https://ojs.aaai.org/index.php/AAAI/article/view/4164
Suchanek FM, Kasneci G, Weikum G (2007) Yago: a core of semantic knowledge. In: Proceedings of the 16th international conference on world wide web, WWW ’07. https://doi.org/10.1145/1242572.1242667. Association for Computing Machinery, New York, pp 697–706
Sun Z, Deng Z, Nie J, Tang J (2019) Rotate: Knowledge graph embedding by relational rotation in complex space. In: 7th International Conference on Learning Representations, ICLR 2019, New Orleans, LA, USA, May 6-9, 2019. OpenReview.net. https://openreview.net/forum?id=HkgEQnRqYQ
Sun Z, Vashishth S, Sanyal S, Talukdar PP, Yang Y (2020) A re-evaluation of knowledge graph completion methods. In: Jurafsky D, Chai J, Schluter N, Tetreault JR (eds) Proceedings of the 58th annual meeting of the association for computational linguistics, ACL 2020, Online, July 5-10, 2020, pp. 5516–5522. association for computational linguistics. https://www.aclweb.org/anthology/2020.acl-main.489/
Toutanova K, Chen D (2015) Observed versus latent features for knowledge base and text inference. In: Proceedings of the 3rd Workshop on Continuous Vector Space Models and their Compositionality, pp. 57–66. Association for Computational Linguistics, Beijing, China. https://doi.org/10.18653/v1/W15-4007. https://www.aclweb.org/anthology/W15-4007
Trouillon T, Welbl J, Riedel S, Gaussier É, Bouchard G (2016) Complex embeddings for simple link prediction. In: International Conference on Machine Learning, pp 2071–2080. 1606.06357
Vashishth S, Sanyal S, Nitin V, Talukdar PP (2020) Composition-based multi-relational graph convolutional networks. In: 8th international conference on learning representations, ICLR 2020, Addis Ababa, Ethiopia, April 26-30, 2020. OpenReview.net. https://openreview.net/forum?id=BylA_C4tPr
Vashishth S, Yadav P, Bhandari M, Talukdar PP (2019) Confidence-based graph convolutional networks for semi-supervised learning. In: Chaudhuri K, Sugiyama M (eds) The 22nd International Conference on Artificial intelligence and statistics, AISTATS 2019, 16-18 April 2019, Naha, Okinawa, Japan, Proceedings of machine learning research, vol 89, PMLR, pp 1792–1801. http://proceedings.mlr.press/v89/vashishth19a.html
Velickovic P, Cucurull G, Casanova A, Romero A, Liò P, Bengio Y (2018) Graph attention networks. In: 6th International Conference on Learning Representations, ICLR 2018, Vancouver, BC, Canada, April 30 - May 3, 2018, Conference Track Proceedings. OpenReview.net. https://openreview.net/forum?id=rJXMpikCZ
Wang Q, Huang L, Jiang Z, Knight K, Ji H, Bansal M, Luan Y (2019) Paperrobot: Incremental draft generation of scientific ideas. In: Korhonen A, Traum DR, Márquez L (eds) Proceedings of the 57th conference of the association for computational linguistics, ACL 2019, Florence, Italy, July 28- August 2, 2019, Volume 1: long papers, association for computational linguistics, pp 1980–1991. https://doi.org/10.18653/v1/p19-1191
Wang Z, Li J (2016) Text-enhanced representation learning for knowledge graph. In: Proceedings of the Twenty-Fifth International Joint Conference on Artificial Intelligence, IJCAI’16, AAAI Press, pp 1293–1299
Wang Z, Zhang J, Feng J, Chen Z (2014) Knowledge graph embedding by translating on hyperplanes. In: Twenty-Eighth AAAI conference on artificial intelligence. https://ojs.aaai.org/index.php/AAAI/article/view/8870
Xie R, Liu Z, Jia J, Luan H, Sun M (2016) Representation learning of knowledge graphs with entity descriptions. In: Proceedings of the AAAI Conference on Artificial Intelligence, vol 30. https://ojs.aaai.org/index.php/AAAI/article/view/10329
Yadav P, Nimishakavi M, Yadati N, Vashishth S, Rajkumar A, Talukdar PP (2019) Lovasz convolutional networks. In: Chaudhuri K, Sugiyama M (eds) The 22nd international conference on artificial intelligence and statistics, AISTATS 2019, 16-18 April 2019, Naha, Okinawa, Japan, Proceedings of Machine Learning Research, vol 89, pp 1978–1987. PMLR. http://proceedings.mlr.press/v89/yadav19a.html
Yang B, Yih W, He X, Gao J, Deng L (2015) Embedding entities and relations for learning and inference in knowledge bases. In: Bengio Y, LeCun Y (eds) 3rd International Conference on Learning Representations, ICLR 2015, San Diego, CA, USA, May 7-9, 2015, Conference Track Proceedings. 1412.6575
Acknowledgements
This work was supported by the National Natural Science Foundation of China under Grant 62077015, No. 62177015, and the Key Laboratory of Intelligent Education Technology and Application of Zhejiang Province, Zhejiang Normal University, Jinhua, Zhejiang, China.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Huang, J., Lu, T., Zhu, J. et al. Multi-relational knowledge graph completion method with local information fusion. Appl Intell 52, 7985–7994 (2022). https://doi.org/10.1007/s10489-021-02876-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-021-02876-4