Abstract
Peripheral artery disease (PAD) is an occlusive disease of limb arteries. Critical limb ischemia (CLI) is an advanced form of PAD that is prognostically worse in subjects with diabetes and can result in limb loss, gangrene, and death, although the underlying signaling mechanisms that contribute to its development remain poorly understood. By comparing plasma samples from diabetic humans with PAD and mouse models of PAD, we identified miR-375 to be significantly downregulated in humans and mice during progression to CLI. Overexpression of miR-375 was pro-angiogenic in endothelial cells in vitro and induced endothelial migration, proliferation, sprouting, and vascular network formation, whereas miR-375 inhibition conferred anti-angiogenic effects. Intramuscular delivery of miR-375 improved blood flow recovery to diabetic mouse hindlimbs following femoral artery ligation (FAL) and improved neovessel growth and arteriogenesis in muscle tissues. Using RNA-sequencing and prediction algorithms, Kruppel-like factor 5 (KLF5) was identified as a direct target of miR-375 and siRNA knockdown of KLF5 phenocopied the effects of miR-375 overexpression in vitro and in vivo through regulatory changes in NF-kB signaling. Together, a miR-375-KLF5-NF-kB signaling axis figures prominently as a potential therapeutic pathway in the development CLI in diabetes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Peripheral artery disease (PAD), a gradual narrowing or occlusion of the peripheral arteries, impairs blood flow to the lower extremities [1]. Although PAD is estimated to affect over 200 million people worldwide. A subset of patients with PAD will experience worsening symptoms and develop critical limb ischemia (CLI) [2]. CLI is defined by rest pain and is frequently coupled with poor wound healing, ulceration, and gangrene which can lead to limb amputation and is associated with increased all-cause mortality [3]. While there are several clinical risk factors associated with PAD and CLI, patients with diabetes are disproportionally represented among those at the highest risk for CLI in part due to micro-angiopathy and impaired angiogenic responses [4,5,6]. Numerous therapeutic approaches have attempted to enhance angiogenesis to restore blood flow and improve outcomes, yet these efforts have been largely unsuccessful [7]. This lack of progress in clinical treatment reflects in part a poor understanding of the molecular mechanisms with which transcriptional drift in affected individuals results in cell- and tissue-level dysfunction.
MicroRNAs (miRNAs) are short, conserved single-stranded noncoding RNAs with seed sequences that post-transcriptionally bind the 3’ untranslated region (3′UTR) of messenger RNAs (mRNAs) to repress protein translation [8]. Currently, over 2000 human miRNAs have been identified and likely regulate over one-third of the genome. Endothelial-enriched miRNAs such as miR-26a, miR-145, and miR-615-5p have emerged as critical angiogenic modulators that are frequently dysregulated in vasculoproliferative and inflammatory diseases that include CLI, atherosclerosis, and tumor angiogenesis [9,10,11]. Indeed, preliminary investigations of miRNA profiles in individuals with cardiometabolic diseases such as diabetes highlight changes in miRNA expression that contribute to disease pathogenesis [12, 13]. While identification of these miRNAs has made significant advances due to high-throughput deep sequencing, putative miRNA networks which contribute to CLI are still largely unexplored [14].
Because miRNAs exhibit high conservation across species, we hypothesized that it was possible to identify key dysregulated endothelial cell (EC)-enriched miRNAs that were commonly affected in human and mouse CLI. We sequenced and compared miRNAs isolated from the plasma of human patients with diabetes and PAD who progressed to CLI and compared these to differentially expressed miRNAs between two diabetic mouse limb ischemia models, sub-acute and acute limb ischemia (SLI and ALI, respectively). Of the 22 overlapping miRNAs identified, we observed significant downregulation of miR-375-3p (referred hereafter as miR-375) as a potential therapeutic target. Overexpression of miR-375 promoted angiogenesis and an analysis of potential targets identified Kruppel-like factor 5 (KLF5) as an anti-angiogenic target of miR-375 in ECs. Furthermore, in vivo studies indicated that therapeutic intramuscular delivery to overexpress miR-375 or knockdown KLF5 following femoral artery ligation (FAL) improved blood flow recovery in part through interactions with subunits of the NF-kB pathway.
Methods
Identification of miRNAs from human and mouse plasma samples
Plasma for human miRNA-sequencing was collected from the previously described Thrombin Receptor Antagonist in Secondary Prevention of Atherothrombotic Ischemic Events (TRA 2°P)-TIMI 50 trial [15]. Briefly, patients with symptomatic PAD were eligible for the trial on the basis of ongoing symptoms and an ABI < 0.85 or prior amputation or revascularization procedure for ischemia. At baseline, all patients had an ABI performed using a standardized protocol and had their Fontaine class (I–IV) (Table S1) assessed by trained investigators in the context of their baseline assessment (including ABI). Patients selected for analysis in the current study were the subgroup of PAD patients who were determined at this baseline assessment to have Fontaine III or IV class disease. Diabetic Db/Db mice were subjected to two different surgeries to replicate CLI: (1) FAL, which causes immediate cessation of blood flow hence (Acute ischemia); and (2) ameroid constrictors which gradually expands from fluid adsorption inducing artery occlusion (Sub-acute ischemia). Acute limb ischemia was performed via ligation of the distal and proximal ends of the femoral artery followed by cauterization. Sub-acute hindlimb ischemia model was performed using ameroid constrictors, which induce gradual femoral artery occlusion over 1–3 days. Two ameroid constrictors were placed on the femoral artery, one proximal to the lateral circumflex femoral artery and the second proximal to the bifurcation of the popliteal and saphenous arteries.
The EdgeSeq miRNA Whole Transcriptome Assay from HTG Molecular Diagnostics, Inc. (AZ, USA) was used to measure miRNA expression in plasma from human donors and mice. The HTG EdgeSeq system combines quantitative nuclease protection assay chemistry with a next-generation sequencing platform to enable the semi-quantitative analysis of 2083 human miRNA transcripts in a single assay. Fifteen microliters of plasma were used for extraction-free sample processing and quantitative nuclease protection assay using the EdgeSeq processor (HTG Molecular Diagnostics, Inc.). The libraries were sequenced using Illumina NextSeq, and data were parsed through HTG EdgeSeq before count data were assessed for quality and analyzed using R.
Cell culture and transfection
Human Umbilical Vein Endothelial Cells (HUVECs, Lonza), Coronary Artery Endothelial Cells (CAECs, Lonza), and Human Aortic Endothelial Cells (HAECs, Lonza) were cultured in endothelial cell growth media (EGM-2, Lonza). Mouse brain endothelial cells (B.End, Lonza) were cultured in DMEM with 10% FBS. Human Coronary Artery Smooth Muscle Cells (CASMCs, Lonza) and mouse aortic vascular smooth muscle cells (MOVAS, Lonza) were cultured in smooth muscle cell growth media (SMGM, Lonza) prior to use. Mouse 3T3 Fibroblasts were grown in DMEM with 10% FBS. Non-specific and miR-375 mimics (50 µM, Life Technologies) were mixed with Lipofectamine 2000 prior to transfection. Non-specific and miR-375 inhibitors (100 µM, Life Technologies) were mixed with Lipofectamine 2000 prior to transfection. Silencer Select scramble and KLF5 (10 µM, Life Technologies) were mixed with Lipofectamine 2000 prior to transfection. All transfections were performed according to manufacturer’s specifications.
RT-qPCR
Total RNA was extracted with QIAzol or TRIzol reagent and miRNeasy micro kit (Qiagen) per manufacturer protocol. RNA concentration and quality were assessed using Qubit (ThermoFisher) and Bioanalyzer (Agilent). Isolated RNA was reverse transcribed using a High-Capacity cDNA Reverse Transcription kit (Applied Biosystems) or miRCURY LNA miRNA RT kit (Qiagen) according to manufacturer specifications. Real-time quantitative PCR (RT-qPCR) was conducted on the AriaMx RT-qPCR system (Agilent).
Specific human primers include KLF5, ICAM1, VCAM1, SELE, IL-1β, HPRT, and GAPDH were purchased (Life Technologies). All expression values were normalized to GAPDH or HPRT and represent 2−ΔCt. The primers used are as follows: KLF5 (5′-CCTGGTCCAGACAAGATGTGA-3′ and 5′-GAACTGGTCTACGACTGAGGC-3′), ICAM1 (5′-ATGCCCAGACATCTGTGTCC-3′ and 5′-GGGGTCTCTATGCCCAACAA-3′), VCAM1 (5′-GGGAAGATGGTCGTGATCCTT-3′ and 5′-TCTGGGGTGGTCTCGATTTTA-3′), SELE (5′-ATGATGGCTTATTACAGTGGCAA-3′ and 5′-GTCGGAGATTCGTAGCTGGA-3′), IL-1β (5′-AGAGTGGAGCCTGGTCTTACA-3′ and 5′-CCTTTGCTGACAATAAGCACTGG-3′), HPRT (5′-CCTGGCGTCGTGATTAGTGAT-3′ and 5′-AGACGTTCAGTCCTGTCCATAA-3′), and GAPDH (5′-GGAGCGAGATCCCTCCAAAAT-3′ and 5′-GGCTGTTGTCATACTTCTCATGG-3′). For miRNAs, miRCURY LNA miRNA PCR primers (Qiagen) for miR-375 and U6 were used.
Specific mouse primers include klf5, icam1, vcam1, sele, il-1β, tnfα, and gapdh were purchased (Life Technologies). All expression values were normalized to gapdh and represent 2−ΔCt. The primers used are as follows: klf5 (5′-CCGGAGACGATCTGAAACACG-3′ and 5′-GTTGATGCTGTAAGGTATGCCT-3′), icam1 (5′-GTGATGCTCAGGTATCCATCCA-3′ and 5′-CACAGTTCTCAAAGCACAGCG-3′), vcam1 (5′-AGTTGGGGATTCGGTTGTTCT-3′ and 5′-CCCCTCATTCCTTACCACCC-3′), sele (5′-ATGCCTCGCGCTTTCTCTC-3′ and 5′-GTAGTCCCGCTGACAGTATGC-3′), il-1β (5′-GAAATGCCACCTTTTGACAGTG-3′ and 5′-TGGATGCTCTCATCAGGACAG-3′), tnfα (5′-CCCTCACACTCAGATCATCTTCT-3′ and 5′-GCTACGACGTGGGCTACAG-3′), and gapdh (5′-AGGTCGGTGTGAACGGATTTG-3′ and 5′-TGTAGACCATGTAGTTGAGGTCA-3′). For miRNAs, miRCURY LNA miRNA PCR primers (Qiagen) for miR-375 and U6 were used.
For chromatin immunoprecipitation (ChIP) analysis, nuclear lysates were collected and DNA precipitated for p65 and extracted by manufacturers protocol (Cell Signaling, 9003). All expression was normalized to the input. The primers used for the p65 promoter region are as follows: IL-1β (5′-TCCCTCCCTTAGTGCCAACTATGT-3′ and 5′-ACAGTCTCCACAGTTCTGCCATGT-3′), IL-6 (5′-TAGAGCTTCTCTTTCGTTCCCGGT-3′ and 5′-TGTGTCTTGCGATGCTAAAGGACG-3′), and TNFα (5′-GCTTCCTCCAGATGAGCTT-3′ and 5′-TGCTGTCCTTGCTGAGGGA-3′).
Western blot and immunoprecipitation
Cultured cells were harvested and lysed in RIPA buffer (50mM Tris–HCl pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) in combination with protease inhibitor cocktail tablets (Roche) and phosphatase inhibitors (Invitrogen). Protein concentrations were determined using a Pierce BCA Assay (ThermoFisher) and protein was loaded on gradient 4–20% Mini-PROTEAN TGX precast gels (Bio-Rad). Separated proteins were transferred to PVDF membranes using the Transfer Turbo Blot System (Bio-Rad) and Trans-Blot Turbo RTA Transfer kit (Bio-Rad) for 2.5 min. Membranes were then blocked with 2.5% BSA in TBST for 1 h at room temperature before being incubated at 4 °C overnight with primary antibodies against KLF5 (D7S3F, Cell Signaling, 1:1000), IKKβ (L570, Cell Signaling, 1:1000), phospho-IKKβ (16A6, Cell Signaling, 1:1000), IκBα (44D4, Cell Signaling, 1:000), phospho-IκBα (14D4, Cell Signaling, 1:1000), p65 (D14E12, Cell Signaling, 1:1000), phospho-p65 (93H1, Cell Signaling, 1:1000), and GAPDH (14C10, Cell Signaling, 1:1000). For LPS (R&D Systems), IL-1β (R&D Systems), and TNFα (R&D Systems) treatments, samples were incubated with 1 µg/mL, 10 ng/mL, or 10 ng/mL, respectively, for indicated time points. Membranes were washed in TBST and incubated with HRP-conjugated anti-mouse (Cell Signaling, 1:5000) or anti-rabbit (Cell Signaling, 1:5000) secondary antibodies for 1 h at room temperature. Visualization of protein bands was performed using SuperSignal West Femto Maximum Sensitivity Substrate (Life Technologies) solution and a luminescent image analyzer (Bio-Rad, Chemidoc). Densiometric analysis was analyzed in ImageJ (NIH). All measurements were normalized to GAPDH (Cell Signaling, 1:1000) lanes run for each sample with phosphorylated proteins being normalized to their non-phosphorylated amount. KLF5, p-65, and phospho-p65 were each utilized for immunoprecipitation. Briefly, untreated cell lysate was incubated with each antibody (1:100) for 3 h at room temperature. Suspensions were then incubated with Protein A/G-conjugated Dynabeads for 1 h. Samples were then separated on a Dynamag-2 Magnet rack and processed as described above.
Immunofluorescent staining and imaging
For confocal analysis, each sample was fixed in 4% PFA (Boston Bio Products) for 30 min and then permeabilized with 0.5% Triton-X and blocked with 2.5% BSA for 1 h. Slides were then incubated with primary antibodies against KLF5, phospho-p65, CD31 (WM59, Biolegend, 1:100), or αSMA (EPR5368, ABCAM, 1:100) overnight. After washing, samples were incubated with the appropriate Alexafluor 488-conjugate antibody (1:500) where applicable as well as counter stained with DAPI (Life Technologies, 1:2500). Confocal images were acquired on a Zeiss LSM 880 Confocal inverted microscope at the Beth Israel Deaconess Medical Center confocal imaging and immunohistochemistry core facility) at 20× or 40× magnification.
RNA-Seq analysis and pathway enrichment analysis
RNA library preparation and sequencing analysis was conducted at GENEWIZ, LLC./Azenta US, Inc. (South Plainfield, NJ, USA). The RNA sequencing libraries were prepared using the NEBNext Ultra II RNA Library Prep Kit for Illumina using manufacturer’s instructions (New England Biolabs, Ipswich, MA, USA). The sequencing libraries were multiplexed and clustered onto a flowcell and loaded onto the Illumina HiSeq instrument according to manufacturer’s instructions. The samples were sequenced using a 2 × 150 bp Paired End configuration. Trimmed reads were mapped to the reference genome available on ENSEMBL using the STAR aligner v.2.5.2b. The mean quality score of all samples was 38.61 with a range of 42,000,000–68,000,000 reads per sample. All samples had > 92% of mapped fragments over total fragments. Unique gene hit counts were calculated by using feature Counts from the Subread package v.1.5.2. Total gene hit counts and CPM values were calculated for each gene and downstream differential expression analysis between specified groups was performed using DESeq2. Genes with adjusted P value < 0.05 were labeled as differentially expressed genes for each comparison. Differentially expressed genes were subjected to gene set enrichment analyses by using MetaCoreTM (Clarivate) software. Enrichment analysis for functional ontologies (Process Networks) and analysis using network building tools was performed in MetaCoreTM. A false discovery rate (FDR) < 0.05 was used as threshold for significance in enrichment analysis. Visualization of pathway enrichment analysis were performed as dotplot using (ggplot2 package).
For identification of predicted targets, the top 120 predicated targets from TargetScan, miRDB, and TarBase were queried and downloaded. Samples which overlapped within all data sets were then identified in Excel and individually found within the generated differential expressed genes file.
In Vitro functional assays
All samples were transfected 48 h prior to use.
For scratch assays, 3-chamber molds (Ibidi) in 24-well plates were utilized. Briefly, molds were attached on the bottom of plates and HUVECs were plated at 2 × 105 cells/chamber and allowed to adhere for 6–8 h. Following adhesion, the molds were gently lifted and wound closure was assessed every hour or 16 h. Images were captured and anlyzed with the CytoSMART Omni.
Matrigel™ basement membrane matrix was added to 24-well culture plates and incubate at 37 °C for 30 min to allow for gelation. HUVECs were plated at 2 × 104 cells/well and network formation was assessed every hour for 16 h. Images were captured with a CytoSMART Omni. Network formation was quantitated by counting the number of tubes formed per field of view utilizing the network formation algorithm within the IKOSA machine learning algorithm software. Transwell migration assays were performed by seeding 2.5 × 104 cells/insert (8 μm pore, Corning) in 200 µL of media. The chamber was flushed with 500 µL of basal media overnight prior to treatment with 50 ng/mL of VEGF. Samples were fixed in 4% PFA (Boston Bio Products) and the underside stained with DAPI (Life Technologies, 1:2500).
For spheroid sprouting assay, spheroid formation utilizing the hanging drop method was conducted. Briefly, 1000 cells were plated into 25 µL droplets on non-adherent, plastic Petri dishes overnight in 2% methyl cellulose. Spheroids were then collected and plated into 1.5 collagen in 24 well plates. Collagen gelation was performed according to manufacturer’s specifications. Images were taken on the CytoSMART Omni every 6 h for 18 h.
For BrdU assays, 5 × 103 cells were seeded per 96-well plate and allowed to adhere overnight. Samples were then incubated with BrdU-incorporating reagent for 12 h, with and without 50 ng/mL of VEGF (R&D Systems). Samples were then fixed and quantitated using the Cell Proliferation ELISA BrdU Colorimetric kit accord to manufacturer instructions (Sigma-Aldrich).
Luciferase assay
The wtKLF5 and mutKLF5 promoters were purchased from GeneCopoeia. Each was co-transfected with either NSm or 375m. Transactivation assays were performed with the Secrete-PairTM Dual Luminescence Assay Kit (GeneCopoeia). NF-kB luciferase reporter was co-transfected with NSm, 375m, siScr, or siKLF5 Transfected cells were then stimulated with TNFα and collected in 200 µL Reporter Lysis Buffer (Promega). The activity of luciferase was measured according to manufacturer’s protocol. Each reading of luciferase activity was normalized to the protein content in the sample for the same lysate.
Animal studies
All protocols concerning animal use were approved by the Institutional Animal Care and Use Committee at Brigham and Women’s Hospital and Harvard Medical School, Boston, MA, USA and conducted in accordance with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals.
Ischemic injury of the hindlimb was induced through a unilateral FAL in diabetic (Db/Db) mice. Db/Db mice were utilized for studies unless otherwise indicated. Mice were anesthetized and the femoral artery with surrounding tissue was sutured closed at the proximal and distal ends and cauterized. Immediately following surgery, mice were imaged on a Laser Doppler Imager-2, and were re-imaged at days 4, 8, 12, 16, and 20 unless indicated otherwise. Mice were injected with either NSm, 375m, siScr, or siKLF5 at 1nmol/100µL/injection intramuscular at days—1, 3, 7, 11, and 15. Visual assessment of limb ischemia was assessed at each imaging session. All blood flow recovery values were generated by comparing the injured limb to the contralateral limb at the same time point.
Isolation of primary mouse tissues and cells
For whole tissue isolations, either Db/+ or Db/Db mice were utilized. Whole gastrocnemius muscle was dissected away from the soleus muscle, minced into small pieces, and then further minced in a dounce until a uniform suspension was obtained. Samples were then transferred into TRIzol for further processing.
For EC isolation, magnetic Dynabeads (sheep anti-rat IgG, Invitrogen) were pre-washed 5x in PBS with 0.1% BSA and then incubated with rat anti-mouse CD31 antibody (BD Biosciences) at a ratio of 5:1 Dynabeads/antibody overnight at 4 °C in a tube rotator. A 15 µL mixture was prepared per tissue requiring EC isolation. Gastrocnemius muscle was dissected away from soleus muscle. The tissue was then placed in 1 mL Digestion Buffer (1 mg/mL Collagenase, 4 mg/mL Dispase) and minced with a razor blade for 1 min. Minced tissue was transferred to a tube containing 9 mL additional Digestion Buffer and incubated at 37 °C for 40 min. The slurry was then passed through a 70 μm cell strainers (Corning Falcon) and 10 mL of Wash Buffer 2 (DMEM/F-12 with 10% FBS) was added. The slurry was then centrifuged at 500×g for 10 min at 4 °C. The supernatant was aspirated and discarded. The residual pellet was manually disrupted, and 1 mL RBC Lysis Buffer was added (E-Bioscience) for 5 min at room temperature with occasional gentle shaking. The RBC Lysis reaction was neutralized with 10 mL DMEM with 10% FBS and then passed through a 40 μm cell strainers (Westnet). The eluate was centrifuged at 500×g for 10 min at 4 °C and the resulting pellet was suspended in 1 mL of Wash Buffer 1 (PBS pH 7.2, 0.1% BSA, 2 mM EDTA, 0.5% FBS). The solubilized pellet was added to a 15 µL pre-washed and conjugated Dynabead/CD31 antibody mixture and allowed to tumble at 37 °C for 30 min. The slurry of lysate and Dynabead/antibody mixture was bound on a Dynamag-2 Magnet (Invitrogen) for 1 min and the supernatant was collected as a non-EC (non-endothelial cell) fraction. The beads containing bound ECs were then washed on the Dynamag-2 Magnet 5x and the resultant pellet was eluted in 0.5 mL TRIzol.
Statistics
Statistical analyses were performed using GraphPad Prism 9.1 (GraphPad Software Inc.). A Student’s t test, one-way, or two-way ANOVA with Tukey’s post hoc test was used to determine significance where appropriate. Data and error bars represent mean ± SEM. A P value ≤ 0.05 was considered statistically significant. All data was confirmed for normality with a separate Shapiro-Wilk test.
Results
MiR-375 expression in ECs is correlated with blood flow recovery
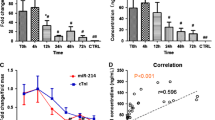
To explore which miRNAs may be commonly dysregulated in diabetic CLI, we sequenced and compared human and mouse plasma samples (Fig. 1A). Briefly, human plasma samples were acquired from two patient cohorts: 1) human subjects with PAD and diabetes who did (Fontaine III/IV) and did not (Fontaine I/II) experience PAD progression into CLI with at least one critical limb event, defined as either revascularization or limb amputation. Samples were acquired from patients enrolled in the Thrombin Receptor Antagonist in Secondary Prevention of Atherothrombotic Ischemic Events (TRA-2P)-Thrombolysis in Myocardial Infarction (TIMI) 50 study. Mouse plasma samples were acquired from the diabetic (Leprdb, Db/Db) mouse model for PAD with and without CLI. Two different surgical models were used for modeling CLI: (1) FAL which mimics CLI (termed acute limb ischemia (ALI)) and (2) implantation of ameroid constrictors which progressively narrows the femoral artery without total blood flow loss (termed sub-acute ischemia (SLI)). Overlapping of human and mouse groups identified 22 downregulated miRNAs, of which miR-375 was identified from this set as a miRNA not previously investigated in the context of diabetic CLI (Figs. 1B, S1). RT-qPCR analysis of whole gastrocnemius muscle in non-diabetic (Db/+) and Db/Db mice over the course of FAL recovery revealed a nearly 10-fold induction of miR-375 in non-diabetic mice while diabetic mice exhibited no changes in miR-375 expression (Fig. 1C). Next, to investigate if the expression profile in ECs changed over time in ischemic limbs, we isolated the EC portion of the gastrocnemius muscle via CD31 antibody-conjugated magnetic bead pulldown in Db/+ mice, collecting whole-cell RNA. We observed a 3- and 7-fold increase in miR-375 in ECs at days 7 and 11, respectively, mirroring changes that occurred in whole gastrocnemius muscle (Fig. 1D). Cell profiling confirmed that endothelial cells were enriched for miR-375 when compared to other cell types found within the skeletal muscle tissue compartment (Fig. 1E). Transfection of human umbilical vein endothelial cells (HUVECs) with miR-375 mimics (375m) increased proliferation while transfection with miR-375 inhibitors (375i) exhibited anti-proliferative effects when compared to their respective controls (non-specific mimic, NSm; non-specific inhibitor, NSi) in BrdU assays (Fig. 1F). Overexpression and inhibition of miR-375 in transfected HUVECs was confirmed via RT-qPCR (Fig. S2).
Identification of miR-375 in CLI. A Schematic of identification of dysregulated miRNAs in CLI. Briefly, RNA-sequencing of miRNAs in diabetic subacute limb ischemia (SLI) and acute limb ischemia (ALI) in mice was compared to Fontaine I/II and Fontaine III/IV diabetic PAD patients to identify miRNAs which were commonly dysregulated in ALI and CLI. B Individual counts of miR-375 in mouse and human serum samples. C RT-qPCR profiling of miR-375 of whole gastrocnemius muscle in Db/+ and Db/Db mice isolated at 0, 3, 11, and 31 days following FAL.N = 6 biological replicates per time point per condition. D RT-qPCR profiling miR-375 in EC and non-EC fractions of the gastrocnemius in Db/+ following FAL at 0, 3, 7, and 11 days. N = 3 biological replicates per condition per time point. E RT-qPCR profiling of miR-375 in endothelial and non-endothelial cells. N = 3 technical replicates. F Cell proliferation analysis by BrdU assay in HUVECs transfected with non-specific mimics (NSm), miR-375 mimics (375m), non-specific inhibitors (NSi), and miR-375 inhibitors (375i). N = 3 technical replicated. All values are mean ± SEM. P values are indicated by *, **, ***, and **** and indicate a value of < 0.05, < 0.01, < 0.001, and < 0.0001, respectively. Data was analyzed using a Student’s t test
Overexpression of miR-375 promotes angiogenesis in functional assays in vitro
To assess the functional impact of miR-375 on endothelial behavior, a series of functional assays were conducted. In a transwell migration assay, 375m increased HUVEC migration across the transwell membrane while 375i had a non-significant trend in decreasing migration as quantified by nuclei count (Fig. 2A). In an endothelial wound scratch assay transfection with 375m significantly accelerated wound closure both in speed and total amount over time compared to non-specific controls (Fig. 2B). Similarly, in a spheroid sprouting assay, 375m markedly increased the number of sprouts per spheroid compared to controls (Fig. 2C). In contrast, treatment of HUVECs with 375i significantly abrogated scratch wound closure and spheroid sprouting when compared to controls (Fig. 2D, E). Lastly, using a Matrigel network tube formation assay, 375m significantly enhanced tube formation in frequency and total length, whereas 375i exhibited the opposite effects (Fig. 2F, G).
Functional in vitro assays in HUVECs reveal the pro-angiogenic effects of miR-375 overexpression and anti-angiogenic effects of miR-375 inhibition. A Quantification and confocal micrographs of transwell migration assay following HUVEC transfection with NSm, 375m, NSi, and 375i. Scale bar is 50 μm. N = 5 technical replicates. B, C Time-lapse images obtained by CytoSMART Omni device and quantification of endothelial scratch assay (t = 1 h increments) wound closure and spheroid sprouting (t = 6 h increments) in HUVECs transfected with NSm and 375m. N = 4 and 5 technical replicates for scratch and spheroid assays, respectively. D, E Time-lapse images obtained by CytoSMART Omni device and quantification of endothelial scratch assay (t = 1 h increments) wound closure and spheroid sprouting (t = 6 h increments) in HUVECs transfected with NSi and 375i. N = 4 and 5 technical replicates for scratch and spheroid assays, respectively. F Time-lapse images and quantification obtained by CytoSMART Omni device of Matrigel tube and network formation (t = 1 h increments) in HUVECs transfected with NSm and 375m. N = 5 technical replicates. G Time-lapse images and quantification obtained by CytoSMART Omni device of Matrigel tube and network formation (t = 1 h increments) in HUVECs transfected with NSi and 375i. N = 5 technical replicates. All values are mean ± SEM. P values are indicated by *, **, ***, and **** and indicate a value of < 0.05, < 0.01, < 0.001, and < 0.0001, respectively. Data was analyzed using a Student’s t test
miR-375 targets pro-angiogenic signaling pathways and KLF5
To explore the mechanisms by which miR-375 regulates endothelial function, RNA from HUVECs transfected with 375m or NSm control was processed for RNA-sequencing. Top upregulated genes, including pro-angiogenic genes CXCL8 and IL11, and top downregulated genes were identified based on differential expression analysis as shown by heatmap and volcano plot (Fig. 3A, B). Sample group homogeneity was confirmed by PCA analysis (Fig. S3A). Organization of identified genes by Ingenuity Pathway Analysis (IPA) enumerated several important angiogenic pathways such as IL-8 signaling, wound healing, HIF1α signaling, and endothelin-1 signaling which were overexpressed in 375m HUVEC data sets (Figs. 3C, D, S3B). Lastly, to identify a potential gene target of miR-375, the top 120 genes in 3 separate predictive databases (TargetScan, miRDB, and TarBase) were queried and 10 overlapping genes identified as potential targets (Figs. 3E, Table S2) [16,17,18]. These genes were then located within the dataset and relative gene expression was analyzed (Fig. 3 F). Of these 10 genes, CHYS1, RBPJ, KLF5, and QKI, exhibited the highest degree of repression in the 375m group. Briefly, each potential target was investigated by conducting a query with the NCBI database and identifying the gene ontology annotations (GOA) under both process and function. KLF5 was identified as a critical potential target of miR-375 since it has been implicated in the regulation of angiogenesis and skeletal muscle repair.
RNA-sequencing of miR-375 overexpression in HUVECs identify pro-angiogenic pathway enrichment and identify potential gene targets. A Heatmap of the top 20 upregulated and downregulated genes in HUVECs transfected with NSm or 375m. B Volcano plot demonstrating the most differentially regulated genes plotted against the Log10(P value) with specific genes identified. C Ingenuity Pathway Analysis (IPA) and gene networks affected in the top differentially regulated genes alongside z-score to indicate activation or inhibition of pathways. D Chord plot mapping of highlighted endothelial pathways and the expression of genes within those pathways. E Schematic of the identification of miR-375 targets. Briefly, the top 120 genes identified in a database query of TargetScan, miRDB, and TarBase were overlapped with top downregulated transcripts from (B) and 10 genes were identified. F Expression of identified targets and the identification of KLF5 as an angiogenic target
To assess if KLF5 was a validated target of miR-375, a potential binding site for miR-375 was identified in the 3’UTR of KLF5 (Fig. 4A). Significant repression of KLF5 was confirmed in 375m-treated HUVECs by RT-qPCR and confocal microscopy while no significant overexpression of KLF5 was observed in 375i-treated HUVECs (Fig. 4B). Western blot analysis of HUVEC lysates verified significant protein repression of KLF5 following transfection with 375m while a modest non-significant increase in KLF5 expression was detected in response to 375i (Fig. 4C, D). Lastly, to exclude any indirect effects of miR-375 on KLF5 expression, we assessed miR-375-mediated repression using either a wildtype or mutated KLF5 3′UTR reporter in HUVECs and HEK293T cells (Fig. 4E). Significant repression of luciferase activity was only observed in 375m-transfected, wildtype 3′UTR cells, confirming miR-375’s ability to directly repress KLF5.
KLF5 is validated as a target of miR-375 in ECs. A Schematic of the miR-375 target site in the 3′ untranslated region (3′UTR) of KLF5 mRNA. B RT-qPCR and confocal micrograph analysis of KLF5 expression in HUVECs following transfection with NSm, 375m, NSi, and 375i. Scale bar is 50 μm. N = 4 technical replicates. C Western blot analysis of KLF5 expression in whole cell lysate of HUVECs transfected with NSm and 375m. N = 4 technical replicates. D Western blot analysis of KLF5 expression in whole cell lysate of HUVECs transfected with NSi and 375i. N = 4 technical replicates. E Schematic of the wild type KLF5 (wtKLF5) and mutated KLF5 (mutKLF5) 3′UTR used for luciferase reporter assays and quantification of luciferase activity in HUVECs and HEK-293T cells co-transfected with wtKLF5 or mutKLF5 luciferase plasmid and NSm or 375m. N = 4 technical replicates. All values are mean ± SEM. P values are indicated by *, **, ***, and **** and indicate a value of < 0.05, < 0.01, < 0.001, and < 0.0001, respectively. Data was analyzed using a Student’s t test
siRNA knockdown of KLF5 phenocopies overexpression of miR-375 in ECs in vitro
Given the effects of miR-375 on KLF5 expression, we next sought to determine if knockdown of KLF5 via siRNA (siKLF5) was capable of phenocopying the 375m-mediated effects on EC function. The siRNA-mediated repression of KLF5 was first confirmed by RT-qPCR and Western blot analysis (Fig. 5A, B). A scratch assay of HUVECs transfected with siKLF5 exhibited accelerated wound closure when compared to the scrambled siRNA (siScr) control (Fig. 5C). Similar to 375m-treated HUVECs, siKLF5 transfection of spheroids increased spheroid sprouting and significantly accelerated tube formation in total count and length in a Matrigel tube formation assay (Fig. 5D, E).
Silencing of KLF5 in HUVECs phenocopies miR-375 overexpression in vitro. A RT-qPCR confirmation of KLF5 inhibition HUVECs transfected with either scramble (siScr) and KLF5 (siKLF5) siRNA. N = 4 technical replicates. B Western blot analysis and quantification of whole cell lysates transfected with siScr or siKLF5 in HUVECs. N = 4 biological replicates. C Time-lapse images obtained by CytoSMART Omni device and quantification of endothelial scratch assay (t = 1 h increments) wound closure in HUVECs transfected with siScr and siKFL5 3. N = 4 technical replicates. D Time-lapse images obtained by CytoSMART Omni device and quantification of spheroid sprouting (t = 12 h) in HUVECs transfected with siScr and siKLF5 3. N = 5 technical replicates. E Time-lapse images and quantification obtained by CytoSMART Omni device of Matrigel tube and network formation (t = 1 h increments) in HUVECs transfected with siScr and siKLF5. N = 5 technical replicates. All values are mean ± SEM. P values are indicated by * and *** indicate a value of < 0.05 and < 0.001, respectively. Data was analyzed using a Student’s t test
miR-375 overexpression disrupts phospho-p65 signaling
Ischemic tissues and vasculature in diabetic limbs are dysfunctional in part due to a sustained pro-inflammatory burden. Because miR-375 was observed to be downregulated in the plasma of CLI patients, miR-375 expression in response to the pro-inflammatory cytokines LPS, IL-1β, and TNFα was assessed and found to be markedly repressed following treatment (Fig. 6A). Because pro-inflammatory signaling in ECs is routed primarily through the NF-kB pathway, miR-375’s potential effects on the individual components of this pathway were investigated [19]. Western blot analysis of whole cell lysates of 375m transfected HUVECs treated without and with TNFα found a 32% reduction in phospho-IKKβ activation, no changes in phospho-IkBα, and a 74% reduction in phospho-p65 protein content (Fig. 6B, C). No differences were noted in total unphosphorylated protein levels. No significant differences were found in HUVECs transfected with 375i and treated with TNFα. Confocal micrographs of HUVECs transfected and treated with TNFα demonstrated that 375m reduced phospho-p65 in the nucleus (Fig. 6D). Interestingly, while no differences in total phospho-p65 was observed in whole cell lysates from 375i-transfected HUVECs, a significant increase in nuclear phospho-p65 was observed by confocal microscopy (Fig. 6D). Lastly, in 375m-transfected ECs treated with TNFα, KLF5 and target NF-kB endothelial genes SELE, ICAM1, VCAM1, and IL-1B were quantified by RT-qPCR and found to be repressed (Fig. 6E). These data raise the possibility that miR-375 via KLF5 may inhibit NF-kB signaling. Chromatin immunoprecipitation analysis for IL-1β, IL-6, and TNFα confirm that overexpression of miR-375 inhibited p65 binding to pro-inflammatory promoters (Fig. S4).
miR-375 overexpression suppresses phospho-p65 signaling. A RT-qPCR analysis of miR-375 expression in HUVECs following treatment with LPS, IL-1β, and TNFα. B, C Western blots and quantification of whole cell lysates for IKKβ, phospho-IKKβ, IκBα, phospho-IκBα, p65, and phospho-p65 in HUVECs transfected for NSm, 375m, NSi, and 375i following a 1 h TNFα treatment. N = 4 technical replicates. D Confocal micrographs and quantification of nuclear phospho-p65 in HUVECs transfected for NSm, 375m, NSi, and 375i following a 1 h TNFα treatment. Scale bar is 50 μm. N = 4 technical replicates. E RT-qPCR analysis KLF5, SELE, ICAM1, VCAM1, and IL-1β expression in HUVECs transfected with NSm and 375m following treatment with TNFα for 8 h. N = 4 technical replicates. All values are mean ± SEM. P values are indicated by *, **, ***, and **** and indicate a value of < 0.05, < 0.01, < 0.001, and < 0.0001, respectively. Data was analyzed using a one-way ANOVA or Student’s t test, where appropriate
Knockdown of KLF5 inhibits phospho-p65 signaling
The above data identifying repression of inflammatory signaling by 375m may be congruent with repression of KLF5 as KLF5 is known to associated with the NF-kB pathway. Indeed, Western blot analysis of HUVECs transfected with siKLF5 phenocopied the repression of phospho-p65, though similar trends in phospho-IKKB were also observed (Fig. 7A, B). RT-qPCR of KLF5, SELE, ICAM1, VCAM1, and IL-1B further confirmed that the EC inflammatory response were significantly blunted in response to TNFα (Fig. 7C). Immunofluorescent analysis of HUVECs transfected with either 375m or siKLF5 and TNFα treatment reduced ICAM1 and VCAM1 expression by confocal microscopy (Fig. S5). To further investigate if KLF5 was directly associated with phospho-p65, whole-cell lysates were immunoprecipitated against antibodies for p65 and phospho-p65 under basal conditions and analyzed by Western blot. Pulldown assays confirmed a physical interaction between KLF5 and p65, as well as phospho-p65; however, KLF5 abundance was significantly decreased in 375 m transfected ECs (Fig. 7D, E). We confirmed through a subsequent immunoprecipitation and Western blot analysis of a time-course treatment of HUVECs with TNFα that siKLF5 transfected HUVECs show a compromised phospho-p65 response (Fig. 7F, G). Lastly, a NF-kB luciferase reporter assay confirmed a significant reduction in luciferase activity with either the transfection of 375m or siKLF5 in HUVECs following TNFα treatment (Fig. S6). Together, these data indicate that miR-375 may alter endothelial function by repression of KLF5 and its association with phospho-p65.
Silencing of KLF5 suppresses pro-inflammatory stimulation of HUVECs. A, B Western blot analysis and quantification of whole cell lysates for IKKβ, phospho-IKKβ, IκBα, phospho-IκBα, p65, and phospho-p65 in HUVECs transfected for siScr and siKLF5 following treatment with TNFα for 1 h. N = 4 technical replicates. C RT-qPCR analysis of KLF5, SELE, ICAM1, VCAM1, and IL-1β expression in HUVECs transfected with siScr and siKLF5 following treatment with TNFα for 8 h. D, E Western blot analysis and quantification of whole protein input and immunoprecipitation of p65 and phospho-p65 in whole cell lysates of HUVECs transfected with NSm and 375m for KLF5, p65, and phospho-p65. N = 4 technical replicates. F, G Western blot analysis and quantification of immunoprecipitation of KLF5, p65, and phospho-p65 of whole cell lysates in HUVECs transfected with siScr and siKLF5 and treatment with TNFα for 0, 10, and 60 min. N = 3 technical replicates. All values are mean ± SEM. P values are indicated by *, **, ***, and **** and indicate a value of < 0.05, < 0.01, < 0.001, and < 0.0001, respectively. Data was analyzed using a one-way Student’s t test
Treatment of Db/Db mice with 375m or siKLF5 improves revascularization after limb ischemia
To connect our in vitro observations to potential similar mechanisms in vivo, diabetic mice, which exhibited reduced miR-375 levels during FAL recovery, were found to exhibit similar trends in EC pro-inflammatory genes (ICAM1, VCAM1, SELE, IL1b, TNFa) by RT-qPCR. Results indicate that several pro-inflammatory genes were upregulated at time points overlapping with increased KLF5 expression (and reduced miR-375 expression) in Db/Db mice during recovery (Fig. S7). Because of these data coupled with the findings that miR-375 confers pro-angiogenic properties yet its expression is reduced in response to CLI, the potential therapeutic effects of 375m in limb ischemia was investigated by intramuscular (IM) delivery of 375 m during FAL in Db/Db mice (Fig. 8A). Local IM delivery of miR-375 significantly improved blood flow recovery by 12 days and overexpression of miR-375 in CD31+ freshly isolated gastrocnemius mouse ECs was confirmed by RT-qPCR (Fig. 8B). Consistent with the in vitro findings, miR-375-mediated IM delivery also markedly repressed Klf5, Sele, Vcam1, Il-1β, and Tnfα in mouse ECs while icam1 trended lower in 375m treated Db/Db mice. After 21 days of recovery, mice were sacrificed and the gastrocnemius muscle was isolated, fixed, embedded into paraffin, and then axially sectioned for immunofluorescent staining against CD31 and αSMA to assess for differences in blood vessel growth in ischemic muscle (Fig. 8C). Quantification of CD31+ area links an increase in blood flow in 375m-injected mice to an increase in over blood vessel coverage (Fig. 8D). Furthermore, it was also discovered that not only is there an increase in vessel density within muscle tissues, but that arterialization (αSMA+) was significantly increased in both overall coverage and frequency in 375m-injected mice.
To further investigate whether therapeutic silencing of KLF5 recapitulated the effects of miR-375 in vivo, Db/Db mice underwent FAL with the use of local IM delivery of siKLF5 instead of 375m. Compared to the siScr control group, a significant increase in blood flow recovery was observed by day 4 in siKLF5-treated mice with a steady improvement in blood flow over the course of the study (Fig. 9A). KLF5 expression was reduced in gastrocnemius ECs from the the siRNA KLF5 group. In contrast to miR-375, only Vcam1 and Sele were found to be repressed in CD31+ isolated gastrocnemius mouse ECs (Fig. 9B). Analogous to the approach above, mice were sacrificed, and the gastrocnemius muscle was isolated and axially sectioned for immunofluorescent staining against CD31 and αSMA to assess for differences in blood vessel growth, however this was completed at 12 days rather than 21 (Fig. 9C). Quantification of CD31+ area found a similar significant increase in overall blood vessel coverage and vessel frequency in siKLF5-injected mice (Fig. 9D). It is also noted that while arterialization trended upwards without statistical significance in coverage, a significant increase in artery frequency was found in siKLF5-injected mice. Taken together, these data indicate that endothelial miR-375 dysregulation in Db/Db mice following HLI likely contributes to impaired blood flow recovery through increased KLF5 and pro-inflammatory signaling.
Overexpression of miR-375 in the gastrocnemius of Db\Db mice following HLI accelerates blood flow recovery. A Schematic representation of experimental design for miR-375 delivery. Briefly, HLI was performed on Db/Db mice and Doppler images acquired at days 0, 4, 8, 12, 16, and 20. Intramuscular injections of NSm or 375m was performed at days—1, 3, 7, 11, and 15. The ischemic gastrocnemius was then isolated and processed for further analysis. Doppler images represent days 0 and 20 of the ischemic and contralateral paw of mice during HLI recovery. Quantification represents the normalized blood flow of the ischemic limb to the contralateral limb at indicated time points. N = 9 biological replicates. B RT-qPCR analysis of klf5, icam1, vcam1, sele, il-1β, and tnfα expression in the EC portion of isolated gastrocnemius muscle. N = 5 biological replicates. C Confocal micrographs of gastrocnemius muscle immunostained with CD31 (red) and αSMA (green). DAPI was used a nuclear counterstain. D Quantification of total CD31+ area and individual vessel frequency as well as αSMA+ coverage and perivascular αSMA+ vessel frequency. N = 4 biological replicates. Scale bars are 150 μm. All values are mean ± SEM. P values are indicated by *, **, ***, and **** and indicate a value of < 0.05, < 0.01, < 0.001, and < 0.0001, respectively. Data was analyzed using a one-way Student’s t test
Silencing of KLF5 improves blood flow recovery in Db/Db mice following HLI. A Doppler images represent days 0, 8, and 12 of the ischemic and contralateral paw of mice during HLI recovery. Quantification represents the normalized blood flow of the ischemic limb to the contralateral limb at indicated time points. N = 9 biological replicates. B RT-qPCR analysis of klf5, icam1, vcam1, sele, il-1β, and tnfα expression in the EC portion of isolated gastrocnemius muscle. N = 4 biological replicates. C Confocal micrographs of gastrocnemius muscle immunostained with CD31 (red) and αSMA (green). DAPI was used a nuclear counterstain. D Quantification of total CD31+ area and individual vessel frequency as well as αSMA+ coverage and perivascular αSMA+ vessel frequency. N = 5 biological replicates. Scale bars are 150 μm. All values are mean ± SEM. P values are indicated by *, **, and ***, and indicate a value of < 0.05, < 0.01, and < 0.001, respectively. Data was analyzed using a one-way Student’s t test
Discussion
Impaired angiogenesis is a hallmark of PAD and contributes to the progression to CLI, especially in subjects with diabetes [20]. CLI manifests as rest pain and/or tissue loss due to impaired tissue perfusion. Recently, the syndrome of Chronic Limb Threatening Ischemia (CLTI), has been defined similarly and is manifested by rest pain and/or tissue loss but includes perfusion parameters. In addition, CLTI and the associated treatment concepts recognize the complex interplay between ischemia, wounds, and infection and may better reflect the broad range of patients who present with impaired perfusion that can delay wound healing and increase amputation risk [21]. In this study, miR-375 and its target KLF5 were identified as a key angiogenic miRNA-mRNA pair in the progression of diabetic CLI through altered phospho-p65 NF-kB signaling. In ECs, when miR-375 is overexpressed, proliferation, wound closure, spheroid sprouting, and tube formation are augmented while miR-375 inhibition exhibits the opposite effects. Findings from the in vitro and in vivo data identified KLF5 in ECs as a key target of miR-375. In support, inhibition of KLF5 phenocopied the functional effects of miR-375 overexpression. While the regulation of miR-375 expression has been studied in diabetes, where it is critical in the development and homeostasis of pancreatic islet cells, or in cancer, where it appears to be anti-tumorigenic, it has yet to be explored in endothelial cells [22,23,24,25]. Indeed, the lack of investigation into miR-375 expression in endothelial function may be attributed to many factors as our data suggests its expression appears to be regulated mainly in response to injury or disease states.
Intriguingly, our data observed an apparent spike in miR-375 expression during the most-angiogenic phase of HLI recovery in Db/+ mice, yet no significant changes were observed in Db/Db mice. In phase with this, an increase in miR-375 expression in Db/+ mice coincided with a decrease in KLF5 expression and a repression of pro-inflammatory gene activation. In contrast, in Db/Db mice, the repression of miR-375 at all time points coincided with increased KLF5 expression in endothelial cells compared to non-endothelial cells as well as a sustained upregulation of the expression of pro-inflammatory genes. As a result, we considered miR-375 to exhibit pro-angiogenic and anti-inflammatory effects on endothelial cells. This hypothesis was mirrored by our RNA-sequencing data in response to miR-375 overexpression in ECs which found an upregulation in numerous pro-angiogenic gene ontology pathways yet a downregulation in specific pro-inflammatory genes such as ICAM1, VCAM1, SELE, and CCL2. Thus, we hypothesized that the repression of miR-375 in Db/Db mice when compared to Db/+ mice may be linked to reduced angiogenesis and increased inflammation. The hyper-inflammatory vascular state associated with diabetes is known to play a role in the disease pathology and sustained inflammatory signals are known to contribute to the inability of tissues to heal in otherwise healthy tissues [26,27,28]. For instance, the angiogenic threshold is known to be higher in individuals with diabetes due to the presence of ineffective pro-angiogenic signaling pathways [29]. Indeed, considerable investigative interest has aimed to assess the potential of anti-inflammatory drugs such as salicylates, methotrexate, and various IL-1β- and TNFα-blocking drugs in treating the inflammation-related effects of diabetes [30,31,32]. Thus, our in vivo findings support that miR-375 may facilitate vessel growth and angiogenesis by regulating both angiogenesis and inflammation, and consequently, tissue repair and regeneration.
The investigated miR-375 target, KLF5, is more widely studied in cardiovascular and cardiometabolic disease [33, 34]. It is important to consider that its contributions to angiogenesis appears to be dependent on tissue and cell context with previous studies largely focusing on smooth muscle cell function [35,36,37]. Accumulating studies indicate that KLF5 expression increases in diabetes, hyperinsulinemia, and hyperlipidemia as well as other cardiovascular disease states and is largely deleterious in vascular injury [38, 39]. Indeed, overexpression of KLF5 is known to inhibit eNOS signaling, inhibit IL-8 production, and alter IL-6 signaling in the vascular endothelium [40, 41]. Interestingly, heterozygous KLF5 mice (KLF5+/−) are athero-resistant while global knockouts are embryonically lethal—highlighting the need to consider its cell-specific context [35]. While KLF5 is also known to facilitate phospho-p65 signaling in some capacity through either modification of p65 phosphorylation stability or facilitating nuclear translocation, our studies highlight for the first time the link between KLF5, phospho-p65, and endothelial inflammation [42, 43]. We speculate that the abnormal inflammatory state of the diabetic endothelium may be a plausible context by which miR-375 overexpression is therapeutic. That is, miR-375 promotes angiogenesis by inhibiting KLF5 in Db/Db HLI mice because of their disease- driven inflammatory burden and likely targets the tissue regeneration phase of wound healing, though the optimal delivery time and kinetics of this process will require future studies. One potential confounding aspect of these data is that while our acute model for HLI in Db/Db mice is meant to model critical limb-threatening events, the disease pathophysiology in humans is a chronic condition rather than acute. Indeed, future studies would require a more suitable model system that would integrate the gradual occlusion and narrowing of the femoral artery in Db/Db mice in the presence of other co-existing conditions (e.g. drug treatment, atherosclerosis, etc.), potentially increasing the clinical translatability.
These data underscore the importance of considering tissue and/or disease state in regulatory signaling networks and highlight a few limitations of our study. Certainly, our delivery mechanism through intramuscular injection of miR-375 achieves overexpressing in both ECs and non-EC cell subtypes. As a result, while we separated out different compartments of the muscle tissue itself to confirm our in vitro findings, the contributions to HLI recovery in Db/Db mice is likely due to multiple contributing cell types [44]. This appears to be true in analogous paradigms for smooth muscle cell fractions of tissues as KLF5 signaling is critical to their function [45]. Investigation of miR-375’s effects on smooth muscle cell function or targeted delivery to ECs in vivo will be of interest to consider in future studies. Another potential confounding issue is to consider the potential contributions of KLF4. As KLF5 and KLF4 are frequently considered to be antagonistic transcription factors, one balancing out the effects of the other in many tissues, the inhibition of KLF5 may tip the activity of this other transcription factor and affect HLI recovery indirectly [46, 47]. In addition, while the overexpression of miR-375 is therapeutic physiologically in Db/Db mice, its utility as a potential biomarker remains unclear to identify patients at risk for the development of CLI in human subjects with diabetes and will require future studies, potentially with other miRNAs identified from our cohort (Table S3). Finally, as advances in RNA-based therapeutics have exploded in various disease contexts, future studies in large animal models will be needed to assess the potential use of miR-375 mimics including durability, efficacy, and safety over time.
In conclusion, we have demonstrated in this study that miR-375 is downregulated in diabetic humans who develop CLI as well as Db/Db mice following HLI (Fig. 10A). We identify KLF5 as an unrecognized EC target of miR-375 and demonstrate that overexpression of miR-375 and inhibition of KLF5 is pro-angiogenic and anti-inflammatory (Fig. 10B). Lastly, we demonstrate that the delivery of miR-375 or siKLF5 into the muscle of Db/Db mice improves HLI recovery (Fig. 10C). These studies provide a foundation for future studies to investigate potential therapies in human patients afflicted with diabetes at risk for CLI.
Proposed role of miR-375 in diabetic CLI. A Progression of PAD into CLI represses miR-375 expression in diabetes. B MiR-375 binds to and inhibits translation of KLF5 expression, resulting in inhibition of phospho-p65 and suppression of inflammatory genes. C Overexpression of miR-375 or inhibition of KLF5 improves blood flow recovery after limb ischemia in diabetic mice
Data availability
All relevant data are available from the authors. The RNA-seq data are accessible at: GSE210778. Source data are provided with this paper.
Abbreviations
- 3′UTR:
-
3′ untranslated region
- ALI:
-
Acute limb ischemia
- CAECS:
-
Coronary artery endothelial cells
- CASMCs:
-
Coronary artery smooth muscle cells
- CD31:
-
Cluster of differentiation 31; PECAM
- ChIP:
-
Chromatin immunoprecipitation
- CLI:
-
Critical limb ischemia
- Db/+:
-
Heterozygous leptin receptor deficient, non-diabetic
- Db/Db:
-
Homozygous leptin receptor deficient, diabetic
- EC:
-
Endothelial cell
- FAL:
-
Femoral artery ligation
- HAECs:
-
Human aortic endothelial cells
- HIF1α:
-
Hypoxia-inducible factor alpha
- HLI:
-
Hind limb ischemia
- HUVECs:
-
Human umbilical vein endothelial cells
- IKKB:
-
Inhibitor of Nuclear factor kappa-B kinase subunit
- Beta IL-1β:
-
Interleukin-1 Beta
- IL-8:
-
Interleukin-8
- IPA:
-
Ingenuity pathway analysis
- IkBα:
-
Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha
- KLF4:
-
Kruppel-like factor 4
- KLF5:
-
Kruppel-like factor 5
- LPS:
-
Lipopolysaccharide
- miRNA:
-
microRNA
- miR-375:
-
microRNA-375-3p
- MOVAS:
-
Mouse vascular smooth muscle cells
- NF-kB:
-
Nuclear factor kappa-light-chain-enhancer of activated B cells
- Non-EC:
-
Non-endothelial cell
- p65:
-
Nuclear factor NF-kappa-B p65 subunit
- PAD:
-
Peripheral artery disease
- RT-qPCR:
-
Real-time quantitative polymerase chain reaction
- siRNA:
-
Short Interfering RNA
- SLI:
-
Sub-acute limb ischemia
- TNFα:
-
Tumor necrosis factor alpha
- αSMA:
-
Smooth muscle actin alpha
References
Criqui MH (2001) Systemic atherosclerosis risk and the mandate for intervention in atherosclerotic peripheral arterial disease. Am J Cardiol 88(7B):43J–47J. https://doi.org/10.1016/s0002-9149(01)01881-1
Hardman RL, Jazaeri O, Yi J, Smith M, Gupta R (2014) Overview of classification systems in peripheral artery disease. Semin Intervent Radiol 31(4):378–388. https://doi.org/10.1055/s-0034-1393976
Fowkes FG, Rudan D, Rudan I, Aboyans V, Denenberg JO, McDermott MM, Norman PE, Sampson UK, Williams LJ, Mensah GA, Criqui MH (2013) Comparison of global estimates of prevalence and risk factors for peripheral artery disease in 2000 and 2010: a systematic review and analysis. Lancet 382(9901):1329–1340. https://doi.org/10.1016/S0140-6736(13)61249-0
Marso SP, Hiatt WR (2006) Peripheral arterial disease in patients with diabetes. J Am Coll Cardiol 47(5):921–929. https://doi.org/10.1016/j.jacc.2005.09.065
Bonaca MP, Creager MA (2015) Pharmacological treatment and current management of peripheral artery disease. Circ Res 116(9):1579–1598. https://doi.org/10.1161/CIRCRESAHA.114.303505
Cooke JP, Losordo DW (2015) Modulating the vascular response to limb ischemia: angiogenic and cell therapies. Circ Res 116(9):1561–1578. https://doi.org/10.1161/CIRCRESAHA.115.303565
Beach JM (2021) Revascularization strategies for acute and chronic limb ischemia. Cardiol Clin 39(4):483–494. https://doi.org/10.1016/j.ccl.2021.06.006
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136(2):215–233. https://doi.org/10.1016/j.cell.2009.01.002
Icli B, Wara AK, Moslehi J, Sun X, Plovie E, Cahill M, Marchini JF, Schissler A, Padera RF, Shi J, Cheng HW, Raghuram S, Arany Z, Liao R, Croce K, MacRae C, Feinberg MW (2013) MicroRNA-26a regulates pathological and physiological angiogenesis by targeting BMP/SMAD1 signaling. Circ Res 113(11):1231–1241. https://doi.org/10.1161/CIRCRESAHA.113.301780
Icli B, Wu W, Ozdemir D, Li H, Cheng HS, Haemmig S, Liu X, Giatsidis G, Avci SN, Lee N, Guimaraes RB, Manica A, Marchini JF, Rynning SE, Risnes I, Hollan I, Croce K, Yang X, Orgill DP, Feinberg MW (2019) MicroRNA-615-5p regulates angiogenesis and tissue repair by targeting AKT/eNOS (protein kinase b/endothelial nitric oxide synthase) signaling in endothelial cells. Arterioscler Thromb Vasc Biol 39(7):1458–1474. https://doi.org/10.1161/ATVBAHA.119.312726
Climent M, Quintavalle M, Miragoli M, Chen J, Condorelli G, Elia L (2015) TGFbeta triggers miR-143/145 transfer from smooth muscle cells to endothelial cells, thereby modulating vessel stabilization. Circ Res 116(11):1753–1764. https://doi.org/10.1161/CIRCRESAHA.116.305178
Liang YZ, Li JJ, Xiao HB, He Y, Zhang L, Yan YX (2020) Identification of stress-related microRNA biomarkers in type 2 diabetes mellitus: a systematic review and meta-analysis. J Diabetes 12(9):633–644. https://doi.org/10.1111/1753-0407.12643
Zhou H, Peng C, Huang DS, Liu L, Guan P (2020) microRNA expression profiling based on microarray approach in human diabetic retinopathy: a systematic review and meta-analysis. DNA Cell Biol 39(3):441–450. https://doi.org/10.1089/dna.2019.4942
Perez-Cremades D, Cheng HS, Feinberg MW (2020) Noncoding RNAs in critical limb ischemia. Arterioscler Thromb Vasc Biol 40(3):523–533. https://doi.org/10.1161/ATVBAHA.119.312860
Morrow DA, Braunwald E, Bonaca MP, Ameriso SF, Dalby AJ, Fish MP, Fox KA, Lipka LJ, Liu X, Nicolau JC, Ophuis AJ, Paolasso E, Scirica BM, Spinar J, Theroux P, Wiviott SD, Strony J, Murphy SA, Committee TPTS, Investigators (2012) Vorapaxar in the secondary prevention of atherothrombotic events. N Engl J Med 366(15):1404–1413. https://doi.org/10.1056/NEJMoa1200933
Vlachos IS, Paraskevopoulou MD, Karagkouni D, Georgakilas G, Vergoulis T, Kanellos I, Anastasopoulos IL, Maniou S, Karathanou K, Kalfakakou D, Fevgas A, Dalamagas T, Hatzigeorgiou AG (2015) DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res 43 (Database issue):D153–159. https://doi.org/10.1093/nar/gku1215
McGeary SE, Lin KS, Shi CY, Pham TM, Bisaria N, Kelley GM, Bartel DP (2019) The biochemical basis of microRNA targeting efficacy. Science. https://doi.org/10.1126/science.aav1741
Chen Y, Wang X (2020) miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res 48(D1):D127–D131. https://doi.org/10.1093/nar/gkz757
Mussbacher M, Salzmann M, Brostjan C, Hoesel B, Schoergenhofer C, Datler H, Hohensinner P, Basilio J, Petzelbauer P, Assinger A, Schmid JA (2019) Cell type-specific roles of NF-kappaB linking inflammation and thrombosis. Front Immunol 10:85. https://doi.org/10.3389/fimmu.2019.00085
Martin A, Komada MR, Sane DC (2003) Abnormal angiogenesis in diabetes mellitus. Med Res Rev 23(2):117–145. https://doi.org/10.1002/med.10024
Mills JL, Sr., Conte MS, Armstrong DG, Pomposelli FB, Schanzer A, Sidawy AN, Andros G, Society for Vascular Surgery Lower Extremity Guidelines C (2014) The society for vascular surgery lower extremity threatened limb classification system: risk stratification based on wound, ischemia, and foot infection (WIfI). J Vasc Surg 59(1):220–234e221–222. https://doi.org/10.1016/j.jvs.2013.08.003
Li X (2014) MiR-375, a microRNA related to diabetes. Gene 533(1):1–4. https://doi.org/10.1016/j.gene.2013.09.105
Avnit-Sagi T, Vana T, Walker MD (2012) Transcriptional mechanisms controlling miR-375 gene expression in the pancreas. Exp Diabetes Res 2012:891216. https://doi.org/10.1155/2012/891216
Ding L, Xu Y, Zhang W, Deng Y, Si M, Du Y, Yao H, Liu X, Ke Y, Si J, Zhou T (2010) MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res 20(7):784–793. https://doi.org/10.1038/cr.2010.79
Higuchi C, Nakatsuka A, Eguchi J, Teshigawara S, Kanzaki M, Katayama A, Yamaguchi S, Takahashi N, Murakami K, Ogawa D, Sasaki S, Makino H, Wada J (2015) Identification of circulating miR-101, miR-375 and miR-802 as biomarkers for type 2 diabetes. Metabolism 64(4):489–497. https://doi.org/10.1016/j.metabol.2014.12.003
Wellen KE, Hotamisligil GS (2005) Inflammation, stress, and diabetes. J Clin Invest 115(5):1111–1119. https://doi.org/10.1172/JCI25102
Esposito K, Nappo F, Marfella R, Giugliano G, Giugliano F, Ciotola M, Quagliaro L, Ceriello A, Giugliano D (2002) Inflammatory cytokine concentrations are acutely increased by hyperglycemia in humans: role of oxidative stress. Circulation 106(16):2067–2072. https://doi.org/10.1161/01.cir.0000034509.14906.ae
Quagliaro L, Piconi L, Assaloni R, Da Ros R, Maier A, Zuodar G, Ceriello A (2005) Intermittent high glucose enhances ICAM-1, VCAM-1 and E-selectin expression in human umbilical vein endothelial cells in culture: the distinct role of protein kinase C and mitochondrial superoxide production. Atherosclerosis 183(2):259–267. https://doi.org/10.1016/j.atherosclerosis.2005.03.015
Findley CM, Mitchell RG, Duscha BD, Annex BH, Kontos CD (2008) Plasma levels of soluble Tie2 and vascular endothelial growth factor distinguish critical limb ischemia from intermittent claudication in patients with peripheral arterial disease. J Am Coll Cardiol 52(5):387–393. https://doi.org/10.1016/j.jacc.2008.02.045
Quan A, Pan Y, Singh KK, Polemidiotis J, Teoh H, Leong-Poi H, Verma S (2017) Cardiovascular inflammation is reduced with methotrexate in diabetes. Mol Cell Biochem 432(1–2):159–167. https://doi.org/10.1007/s11010-017-3006-0
Rumore MM, Kim KS (2010) Potential role of salicylates in type 2 diabetes. Ann Pharmacother 44(7–8):1207–1221. https://doi.org/10.1345/aph.1M483
Peiro C, Lorenzo O, Carraro R, Sanchez-Ferrer CF (2017) IL-1beta inhibition in cardiovascular complications associated to diabetes mellitus. Front Pharmacol 8:363. https://doi.org/10.3389/fphar.2017.00363
Wang F, Ge J, Huang S, Zhou C, Sun Z, Song Y, Xu Y, Ji Y (2021) KLF5/LINC00346/miR148a3p axis regulates inflammation and endothelial cell injury in atherosclerosis. Int J Mol Med. https://doi.org/10.3892/ijmm.2021.4985
Miyamoto S, Suzuki T, Muto S, Aizawa K, Kimura A, Mizuno Y, Nagino T, Imai Y, Adachi N, Horikoshi M, Nagai R (2003) Positive and negative regulation of the cardiovascular transcription factor KLF5 by p300 and the oncogenic regulator SET through interaction and acetylation on the DNA-binding domain. Mol Cell Biol 23(23):8528–8541. https://doi.org/10.1128/MCB.23.23.8528-8541.2003
Nagai R, Suzuki T, Aizawa K, Shindo T, Manabe I (2005) Significance of the transcription factor KLF5 in cardiovascular remodeling. J Thromb Haemost 3(8):1569–1576. https://doi.org/10.1111/j.1538-7836.2005.01366.x
Ding D, Jiang H, He Y, Li X, Liu X (2021) miR-320-3p regulates the proliferation, migration and apoptosis of hypoxia-induced pulmonary arterial smooth muscle cells via KLF5 and HIF1alpha. Am J Transl Res 13(4):2283–2295
Zhang J, Zheng B, Zhou PP, Zhang RN, He M, Yang Z, Wen JK (2014) Vascular calcification is coupled with phenotypic conversion of vascular smooth muscle cells through Klf5-mediated transactivation of the Runx2 promoter. Biosci Rep 34(6):e00148. https://doi.org/10.1042/BSR20140103
Nan S, Wang Y, Xu C, Wang H (2021) Interfering microRNA-410 attenuates atherosclerosis via the HDAC1/KLF5/IKBalpha/NF-kappaB axis. Mol Ther Nucleic Acids 24:646–657. https://doi.org/10.1016/j.omtn.2021.03.009
Wang J, Zhang L, Wang T, Li C, Jiao L, Zhao Z, Li Y (2021) miRNA-576 alleviates the malignant progression of atherosclerosis through downregulating KLF5. Dis Markers 2021:5450685. https://doi.org/10.1155/2021/5450685
Wang XH, Yan CY, Liu JR (2019) Hyperinsulinemia-induced KLF5 mediates endothelial angiogenic dysfunction in diabetic endothelial cells. J Mol Histol 50(3):239–251. https://doi.org/10.1007/s10735-019-09821-3
Li Y, Li J, Hou Z, Yu Y, Yu B (2016) KLF5 overexpression attenuates cardiomyocyte inflammation induced by oxygen-glucose deprivation/reperfusion through the PPARgamma/PGC-1alpha/TNF-alpha signaling pathway. Biomed Pharmacother 84:940–946. https://doi.org/10.1016/j.biopha.2016.09.100
Ma Y, Wang Q, Liu F, Ma X, Wu L, Guo F, Zhao S, Huang F, Qin G (2018) KLF5 promotes the tumorigenesis and metastatic potential of thyroid cancer cells through the NF-kappaB signaling pathway. Oncol Rep 40(5):2608–2618. https://doi.org/10.3892/or.2018.6687
Chen HL, Chong IW, Lee YC, Tsai JR, Yuan SS, Wang HM, Liu WL, Liu PL (2014) Kruppel-like factor 5 mediates proinflammatory cytokine expression in lipopolysaccharide-induced acute lung injury through upregulation of nuclear factor-kappaB phosphorylation in vitro and in vivo. Mediators Inflamm 2014:281984. https://doi.org/10.1155/2014/281984
Qadura M, Terenzi DC, Verma S, Al-Omran M, Hess DA (2018) Concise review: cell therapy for critical limb ischemia: an integrated review of preclinical and clinical Studies. Stem Cells 36(2):161–171. https://doi.org/10.1002/stem.2751
Khachigian LM (2019) Transcription factors targeted by miRNAs regulating smooth muscle cell growth and intimal thickening after vascular injury. Int J Mol Sci. https://doi.org/10.3390/ijms20215445
Ghaleb AM, Nandan MO, Chanchevalap S, Dalton WB, Hisamuddin IM, Yang VW (2005) Kruppel-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Res 15(2):92–96. https://doi.org/10.1038/sj.cr.7290271
Kinisu M, Choi YJ, Cattoglio C, Liu K, Roux de Bezieux H, Valbuena R, Pum N, Dudoit S, Huang H, Xuan Z, Kim SY, He L (2021) Klf5 establishes bi-potential cell fate by dual regulation of ICM and TE specification genes. Cell Rep 37(6):109982. https://doi.org/10.1016/j.celrep.2021.109982
Acknowledgements
The authors would like to thank Ana Lay-Hong and Aniket P. Gad for their assistance with immunofluorescence imaging (Harvard Digestive Disease Center, NIH P30DK034854). This work was supported by the National Institutes of Health (Grant Nos. HL115141, HL134849, HL148207, HL148355, HL153356 to M.W.F.), and the American Heart Association (Grant Nos. 18SFRN33900144 and 20SFRN35200163 to M.W.F.; Grant No. 907663 to M.G.M.).
Author information
Authors and Affiliations
Contributions
Conceived the hypothesis: MWF and MGM; performed the experiments: MGM, AJ, GS, HSC, DP-C, RZ, JC; designed or interpreted the results: MGM, AJ, GS, HSC, DP-C, RZ, JC, PPG, MAC, MSS, MPB, and MWF; wrote the manuscript: MGM and MWF.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
McCoy, M.G., Jamaiyar, A., Sausen, G. et al. MicroRNA-375 repression of Kruppel-like factor 5 improves angiogenesis in diabetic critical limb ischemia. Angiogenesis 26, 107–127 (2023). https://doi.org/10.1007/s10456-022-09856-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10456-022-09856-3