Abstract
Reduced expression of endothelial nitric oxide synthase (eNOS) is a hallmark of endothelial dysfunction in diabetes, which predisposes diabetic patients to numerous cardiovascular complications including blunted angiogenesis. The Krüppel-like factor (KLF) five has been implicated as a central regulator of cardiovascular remodeling, but its role in endothelial cells (ECs) remains poorly understood. We show here that expression of endothelial KLF5 was significantly increased in the ECs from mouse diabetes mellitus type 2 (T2DM) model, when compared to non-diabetic or T1DM mouse. KLF5 up-regulation by insulin was dependent on activation of multiple pathways, including mammalian target of rapamycin, oxidative stress and Protein kinase C pathways. Hyperinsulinemia-induced KLF5 inhibited endothelial function and migration, and thereby compromised in vitro and in vivo angiogenesis. Mechanistically, KLF5 acted in concert with the MTA1 coregulator to negatively regulate NOS3 transcription, thereby leading to the diminished eNOS levels in ECs. Conversely, potentiation of cGMP content (the essential downstream effector of eNOS signaling) by pharmacological approaches successfully rescued the endothelial proliferation and in vitro tube formation, in the HUVECs overexpressing the exogenous KLF5. Collectively, the available data suggest that the augmentation of endothelial KLF5 expression by hyperinsulinemia may represent a novel mechanism for negatively regulating eNOS expression, and may thus help to explain for the T2DM-related endothelial dysfunction at the transcriptional level.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Despite the prominent progress in diagnosis and endocrine therapy, diabetes mellitus (DM), remains the most common chronic metabolic disorder affecting 366 million people in 2011 worldwide, with incidence rate projected to double by 2030 (Al Ali et al. 2013). DM is currently categorized into two types: insulin-dependent type 1 (T1DM, ~ 5–10%) and insulin-independent type 2 (T2DM, ~ 90–95%). The latter is characterized by inappropriate hyperglycemia due to insulin resistance or relative lack of insulin. Both hyperglycemia and hyperinsulinemia cause endothelial cells (ECs) dysfunction including disturbances in endothelial proliferation and migration, attenuated angiogenic response, blunted tube formation and deterrence of white blood cells from adhesion and diapedesis, leading to numerous cardiovascular complications (Dhananjayan et al. 2016). Multiple mechanisms, including evoked oxidative stress, disruption of inflammation, overactivation of general survival signals such as mitogen-activated protein kinase (MAPK) and AKT and dysfunction of transcription and epigenetic factors, integrate together to regulate this complicated pathogenesis (Muniyappa and Sowers 2013). Nevertheless, most mechanisms still remain obscure despite the fact that investigation of these phenomena remains intensive.
The biosynthesis and release of nitric oxide (NO) by ECs is the main strategy for endothelial vasoprotective functions. Production of NO in ECs is controlled by endothelial nitric oxide synthase (eNOS), an important regulatory enzyme that catalyzes arginine (Dudzinski and Michel 2007; Fish and Marsden 2006) in ECs. The expression and activity of eNOS was regulated at both transcriptional and translational levels by distinct mechanisms. As a core component of caveolae, caveolin inhibits eNOS activity through direct interaction. Multiple protein kinases, such as AMP-activated protein kinase, AKT and calmodulin-dependent kinase II activates eNOS by phosphorylating the Ser-1177 site. By contrast, protein kinase C suppresses eNOS activity by inducing phosphorylation of Thr-495 as well as dephosphorylation of Ser-1177 (Niwano et al. 2003). Recently, advance in this filed shows that transcription of the gene encoding eNOS (NOS3) is modulated fundamentally at the transcriptional level through a variety of cis-acting elements, including AP-1, Sp-1, CRE (Niwano et al. 2003), HIF-2 and NF-κB (Kalinowski et al. 2016).
The KLF family of transcription factors are important regulators of cell differentiation and growth, pathogenesis of atherosclerosis, tumor development and adipogenesis. Of particular interest, acting as a positive regulator of SMemb (a marker gene for activated smooth muscle cells in vascular diseases), KLF5 has been implicated as a central regulator of vascular smooth muscle cell differentiation and cardiovascular remodeling (Haldar et al. 2007). Additionally, KLF5 regulates angiotensin II stimulation in cardiac fibroblasts by directly recruiting onto the PDGF-A promoter (Shindo et al. 2002). An appreciation for the importance of KLF5 in adipocyte differentiation has been elegantly developed principally through KLF5 knockout mice studies. In concert with C/EBPβ/δ to activate the PPARγ2 promoter, KLF5 as a key component of the transcription factor network controlling adipogenesis (Oishi et al. 2005). Given a close association between lipid metabolism and vascular function (Chechi et al. 2009; Ono et al. 2009), KLF5 is emerging as an important transcriptional regulator with great pertinence to vascular dysfunction in diabetes.
Based on this rationale, we studied the expression profile of KLF5 in ECs from diabetic organisms. Using the gain-of-function approaches, we then determined the effects of KLF5 upregulation on the angiogenic dysfunction in ECs. Our results show that KLF5 is induced by insulin and that it then acts in concert with MTA1 coregulator to negatively regulate NOS3 transcription, thereby compromising the eNOS levels in ECs from T2DM. These data provide compelling evidence that KLF5 mediates in part the vascular dysfunction caused by hyperinsulinemia.
Materials and methods
Animal model and human samples
All procedures involved in the animal study, strictly conformed to the NIH guidelines for animal handling, were approved by the IACUC of the 2nd Affiliated Hospital of Xi’an Medical University. Adult male C57B/6 mice were obtained from the animal facility in our university, and were provided food pellets and water ad libitum. Mice were allowed to acclimatize for at least 7 days prior to experiments. T1DM was induced by one single intraperitoneal injection with streptozotocin (STZ, 160 mg/kg/body weight, Sigma-Aldrich, Shanghai, China). Mice with blood glucose ≥ 250 mg/dl, measured by an electronic blood glucose monitor (Ningbo E-Fitness, Ningbo, China) at 2 weeks after STZ injection, were treated as diabetic (Chen et al. 2016). To induce T2DM, mice were fed with high-fat diet (Jiangsu Medicience Biomedicine Co., Ltd., Yangzhou, China) for 3 weeks, followed by one single intraperitoneal injection with STZ (80 mg/kg/body weight). After another 3 weeks, HFD/STZ mice with distinct hyperglycemia concomitant with insulin resistance (beyond 1 – 2 S.E.M. values from the normal mice mean) were considered as T2DM (Chen et al. 2016).
Human CD34+ cells were isolated from peripheral blood samples, which were obtained from healthy volunteers (n = 8) and T2DM donors (n = 12) in our hospital from February 2014 to July 2017, with the aid of EasySep™ Human CD34 Positive Selection Kit (STEMCELL Technologies, Shanghai, China). The human study, strictly conformed to the ethical standards of Helsinki Declaration, was approved by the Clinical Investigation Committee in our hospital. All participants gave their informed consent in writing prior to inclusion in the study.
Immunohistochemistry
Immunostaining was carried out on mouse liver, kidney and pancreas sections following the reported protocols (Liang et al. 2013), with the aid of the Vectastain Elite ABC kit from Vector Laboratories (Burlingame, CA, USA). Sections were incubated with the rabbit polyclonal anti-KLF5 antibody (Zhang et al. 2018) (Abcam, Shanghai, China) at 4 °C overnight. Sections were then treated with biotin-labeled anti-rabbit IgG and incubated with preformed avidin–biotin peroxidase complex. Finally, slides were counterstained with hematoxylin, dehydrated, and mounted.
Cell treatment
ECs were isolated and purified from mouse kidney and liver according to a previously reported protocol (Chen et al. 2016). Briefly, mouse kidney or liver tissues were gently minced with fine scissors and incubated with collagenase (200 U/ml,) at 37 °C for 45 min. The digested homogenates were then filtered through a 70-µm cell strainer (BD Medical Technology, Bedford, MA, USA). Following the centrifugation at 400 ×g at 4 °C for 10 min, cell pellets were resuspended in precoldened Dulbecco’s phosphate-buffered saline, followed by sequential incubation with anti-PECAM-1 or anti-ICAM-2 monoclonal antibody-coated beads (Thermo Fisher Scientific, Shanghai, China) at RT for 15 min. Final separation was achieved on a magnetic separator (Miltenyi Biotec, Shanghai, China). ECs were routinely cultured at 37 °C with 5% CO2 in EGM2-MV media with 5% of FBS (Gibco, Shanghai, China). To represent the maximum angiogenic capacity, isolated ECs were either immediately used for experiments or cultured to passage two or three before being subjected to other assays.
Human umbilical vein endothelial cells (HUVECs) and human dermal fibroblast (hDFs), obtained from American Type Culture Collection (Manassas, VA, USA), were cultured in endothelial basal medium (EBM-2) supplemented with 1 mg/ml hydrocortisone, 12 mg/ml bovine brain extract, 50 mg/ml gentamycine, 50 ng/ml amphothericin-B, 10 ng/ml epidermal growth factor and in 10% FCS and DMEM supplemented with 10% FCS (Invitrogen, Shanghai, China), respectively. To investigate the effects of glucose on KLF5 expression, HUVECs were incubated with 5.5 mmol/L glucose, 25 mmol/L glucose, or 25 mmol/L mannitol (osmolar control group, Sigma-Aldrich, Shanghai, China) for indicated durations. To study the effects of hyperinsulinemia on KLF5 expression, HUVECs were incubated with different doses of insulin (Sigma-Aldrich, Shanghai, China), in the presence or absence of the insulin receptor (IR) inhibitor BMS-754807 (Sigma-Aldrich), for 6 h. To determine the signaling pathways involved in the regulation of KLF5 expression by insulin, HUVECs were incubated with 40 nM of insulin, in the presence of different pathway inhibitors/stimulators including Rapamycin (mTOR, 10 nM), SP600125 (JNK MAPK, 20 µM), SB203580 (p38 MAPK, 20 µM), Ceramide C6 (ERK activator, 10 µM), diphenyleneiodonium chloride (DPI, oxidative stress, 20 µM) and Go6976 (PKC, 1 µM), for 6 h. These inhibitors/stimulators were all purchased from Sigma-Aldrich. To stably express the exogenous KLF5, HUVECs were transfected with pCMV3-His-KLF5 or empty vector (Sino Biological, Beijing, China) using FuGENE® HD (Promega, Beijing, China), followed by selection with 200 µg/ml of Hygromycin (Sigma-Aldrich). To stimulate cGMP activity without affecting eNOS expression, HUVECs were incubated with 2.5 µM of BAY 63-2521 (BioVision, Milpitas, CA, USA) for 6 h prior to other assays.
Human endothelial colony forming cells (ECFCs) were isolated from the mononuclear cell fractions of donors’ cord blood, and were purified using CD31-coated magnetic beads (Thermo Fisher Scientific), as per the manufacturer’s instructions. ECFCs were maintained in EBM-2 supplemented with 20% FCS, 1 × glutamine-penicillin-streptomycin, as described elsewhere. To transiently overexpress KLF5, confluent ECFCs were infected with Ad-h-KLF5, a recombinant adenovirus encoding human KLF5 (Vector BioLabs, Malvern, PA, USA) for 24 h, with the parental adenoviral vector infection as negative control.
Measurement of endothelial proliferation and migration
HUVECs with different transfections were fixed 10% trichloroacetic acid at 4 °C for 1 h. Cells were then incubated with Sulforhodamine-B (Sigma-Aldrich) for another 0.5 h. After thorough rinse in unbuffered Tris Base (10 mM, pH 10.5), cells were harvested and final spectrophotometry was carried out at 492 nm on the NanoDrop™ 8000 Spectrophotometer (Thermo Fisher Scientific).
To assay the endothelial migration, confluent HUVECs were resuspended in DMEM containing 0.4% FBS, and then seeded at the density of 1 × 105 cells/insert onto a 8-µm-pore Transwell pre-coated with gelatin (Sigma-Aldrich). The Transwell was then inserted into a well containing the same media with or without mouse VEGF (10 ng/ml; Sigma-Aldrich). Cells were allowed to migrate for 6 h. The migrated cells on the lower well were then stained with DAPI and calculated in four random fields.
Tube formation assay
HUVECs with different transfections were seeded onto a 24-well plate pre-coated with growth factor-reduced Matrigel (300 µl/well, BD Biosciences, Shanghai, China), at the density of 1 × 105 cells/well. After another 24-h culture at 37 °C, tube formation was observed and branching points were quantified as described elsewhere (Jiang et al. 2009). Briefly, seven randomly chosen fields from each assay were captured under an inverted fluorescence phase-contrast microscope using a 10 × objective (Zeiss, Beijing, China). The lengths of the tubes in each field were measured with Image J software (National Institutes of Health, Bethesda, MD, USA) and calculated accordingly.
In vivo vasculogenesis assay
In vivo assessment of ECFCs angiogenesis was carried out using a xenograft model (Chen et al. 2016). Briefly, one million cells, containing ECFCs and hDFs at 1:1 ratio, were mixed with 100 ml Matrigel containing 500 ng/ml of VEGF-A (Sigma-Aldrich). The mixed cells were then implanted subcutaneously into the dorsal side of 10-week-old nude mice (n = 7). After 14 days of growth, the xenografts were harvested, fixed with formalin and subjected to immunohistochemical staining with anti-CD31 antibody. For angiogenesis contribution, a 4 × image of the entire graft area was assembled using the stitch function on the Zeiss microscope. From the 4 × image, proximal, middle, and distal areas were chosen and each area was imaged for CD31 as serial 60 × images taken randomly. Microvessel density is the percentage by taking the ratio of the CD31-positive vessels to the area of analyzed region (mm2). Totally, in each experimental group (Ad-null and Ad-h-KLF5), at least five sections from each of 4–6 animals were analyzed. The data from all sections were finally pooled to generate the mean for each animal.
Determination of endothelial eNOS activity and cGMP production
The eNOS activity (expressed as NO production in nM/mg/hour) and cGMP production (expressed as fmol/µg protein), were assessed in supernatants from cell culture as instructed by the manufacturer using a NOS Activation Assay Biochem Kit (Genmed Scientifics, Shanghai, China) and a Direct cGMP ELISA kit (Enzo life sciences, Shanghai, China), respectively.
Quantitative RT-PCR
Total RNA was extracted using RNeasy Mini Kit (QIAGEN Inc., Valencia, CA, USA) and purified with DNase (Applied Biosystems/Ambion, Austin, TX) treatment. First-strand cDNA was synthesized using Superscript III (Rnase H-Reverse Transcriptase; Invitrogen) as instructed by the manufacturer. Subsequent PCR reactions were performed according to a QIAGEN protocol on the ABI Prism 7300 system (Applied Biosystems, Foster City, CA, USA). Relative mRNA equivalents for each target were obtained by standardizing to 18S levels according to 2−△△Ct method (Dong et al. 2016). The primers used were: KLF5, 5′-CAGAGGACCTGGTCCAGACAAG-3′ and 5′-GAGGCCAGTTCTCAGGTGAGTG-3′; Klf5, 5′-CACCGGATCTAGACATGCCC-3′ and 5′-ACGTCTGTGGAACAGCAGAG-3′; NOS3, 5′-AAAGACAAGGCAGCAGTGGAAAT-3′ and 5′-TCCACGATGGTGACTTTGGCTA-3′; 18S, 5′-CTCGCCGCGCTCTACCTACCTA-3′ and 5′-ATGAGCCATTCGCAGTTTCACTGTA-3′.
Immunoblotting
Immunoblotting was carried out as described (Zhang et al. 2014). Total protein was isolated using ReadyPrep™ Protein Extraction Kit (Bio-Rad, Guangzhou, China). 50 µg of protein samples were separated by SDS-PAGE and transferred onto PVDF membranes (Thermo Fisher Scientific). Blots were then incubated with different primary antibodies including rabbit-anti KLF5 (Abcam), rabbit-anti eNOS and rabbit-anti β-ACTIN (Abcam) in blocking solution at 4 °C overnight. Final immunocomplexes were developed using a Chemiluminescent Western blots Imager (LI-COR Biotechnology, Lincoln, NE, USA).
Luciferase activity assay
The human NOS3 promoter region (Nucleotides: − 1654 to + 106; Genbank accession number: NC_000007.14) was amplified by PCR and cloned into pGL3-basic reporter plasmids (Promega) using In-Fusion® HD Cloning Kit (Takara, Dalian, China). Site-directed mutagenesis was carried out using QuikChange II Site-Directed Mutagenesis Kit (Agilent, Beijing China). For promoter luciferase assay, pCMV3-His-KLF5 or empty pCMV3 vector, the WT or Mu luciferase reporter plasmids, and pRL-TK Renilla reporter plasmid (Promega) were co-transfected into HEK 293 T cells using FuGENE® HD. 24 h later, cells were stimulated with different doses of insulin for another 6 h, followed by measurement of the relative luciferase activity using the Dual-Luciferase Activity System (Promega).
Chromatin immunoprecipitation (ChIP) and double ChIP
ChIP assays were performed based on published protocols (Hao et al. 2018). HUVECs were challenged with 40 nM insulin or Ctrl vehicle for 6 h, and cells were then cross-linked with 1% formaldehyde and sonicated to fragment the chromatin. The resultant supernatants, diluted in chromatin dilution buffer (Sigma-Aldrich), were incubated at 4 °C overnight with rabbit-anti KLF5 or rabbit IgG (Active Motif, Shanghai, China). The immunoprecipitates were then incubated at 4 °C for 6 h with Protein A-Sepharose beads (Abcam) to form the antibody-bound complexes. After a thorough wash, the eluate was “decross-linked” by heating at 70 °C for 6 h and subjected to PCR amplification. For the double ChIP assay, rabbit-anti KLF5 or rabbit IgG (Active Motif) was added into chromatin extracts and incubated at 4 °C overnight. The immunocomplexes were then recovered by incubation with 60 ml salmon sperm/protein A agarose (Sigma-Aldrich), followed by reimmunoprecipitation using goat-anti MTA1 (Santa Cruz Biotechnology, Shanghai, China). The primers used for ChIP assays were: P1, 5′-CTGAGAGGGAAGGCAGGGGCAAG-3′ and 5′-GTAAGGTGACAGCCTTCTGTG-3′; P2, 5′-GCATAGAGCAGGCCAGCACCAGGCA-3′ and 5′-TCTTGGGCCGTGGCCAGTGTTGTCCG-3′; P3, 5′-AGAGAGGAACTGACCTGTGTCT-3′ and 5′- CTCTGTGAGGTCTTCCTTACGTC-3′.
Statistical analysis
Results are presented as mean ± S.E.M., and were analyzed for statistically significant differences using Student’s t-test or one way analysis of variance (ANOVA) as appropriate. P < 0.05 was considered statistically significant.
Results
Induction of endothelial KLF5 expression by insulin signaling
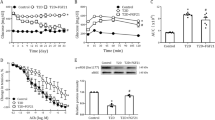
Previous transcriptome profiling analysis (Kulyte et al. 2017) and clinical studies (Ghavami et al. 2018) both suggest an association between KLF5 expression and T2DM. To verify this, we established murine DM models (SFig. 1). Subsequent immunohistochemical analysis on liver, kidney and pancreas sections showed that the endothelial KLF5 deposits were robust in T2DM mice compared to the negligible expression in Ctrl mice (Fig. 1a–c′′). In line with these in vivo data, quantitative analyses using primary ECs isolated from mouse kidney and liver (SFig. 2) revealed that, at both transcriptional and translational levels, the expression of endothelial KLF5 was significantly upregulated in the ECs from T2DM than that from Ctrl mice (Fig. 1d, e). Human circulating CD34+ cells contain large numbers of endothelial progenitor cells (Sawada et al. 2014). Interestingly, KLF5 expression was ∼ 1.86 fold higher in CD34+ cells from T2DM patients, compared to Ctrl patients (SFig. 3 and Fig. 1f). The observation that endothelial KLF5 expression was elevated in T2DM but not in T1DM suggested that hyperinsulinemia, rather than hyperglycemia, stimulates local KLF5. In favor of this assumption, treatment with different doses of glucose failed to affect the expression levels of KLF5 mRNA in HUVECs (Fig. 1g). Contrarily, KLF5 mRNA levels were noticeably stimulated in HUVECs treated with high concentration of insulin in a dose-dependent manner (Fig. 1h). This stimulatory effect was substantially abolished when HUVECs were challenged with 40 nM of insulin, together with the IR inhibitor BMS-754807, for 6 h (Fig. 1i). Together, these results are indicate of a close association between endothelial KLF5 upregulation and T2DM.
T2DM induces KLF5 expression in ECs. Immunohistochemical staining of KLF5 (black arrows) in liver (a–a′′′), kidney (b–b′′′) and pancreas (c–c′′′) from Ctrl (a–c), T1DM (a′–c′) and T2DM (a′′–c′′) mouse models. IgG staining was included as a negative control in a′′′–c′′′. The asterisks denote the microvascular. Bar = 25 µm d Relative Klf5 mRNA abundance in ECs freshly isolated from kidney and liver from different models. e KLF5 protein abundance in ECs freshly isolated from kidney and liver from different models was assayed using immunoblotting. f Relative KLF5 mRNA abundance in vasculogenic circulating CD34+ cells isolated from patients with T2DM, versus matched normal subjects. g Relative KLF5 mRNA abundance in human HUVECs 4 or 12 h after exposing cells to hyperglycemia or hyperosmolarity (mannitol). h Relative KLF5 mRNA abundance in human HUVECs 6 h after exposing cells to different doses of insulin (*P < 0.05 and **P < 0.01 when compared to the value in 0 nM of insulin group). i Relative KLF5 mRNA abundance in insulin-challenged HUVECs in absence or presence of the IR inhibitor BMS-754807 for 6 h
Involvement of multiple pathways in the endothelial KLF5 upregulation by insulin signaling
To gain mechanistic insight into the regulation of KLF5 expression by insulin signaling, we treated challenged HUVECs with 40 nM of insulin, together with different pathway inhibitors/stimulators including Rapamycin (mTOR), SP600125 (JNK MAPK), SB203580 (p38 MAPK), Ceramide C6 (ERK activator), DPI (oxidative stress) or Go6976 (PKC), for 6 h. Incubation with insulin alone caused a ~ 2.3-fold increase in KLF5 mRNA expression, and this stimulatory effect was partially but effectively reversed by concomitant incubation with Rapamycin, DPI or Go6976 (Fig. 2). The endothelial KLF5 upregulation by insulin signaling, at least in part, may be mediated through the activation of mTOR, oxidative stress and PKC pathways.
Involvement of multiple pathways in the endothelial KLF5 upregulation by insulin signaling. RT-qPCR showing KLF5 mRNA expression in HUVECs induced with insulin (40 nM) for 6 h in the presence of different pathway inhibitors, as described in detail in “Materials and methods”. §P < 0.05 and §§P < 0.01 when compared to the value in naïve cells; *P < 0.05 and **P < 0.01 when compared to the value in insulin-challenged cells
Impairment of angiogenic function by endothelial KLF5 overexpression
To directly ask whether endothelial KLF5 upregulation bears any biological effect, we established the HUVECs/KLF5 which stably expressed exogenous KLF5 (Fig. 3a). Subsequent angiogenic function analysis revealed that KLF5 overexpression significantly reduced endothelial proliferation (Fig. 3b), attenuated VEGF-induced endothelial migration (Fig. 3c) and blunted the formation of new blood vessels in the in vitro model (Fig. 3d). To further test the inhibitory effect of KLF5 on the in vivo angiogenesis, we isolated ECFCs from human cord blood samples (SFig. 4) and transfected these cells with Ad-h-KLF5 or empty Ad-null vector. After confirmation of KLF5 overexpression in the ECFCs by immunoblotting (Fig. 3e, f), we mixed these ECFCs with hDFs and coinjected them into nude mice. Injection of ECFCs overexpressing KLF5 noticeably blunted the formation of new blood vessels in the xenografts after a 14-day in vivo angiogenesis, compared to the use of the ECFCs transfected with parental adenoviral vector (Fig. 3g–g′′). To further verify the effects of KLF5 on angiogenesis, we knocked down the expression of KLF5 in HUVECs (SFig. 5a). Interestingly, ablation of endogenous KLF5 did not affect endothelial proliferation (SFig. 5b) but significantly enhanced VEGF-induced endothelial migration (SFig. 5c) and in vitro tube formation (SFig. 5d). Moreover, using endothelial migration and tube formation assays, we observed that KLF5 overexpression significantly impaired insulin-stimulated endothelial migration and tube formation (SFig. 6). Together, we have validated the detrimental effects of KLF5 upregulation on angiogenic function using a gain-of-function approach, and the effects of KLF5 on angiogenesis appears to be more prominent under angiogenic stimulation (VEGF or insulin).
Endothelial KLF5 induces angiogenic dysfunction. a Stable expression of the exogenous KLF5 in HUVECs was validated using immunoblotting. b Effects of KLF5 upregulation on endothelial proliferation was determined using Sulforhodamine-B (SRB) absorption assay. c Stable expression of KLF5 inhibits migration of HUVECs in Transwell assays stimulated by 10 ng/ml VEGF. d Stable expression of KLF5 inhibits the tube-forming activity of HUVECs. e Diagram of in vivo vasculogenesis assay. f Effects of adenovirus overexpression of exogenous KLF5 in ECFCs were confirmed by immunoblotting. In vivo vasculogenesis assay using hDFs and human ECFCs expressing control vector g versus Ad-h-KLF5 g′. IgG staining was included as a negative control in g′′. g′′′ Quantification of vessel density in the in vivo vasculogenesis assay
Endothelial KLF5 represses eNOS to impair angiogenic function
Because eNOS is essentially involved in endothelial motility, proliferation and permeability (Niwano et al. 2003), we were curious about the possible link between KLF5 deregulation and endothelial eNOS activity. Stable expression of the exogenous KLF5 caused a decrease in NOS3 mRNA by ~ 57.2% in HUVECs (Fig. 4a). This inhibitory effect of KLF5 on eNOS expression was further confirmed at the protein level by immunoblotting (Fig. 4b). Consistent with the impaired eNOS expression, the eNOS activity and levels of cGMP (the downstream effector of eNOS signaling) were reduced by ~ 52.3% (Fig. 4c) and ~ 28.6% (Fig. 4d) in HUVECs/KLF5, respectively. To further corroborate the role of eNOS in KLF5 signaling, we tried to activate cGMP production without affecting eNOS expression in HUVECs/KLF5. Stimulation with BAY 63-2521, a potent activator of cGMP synthesis, reversed KLF5-impaired cGMP production (Fig. 4e) without affecting eNOS expression (Fig. 4f) in HUVECs/KLF5. Importantly, potentiation of cGMP content successfully rescued the endothelial proliferation (Fig. 4g) and in vitro tube formation (Fig. 4h) in HUVECs/KLF5. Thus, deregulation of endothelial KLF5 diminishes neovascularization, at least in part through inhibition of eNOS synthesis.
Repression of eNOS expression mediates the detrimental effects of endothelial KLF5 on angiogenic functions. a Effect of KLF5 overexpression on eNOS mRNA levels in HUVECs was assessed by RT-qPCR. b Effect of KLF5 overexpression on eNOS protein levels in HUVECs was assessed by immunoblotting. c The eNOS activity (expressed as NO production in nM/mg/hour) was evaluated in supernatants from HUVECs culture as instructed by the manufacturer using a NOS Activation Assay Biochem Kit. d The cGMP production (expressed as fmol/µg protein) was assessed in supernatants from HUVECs culture as instructed by the manufacturer using a Direct cGMP ELISA kit. Quantitative values are the mean ± S.E.M. of at least three independent determinations. e HUVECs with different transfections were incubated with 2.5 µM of BAY 63-2521 (a potent activator of cGMP synthesis) for 6 h, followed by measurement of cGMP production in supernatants from HUVECs culture. f HUVECs with different transfections were incubated with 2.5 µM of BAY 63-2521 (a potent activator of cGMP synthesis) for 6 h, followed by measurement of eNOS expression using immunoblotting. g HUVECs with different transfections were incubated with 2.5 µM of BAY 63-2521 (a potent activator of cGMP synthesis) for 6 h, followed by measurement of endothelial proliferation using SRB absorption assay. h HUVECs with different transfections were incubated with 2.5 µM of BAY 63-2521 (a potent activator of cGMP synthesis) for 6 h, followed by measurement of the tube-forming activity of HUVECs
Direct regulation of NOS3 transcription by KLF5
To further ascertain the specificity of our finding at the molecular level, we investigated the effects of KLF5 on the NOS3 promoter activity. Treatment with different doses of insulin notably stimulated the NOS3 promoter activity. Interestingly, co-transfection with pCMV3-His-KLF5 could only effectively suppress the NOS3 promoter activity in the presence of co-treatment with high doses of insulin (40 and 60 nM) (Fig. 5a). Sequence analysis of the NOS3 major promoter revealed three potential binding sites, located downstream from the start codon (Fig. 5b). Subsequent ChIP assay showed that KLF5 was recruited to the − 1499 to − 1364 bp region of the NOS3 promoter (Fig. 5c). To further solid this observation, we mutated the binding site that was responsible for the binding of KLF5 onto NOS3 promoter (Fig. 5d). As expected, co-transfection with pCMV3-His-KLF5 completely inhibited insulin (40 nM)-elicited NOS3 promoter activity and this inhibitory effect was substantially abolished when WT pGL3-NOS3 reporter plasmid was replaced with Mu pGL3-NOS3 reporter plasmid (Fig. 5e). To extend the mechanistic understanding of how KLF5 negatively regulates the transcription of NOS3 upon insulin stimulation, we performed a double ChIP assay. Initial ChIP was done with anti-KLF5 antibody, and the second ChIP was carried out using the anti-MTA1 antibody. The results demonstrated a simultaneous co-association of KLF5 and MAT1 with the NOS3 chromatin, and this association was further enhanced by supplement with 40 nM of insulin (Fig. 5f). Thus, endothelial KLF5, in concert with MTA1, may directly repress the NOS3 transcription in the insulin-challenged ECs.
KLF5 inhibits the expression of NOS3 mRNA at the transcriptional level. a The pGL3-Luc-NOS3 reporter plasmid and pRL-TK Renilla reporter plasmid were co-transfected with pCMV3-His-KLF5 or pCMV3-His vector into HEK 293 T cells using FuGENE® HD. 24 h later, cells were stimulated with different doses of insulin for another 6 h, followed by measurement of the relative luciferase activity. b Schematic of predicted KLF5 binding sites on the NOS3 promoter region. c ChIP analysis showing recruitment of KLF5 onto a specific region of the NOS3 promoter. d Schematic of the mutated KLF5 binding site on the NOS3 promoter region. e The WT or mutated pGL3-Luc-NOS3 reporter plasmid and pRL-TK Renilla reporter plasmid were co-transfected with pCMV3-His-KLF5 or pCMV3-His vector into HEK 293 T cells using FuGENE® HD. 24 h later, cells were stimulated with 40 nM of insulin for 6 h, followed by measurement of the relative luciferase activity. f Double ChIP analysis of KLF5-MTA1 complex onto the NOS3 promoter in HUVECs/KLF5 cells upon challenge for 6 h with 40 nM of insulin
Discussion
Transcriptional regulation mechanisms function as a crucial interface between the effects of T2DM and vascular endothelial dysfunction, and deregulation of them lead to significant cardiovascular complications (Moradipoor et al. 2016). The data collected here reveal that KLF5 expression is upregulated in ECs from T2DM organisms, that this elevated KLF5 expression is controlled by insulin through regulation of multiple signaling pathways, and that KLF5 upregulation predisposes ECs to vascular dysfunctions including impaired proliferation, diminished migration, and blunted tube formation potential. We therefore propose that endothelial KLF5 acts as a negative feedback loop of insulin signaling that normally stimulates endothelial activation. In the presence of hyperinsulinemia, however, sustained KLF5 hyperactivation damages ECs and inhibits vessel re-endothelialization (Fig. 6). Deregulation of endothelial KLF5 thus mediates, at least in part, the vascular dysfunction in T2DM.
Our pharmacological approaches reveal that multiple signaling pathways, including activation of mTOR and PKC, and excessive production of oxidative stress, were involved in KLF5 upregulation by insulin in ECs (Fig. 2). Actually, accumulated data evidence a close association between KLF5 expression and these candidate signaling pathways. For example, activation of mTOR pathway by TGFβ directly stimulates KLF5 expression, thus regulating differentiation in smooth muscle cells (Zhu et al. 2017). Using in vitro phosphorylation, Zhang et al. found a functional phosphorylation site by PKC within the CREB-binding protein interaction domain of KLF5 (Zhang and Teng 2003). Likewise, oxidative stress augments the KLF5 expression in lung fibroblasts as well as the translocation of KLF5 into the nuclei and silencing of KLF5 suppresses the stress-augmented differentiation into myofibroblasts, the release of collagens and metalloproteinases (Abe et al. 2016). Based on these reports, along with our findings in ECs, we reason that: (1) Regulation at the different levels by the above-mentioned signaling pathways appears to be an intrinsic feature of KLF5 in both normal and transformed cells. (2) Given that these pathways have been shown to be essentially linked to insulin resistance, the available data suggest that insulin and these pathways coordinate in a reciprocal manner and KLF5 might serve as a critical converging point linking various antiangiogenic factors (mTOR, PKC and ROS) to hyperinsulinemia.
eNOS is a potent regulatory enzyme catalyzing NO production from arginine. With the aid of eNOS, the vascular endothelium keeps normal relaxation and contraction activity and functions as an essential interface between blood and smooth muscle cells (Niwano et al. 2003). Endothelial dysfunction, including impairment of the barrier function and vasodilation, disturbances in proliferative capacities, migratory as well as tube formation properties, and attenuation of angiogenic properties, is frequently observed in T2DM patients and experimental diabetic models of diabetes. One of the key etiologies contributing to this complicated pathogenesis is the impaired endothelium-dependent NO-mediated relaxation caused by either quenching of normally released NO or compromise of eNOS activity/suppression of eNOS expression (Dhananjayan et al. 2016). Emerging data suggest that eNOS expression is tightly controlled at the transcriptional level. For example, beraprost sodium (BPS), an orally active prostacyclin analogue, stimulates endothelial eNOS expression through cAMP-responsive element (CRE)-mediated transcriptional activation (Niwano et al. 2003). Rac1 induces eNOS gene (NOS3) transcription via direct recruitment of PAK-NF-κB coregulators (Rao and Bender 2008). In continuance to these observations, our molecular biological analyses have identified NOS3 chromatin to be a direct target of KLF5 coregulator in insulin-chanllenged ECs. Of note, multiple putative cis-regulatory elements have been reported in the NOS3 promoter region. The CRE responsive sequences is located at − 733 to − 603 bp of NOS3 promoter (Niwano et al. 2003). TRANSFAC analysis indicates that a consensus NF-κB site exists in the human NOS3 promoter at nucleotides − 994 to − 988 bp (Davis et al. 2004). Additionally, during the early hypoxia, the hypoxic induction of NOS3 expression is virtually dependent on hypoxia-responsive elements at position − 5375 to − 5366 bp (Kalinowski et al. 2016). By contrast, the promoter region of NOS3 bound by KLF5 (− 1414 to − 1410 bp) was not included in the above-mentioned cis-acting elements. These data together reveal the diversity of the transcriptional regulation sites of NOS3 expression in ECs.
The finding that KLF5 recruits MTA1 together to repress the NOS3 transcription deserves particular attention. MTA1 is one of the master coregulators in mammalian cells. By recruiting diverse coregulators, MTA1 regulates fundamentally cancerous progression, inflammation, spermatogenesis and neuroprotective effects at the transcriptional level (Sen et al. 2014). More importantly, MTA1 coregulation can be bidirectional. For instance, during oncogenesis, MTA1 mediates the transactivation of tumor suppressor alternative reading frame (ARF), and ARF in turn negatively regulates MTA1 expression independently of p53 and c-Myc (Li et al. 2011). Interestingly, transregulation of NOS3 expression, even in the same cell context, can also be bidirectional. During early hypoxia, HIF-2 directly binds to the NOS3 promoter and induce NOS3 expression in ECs. Under prolonged hypoxia, however, NOS3 expression is significantly down-regulated due to the recruitment of distinct coregulators (e.g. ATG9B, NOS3AS, and APG9L2) (Kalinowski et al. 2016). In this context, we reason that endothelial NOS3 transcription is regulated delicately at the transcriptional level, which results from the simultaneous and synchronized interplay between KLF5-MTA1 and other corepressors. Our lab is currently in process of identification of these core corepressors recruited by KLF5-MTA1 components, in insulin-challenged ECs.
In summary, the present study shows that expression of endothelial KLF5 is significantly increased in T2DM, and hyperinsulinemia-induced KLF5 inhibits endothelial function and compromises angiogenesis. One of the major molecular mechanisms underlying the above-mentioned phenotype is the loss of eNOS expression due to the transcriptional repression of NOS3 by the synergistic action of KLF5 and multiple coregulators (e.g. MTA1). The presence of functional KLF binding site within human NOS3 chromatin may represent a novel mechanism for regulating eNOS expression, and may help to explain for the T2DM-related endothelial dysfunction at the transcriptional level.
References
Abe K, Sugiura H, Hashimoto Y, Ichikawa T, Koarai A, Yamada M, Numakura T, Onodera K, Tanaka R, Sato K, Yanagisawa S, Okazaki T, Tamada T, Kikuchi T, Ichinose M (2016) Possible role of Kruppel-like factor 5 in the remodeling of small airways and pulmonary vessels in chronic obstructive pulmonary disease. Respir Res 17:7. https://doi.org/10.1186/s12931-016-0322-y
Al Ali R, Mzayek F, Rastam S, Fouad MF, O’Flaherty M, Capewell S, Maziak W (2013) Forecasting future prevalence of type 2 diabetes mellitus in Syria. BMC Public Health 13:507. https://doi.org/10.1186/1471-2458-13-507
Chechi K, McGuire JJ, Cheema SK (2009) Developmental programming of lipid metabolism and aortic vascular function in C57BL/6 mice: a novel study suggesting an involvement of LDL-receptor. Am J Physiol Regul Integr Comp Physiol 296:R1029–R1040. https://doi.org/10.1152/ajpregu.90932.2008
Chen RA, Sun XM, Yan CY, Liu L, Hao MW, Liu Q, Jiao XY, Liang YM (2016) Hyperglycemia-induced PATZ1 negatively modulates endothelial vasculogenesis via repression of FABP4 signaling. Biochem Biophys Res Commun 477:548–555. https://doi.org/10.1016/j.bbrc.2016.06.052
Davis ME, Grumbach IM, Fukai T, Cutchins A, Harrison DG (2004) Shear stress regulates endothelial nitric-oxide synthase promoter activity through nuclear factor kappaB binding. J Biol Chem 279:163–168. https://doi.org/10.1074/jbc.M307528200
Dhananjayan R, Koundinya KS, Malati T, Kutala VK (2016) Endothelial dysfunction in type 2 diabetes mellitus. Indian J Clin Biochem 31:372–379. https://doi.org/10.1007/s12291-015-0516-y
Dong YS, Hou WG, Li Y, Liu DB, Hao GZ, Zhang HF, Li JC, Zhao J, Zhang S, Liang GB, Li W (2016) Unexpected requirement for a binding partner of the syntaxin family in phagocytosis by murine testicular sertoli cells. Cell Death Differ 23:787–800. https://doi.org/10.1038/cdd.2015.139
Dudzinski DM, Michel T (2007) Life history of eNOS: partners and pathways. Cardiovasc Res 75:247–260. https://doi.org/10.1016/j.cardiores.2007.03.023
Fish JE, Marsden PA (2006) Endothelial nitric oxide synthase: insight into cell-specific gene regulation in the vascular endothelium. Cell Mol Life Sci 63:144–162. https://doi.org/10.1007/s00018-005-5421-8
Ghavami A, Roshanravan N, Alipour S, Barati M, Mansoori B, Ghalichi F, Nattagh-Eshtivan E, Ostadrahimi A (2018) Assessing the effect of high performance inulin supplementation via KLF5 mRNA expression in adults with type 2 diabetes: a randomized placebo controlled clinical trail. Adv Pharm Bull 8:39–47. https://doi.org/10.15171/apb.2018.005
Haldar SM, Ibrahim OA, Jain MK (2007) Kruppel-like Factors (KLFs) in muscle biology. J Mol Cell Cardiol 43:1–10. https://doi.org/10.1016/j.yjmcc.2007.04.005
Hao JS, Zhu CJ, Yan BY, Yan CY, Ling R (2018) Stimulation of KLF14/PLK1 pathway by thrombin signaling potentiates endothelial dysfunction in type 2 diabetes mellitus. Biomed Pharmacother 99:859–866. https://doi.org/10.1016/j.biopha.2018.01.151
Jiang A, Hu W, Meng H, Gao H, Qiao X (2009) Loss of VLDL receptor activates retinal vascular endothelial cells and promotes angiogenesis. Invest Ophthalmol Vis Sci 50:844–850. https://doi.org/10.1167/iovs.08-2447
Kalinowski L, Janaszak-Jasiecka A, Siekierzycka A, Bartoszewska S, Wozniak M, Lejnowski D, Collawn JF, Bartoszewski R (2016) Posttranscriptional and transcriptional regulation of endothelial nitric-oxide synthase during hypoxia: the role of microRNAs. Cell Mol Biol Lett 21:16. https://doi.org/10.1186/s11658-016-0017-x
Kulyte A, Ehrlund A, Arner P, Dahlman I (2017) Global transcriptome profiling identifies KLF15 and SLC25A10 as modifiers of adipocytes insulin sensitivity in obese women. PLoS ONE 12:e0178485. https://doi.org/10.1371/journal.pone.0178485
Li DQ, Pakala SB, Reddy SD, Ohshiro K, Zhang JX, Wang L, Zhang Y, Moreno de Alboran I, Pillai MR, Eswaran J, Kumar R (2011) Bidirectional autoregulatory mechanism of metastasis-associated protein 1-alternative reading frame pathway in oncogenesis. Proc Natl Acad Sci USA 108:8791–8796. https://doi.org/10.1073/pnas.1018389108
Liang Y, Dong Y, Zhao J, Li W (2013) YES1 activation elicited by heat stress is anti-apoptotic in mouse pachytene spermatocytes. Biol Reprod 89:131. https://doi.org/10.1095/biolreprod.113.112235
Moradipoor S, Ismail P, Etemad A, Wan Sulaiman WA, Ahmadloo S (2016) Expression profiling of genes related to endothelial cells biology in patients with type 2 diabetes and patients with prediabetes. Biomed Res Int 2016:1845638. https://doi.org/10.1155/2016/1845638
Muniyappa R, Sowers JR (2013) Role of insulin resistance in endothelial dysfunction. Rev Endocr Metab Disord 14:5–12. https://doi.org/10.1007/s11154-012-9229-1
Niwano K, Arai M, Tomaru K, Uchiyama T, Ohyama Y, Kurabayashi M (2003) Transcriptional stimulation of the eNOS gene by the stable prostacyclin analogue beraprost is mediated through cAMP-responsive element in vascular endothelial cells: close link between PGI2 signal and NO pathways. Circ Res 93:523–530. https://doi.org/10.1161/01.RES.0000091336.55487.F7
Oishi Y, Manabe I, Tobe K, Tsushima K, Shindo T, Fujiu K, Nishimura G, Maemura K, Yamauchi T, Kubota N, Suzuki R, Kitamura T, Akira S, Kadowaki T, Nagai R (2005) Kruppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab 1:27–39. https://doi.org/10.1016/j.cmet.2004.11.005
Ono Y, Nakaya Y, Bando S, Soeki T, Ito S, Sata M (2009) Telmisartan decreases plasma levels of asymmetrical dimethyl-L-arginine and improves lipid and glucose metabolism and vascular function. Int Heart J 50:73–83
Rao GK, Bender JR (2008) Rac, PAK, and eNOS ACTion. Circ Res 103:328–330. https://doi.org/10.1161/CIRCRESAHA.108.182238
Sawada N, Jiang A, Takizawa F, Safdar A, Manika A, Tesmenitsky Y, Kang KT, Bischoff J, Kalwa H, Sartoretto JL, Kamei Y, Benjamin LE, Watada H, Ogawa Y, Higashikuni Y, Kessinger CW, Jaffer FA, Michel T, Sata M, Croce K, Tanaka R, Arany Z (2014) Endothelial PGC-1alpha mediates vascular dysfunction in diabetes. Cell Metab 19:246–258. https://doi.org/10.1016/j.cmet.2013.12.014
Sen N, Gui B, Kumar R (2014) Physiological functions of MTA family of proteins. Cancer Metastasis Rev 33:869–877. https://doi.org/10.1007/s10555-014-9514-4
Shindo T, Manabe I, Fukushima Y, Tobe K, Aizawa K, Miyamoto S, Kawai-Kowase K, Moriyama N, Imai Y, Kawakami H, Nishimatsu H, Ishikawa T, Suzuki T, Morita H, Maemura K, Sata M, Hirata Y, Komukai M, Kagechika H, Kadowaki T, Kurabayashi M, Nagai R (2002) Kruppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nat Med 8:856–863. https://doi.org/10.1038/nm738
Zhang Z, Teng CT (2003) Phosphorylation of Kruppel-like factor 5 (KLF5/IKLF) at the CBP interaction region enhances its transactivation function. Nucleic Acids Res 31:2196–2208
Zhang C, Lai JH, Hu B, Zhang S, Zhao J, Li W (2014) A chromatin modifier regulates Sertoli cell response to mono-(2-ethylhexyl) phthalate (MEHP) via tissue inhibitor of metalloproteinase 2 (TIMP2) signaling. Biochim Biophys Acta 1839:1170–1182. https://doi.org/10.1016/j.bbagrm.2014.08.006
Zhang C, D’Alessandro A, Wellendorf AM, Mohmoud F, Serrano-Lopez J, Perentesis JP, Komurov K, Alexe G, Stegmaier K, Whitsett JA, Grimes HL, Cancelas JA (2018) KLF5 controls glutathione metabolism to suppress p190-BCR-ABL + B-cell lymphoblastic leukemia. Oncotarget 9:29665–29679. https://doi.org/10.18632/oncotarget.25667
Zhu Y, Takayama T, Wang B, Kent A, Zhang M, Binder BY, Urabe G, Shi Y, DiRenzo D, Goel SA, Zhou Y, Little C, Roenneburg DA, Shi XD, Li L, Murphy WL, Kent KC, Ke J, Guo LW (2017) Restenosis inhibition and re-differentiation of TGFbeta/Smad3-activated smooth muscle cells by resveratrol Sci Rep 7:41916. https://doi.org/10.1038/srep41916
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10735_2019_9821_MOESM1_ESM.ppt
Supplementary material 1—Measurement of diabetes induction was carried out through the assessment of whole blood glucose (a) and serum insulin levels (b) in different experimental groups (PPT 149 KB)
10735_2019_9821_MOESM2_ESM.ppt
Supplementary material 2—Isolation and purification of ECs from mouse liver or kidney. (a) Cultures of ECs isolated from mouse liver or kidney exhibited the classical cobblestone morphology. (b) Representative FACS analyses of VECAM1 and CD144 in isolated ECs. (c) Immunofluorescence images shows a cytoplasm expression of the endothelial cell-specific marker CD34 in isolated ECs, further confirming the cell identity. Nuclear are demonstrated using DAPI staining (PPT 256 KB)
10735_2019_9821_MOESM3_ESM.ppt
Supplementary material 3—CD34+ cells were isolated from the peripheral blood samples of T2DM patients and normal volunteers using FACS method (PPT 134 KB)
10735_2019_9821_MOESM4_ESM.ppt
Supplementary material 4—Human endothelial colony forming cells (ECFCs), isolated from the mononuclear cell fractions of donors’ cord blood, were identified using RT-qPCR method. UN, undetectable (PPT 124 KB)
10735_2019_9821_MOESM5_ESM.ppt
Supplementary material 5—Effects of KLF5 knockdown on endothelial function in HUVECs. (a) HUVECs were transfected with KLF5 siRNA or Ctrl siRNA (Sigma-Aldrich, Shanghai, China) using FuGENE® HD. 48 h later, cells were harvested, and ablation of endogenous KLF5 was validated using immunoblotting. (b) Effects of KLF5 knockdown on endothelial proliferation was determined using Sulforhodamine-B (SRB) absorption assay, as described in Materials and methods section. (c) Knock down of KLF5 promotes migration of HUVECs in Transwell assays stimulated by 10 ng/ml VEGF. (d) Knock down of KLF5 potentiates the tube-forming activity in HUVECs (PPT 216 KB)
10735_2019_9821_MOESM6_ESM.ppt
Supplementary material 6—Effects of KLF5 knockdown on endothelial function in HUVECs. (a) HUVECs were transfected with KLF5 siRNA or Ctrl siRNA (Sigma-Aldrich, Shanghai, China) using FuGENE® HD. 48 h later, cells were harvested, and ablation of endogenous KLF5 was validated using immunoblotting. (b) Effects of KLF5 knockdown on endothelial proliferation was determined using Sulforhodamine-B (SRB) absorption assay, as described in Materials and methods section. (c) Knock down of KLF5 promotes migration of HUVECs in Transwell assays stimulated by 10 ng/ml VEGF. (d) Knock down of KLF5 potentiates the tube-forming activity in HUVECs (PPT 215 KB)
Rights and permissions
About this article
Cite this article
Wang, Xh., Yan, Cy. & Liu, Jr. Hyperinsulinemia-induced KLF5 mediates endothelial angiogenic dysfunction in diabetic endothelial cells. J Mol Hist 50, 239–251 (2019). https://doi.org/10.1007/s10735-019-09821-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10735-019-09821-3