Abstract
A closed-loop device for bladder control may offer greater clinical benefit compared to current open-loop stimulation devices. Previous studies have demonstrated the feasibility of using single-unit recordings from sacral-level dorsal root ganglia (DRG) for decoding bladder pressure. Automatic online sorting, to differentiate single units, can be computationally heavy and unreliable, in contrast to simple multi-unit thresholded activity. In this study, the feasibility of using DRG multi-unit recordings to decode bladder pressure was examined. A broad range of feature selection methods and three algorithms (multivariate linear regression, basic Kalman filter, and a nonlinear autoregressive moving average model) were used to create training models and provide validation fits to bladder pressure for data collected in seven anesthetized feline experiments. A non-linear autoregressive moving average (NARMA) model with regularization provided the most accurate bladder pressure estimate, based on normalized root-mean-squared error, NRMSE, (17 ± 7%). A basic Kalman filter yielded the highest similarity to the bladder pressure with an average correlation coefficient, CC, of 0.81 ± 0.13. The best algorithm set (based on NRMSE) was further evaluated on data obtained from a chronic feline experiment. Testing results yielded a NRMSE and CC of 10.7% and 0.61, respectively from a model that was trained on data recorded 2 weeks prior. From offline analysis, implementation of NARMA in a closed-loop scheme for detecting bladder contractions would provide a robust control signal. Ultimate integration of closed-loop algorithms in bladder neuroprostheses will require evaluations of parameter and signal stability over time.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Loss of bladder control is a highly prevalent condition that can have severe impact on quality of life.22 Typical nerve stimulation approaches to restore function do not utilize direct feedback on the bladder state. Conditional stimulation may provide an advantage over continuous stimulation by minimizing habituation of neural pathways and requiring less battery power. A system that can continuously monitor bladder state and deliver stimulation only as needed may be a tremendous benefit to patients.
A major challenge for developing a robust closed-loop system for bladder control is in determining an effective way to continuously monitor bladder state. Sensors directly implanted in the bladder muscle have been used to measure bladder pressure or bladder muscle activity, but there can be issues with device migration, tissue damage, and obtaining large reliable signals.18,19 A few studies have demonstrated feasibility of closed-loop control by detecting key pressure events, but their methods currently rely on directly measuring bladder pressure using external sensors and catheters,15,17 which can cause urinary tract infections. Tapping into the body’s natural sensors (mechanoreceptors in the bladder wall) via nerves that encode this information may provide increased accuracy and reliability over artificial sensors.20 Researchers have used neural activity to estimate bladder pressure, bladder volume, or onset of bladder contractions.2,6,13,21,30 These approaches target the pelvic nerve,21 pudendal nerve,30 or spinal nerve 6,13 where access can be challenging or may require a separate location to stimulate for bladder control. Our approach interfaces with the sacral dorsal root ganglia (DRG) which contain the cell bodies for afferent pelvic and pudendal fibers that innervate the lower urinary tract entering the spinal cord. Here we can monitor information about the bladder state via pelvic nerve activity 3,16 and also control bladder function by stimulating pudendal nerve inputs through reflex spinal pathways.5,16
Mechanosensitive bladder neurons within the pelvic nerve are quiescent when the bladder is empty and gradually increase in firing as the bladder fills and pressure rises.31 The relationship between bladder pressure and the firing rate of these neurons is nonlinear as there is hysteresis.27 A static linear regression model does not take into account this dynamic relationship between bladder pressure and neural activity, can be susceptible to artifacts, and has yielded poor results previously.2 A basic Kalman filter, though still a linear-based model, can better handle noisy input signals by incorporating noise filtering and a dynamic transfer function.32 A more complex model that takes into account the non-linear relationship between bladder pressure and firing rate may yield a better fit than linear methods. One approach is to use a non-linear auto-regressive moving average (NARMA) model identified through a recursive artificial neural network structure.
In addition to the specific algorithm itself, other input feature selection factors can influence the performance of a decoding model. One important factor is how the input neural signal channels are selected, whether by using all recorded signals or only those channels with specific correlations to bladder activity. Another factor is the approach used to sum the occurrence of neural activity. For microelectrode recordings that yield action potentials, the timing of these events are incorporated into a firing rate estimation, which smooths the time-interval occurrence. The sampling period and smoothing duration can affect the firing rate calculation.
The goal of this study was to evaluate three different algorithms and their parameters to determine the tradeoffs between accuracy of fit and processing load for estimating bladder pressure from DRG signals in anesthetized cats. The models evaluated were simple linear regression, a Kalman filter, and NARMA. We hypothesized that the NARMA model would yield the best bladder pressure estimation and the Kalman filter would provide the second best fit. Our findings inform progress in development of an interface for closed-loop bladder control and provide insights for parameter selection in decoding applications for other physiological signals.
Materials and Methods
Animals
All procedures were approved by the University of Michigan Institutional Animal Care and Use Committee, in accordance with the National Institute of Health’s guidelines for the care and use of laboratory animals. Eight spinal-intact adult male, domestic, short-hair cats (age: 0.9–1.4 years old, 4.2–6.3 kg, Liberty Research, Inc., Waverly, NY) were used in this study. Seven of the cats were acute, non-survival experiments and one cat was a chronic experiment. Cats were used due to their high relevance to human physiology and their long history of study in bladder neurophysiology.11 Prior to use, animals were free-range housed with 0–3 other cats in a 38.4 m2 room with controlled temperature (19–21 °C), relative humidity (35–60%), food and water available ad lib, and a 12 h light/dark cycle. Animals received enrichment via daily staff interaction and toys.
Surgical Procedure
As previously described,27 animals were anesthetized with a mixture of ketamine (6.6 mg/kg)–butorphanol (0.66 mg/kg)-dexmedetomidine (0.033 mg/kg) administered intramuscularly (IM), intubated, and then maintained on isoflurane anesthesia (0.5–4%) during surgical procedures. Respiratory rate, heart rate, end-tidal CO2, O2 perfusion, temperature, and intra-arterial blood pressure were monitored continuously using a Surgivet vitals monitor (Smiths Medical, Dublin, OH). Fluids (1:1 ratio of lactated Ringers solution and 5% dextrose) were infused intravenously via the cephalic vein at a rate of 5–10 mL kg−1 h−1 (increased up to 30 mL kg−1 h−1 during surgery as needed). One or two catheters were inserted into the bladder at the bladder dome via a laparotomy and/or via the urethra for intravesical fluid infusion and pressure monitoring. If the urethra catheter was used during data collection, its size (3.5 Fr) did not prevent urine leakage at a full bladder.
A midline dorsal incision was made to expose the L7 to S3 vertebrae and a laminectomy was performed to access the S1 and S2 sacral DRG.3,27 Penetrating iridium oxide microelectrode arrays (Fig. 1a) with shank length of 0.5 or 1.0 mm and inter-shank spacing of 0.4 mm (Blackrock Microsystems, Salt Lake City, UT) were implanted into DRG (Fig. 1b) using a pneumatic inserter (Blackrock Microsystems). Arrays of various configurations (3 × 8 up to 5 × 10) were inserted unilaterally or bilaterally, as detailed in Table 1. Array reference wires were placed near the spinal cord and ground wires were attached to a stainless steel needle inserted in the skin (lateral and caudal to the laminectomy incision site). At the conclusion of surgical procedures, prior to experimental testing, animals were transitioned to intravenous alpha-chloralose (C0128, Sigma Aldrich; 70 mg/kg induction; 20 mg/kg maintenance). Analgesia was augmented with 0.01 mg/kg buprenorphine every 8–12 h intravenously.
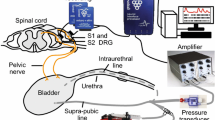
Experimental setup. (a) Blackrock iridium-oxide microelectrode arrays with parylene-C insulation. Shank lengths were either 0.5 mm or 1.0 mm with 0.4 mm inter-shank spacing; (b) Arrays implanted in left S1 and S2 sacral DRG in Experiment 8 (chronic); (c) Illustration of the testing setup (modified from5). Neural recordings were acquired with a Ripple Grapevine system and Trellis software via arrays implanted in S1 and S2 DRG. Trials consisted of recording neural data and bladder pressure (monitored with a pressure transducer and amplifier) during saline infusions at a controlled rate via either a supra-pubic or an intraurethral bladder catheter. Pressure data was also recorded with the Grapevine system after amplification.
Experimental Setup and Data Collection
DRG neural data was sampled at 30 kHz and low-pass filtered (7.5 kHz cutoff frequency) using a Grapevine neural interface processer and Trellis recording system (Ripple, Salt Lake City, UT). Bladder pressure was monitored with a pressure transducer (DPT-100, Utah Medical Products, Midvale, UT) and transducer amplifier (TBM4M, World Precision Instruments, Sarasota, FL). The bladder pressure signal was sampled by the Grapevine system at 1 kHz and all data was stored on a desktop computer. During testing saline was infused into the bladder with a syringe pump (NE-1000, New Era Pump Systems, Inc., Farmingdale, NY or AS50 Infusion Pump, Baxter International, Deerfield, IL). Figure 1c shows the experimental set-up.
Bladder pressure and neural recordings were collected during two bladder infusions at 2 mL/min. The bladder was verified as empty by withdrawing from a catheter prior to each infusion. Fluid infusion was stopped when leakage occurred. After the first infusion, the bladder was emptied and a rest period of at least 30 min occurred before a second bladder infusion. In analyses described below, the first infusion was used to train a given algorithm and the second infusion was used to validate that algorithm. We considered a microelectrode pair on the same side (LS1, LS2 or RS1, RS2) to constitute a set. In total, 12 data pairs of training and validation sets were collected from 12 unique S1, S2 DRG location pairs from 7 non-survival cat experiments. If an experiment had bilateral implant sets then each side was separately analyzed as a distinct data set with unique neural signals. In Experiments 2 and 4, the microelectrode array pairs were removed and physically re-inserted in the same DRGs before two new bladder infusions were performed. We considered this an opportunity to collect a new, unique data set from the new electrode implant locations with a different set of neural recordings (Table 1).
After completion of all testing, animals were euthanized with a 2–3 mL intravenous or intracardiac dose of sodium pentobarbital (390 mg/mL) while under deep isoflurane anesthesia.
Algorithm Development and Analysis
Figure 2 shows the overall algorithm flowchart with the different feature selection methods used. Individual components are described in further detail below. All data processing, model development, and analyses (unless otherwise specified) were performed using MATLAB (Mathworks, Natick, MA). Acute experiment trials were used to train and validate algorithms and system parameters, while the chronic experiment trials were used to train and test the algorithm and model parameters.
Multi-Unit Thresholding
A consistent threshold for identifying neural activity was used across experiments. For each neural channel, the signal was filtered using a 250 Hz high-pass digital filter. Automatic dual thresholds were set at ± 4.5 × the standard deviation of the estimated noise.26 Noise was considered any signal within ± 2.7 × the full signal standard deviation. The thresholding was recalculated periodically to accommodate change of noise level over time. This filtering and thresholding was done offline. In this study, we did not isolate single unit activity, but instead used the unsorted threshold crossings (i.e., multiunit activity) as inputs to our bladder state decode models. As has been demonstrated in brain-computer interface applications7 and DRG recordings for closed-loop control of electrical stimulation to control limb state,4 sorting threshold crossings into individual units can provide a marginal improvement in accuracy, for low channel counts, but requires additional computational load.
Neural Signal Firing Rate
The bladder pressure and thresholded multi-unit activity from each bladder fill sequence were then further processed offline and used to evaluate the performance of different models. Bladder pressure was low-pass filtered at 4 Hz and down-sampled to align with the time intervals used to calculate the firing rate (see below). Three approaches were used to calculate the firing rate for each microelectrode channel, at intervals of 0.1, 0.5, 1, and 2 s, using causal methods. In SmoothBin the number of spikes were counted in interval bin widths and the counts were smoothed over prior time points.4 In TriangleC the instantaneous firing rates were convolved with a triangle kernel of width twice the firing rate interval.29 In Boxcar all spikes within an interval were counted and divided by the bin width to obtain the firing rate. We also considered the use of a firing rate normalization (Eq. 1), to decrease the variance in contributions across channels and to potentially increase the robustness of each algorithm.14 When applied, normalization takes the current firing rate (f) and divides by the sum of the firing rate range in the first 30 s of the trial (r) and a scalar term (v) of 20 spikes/s.
Input Channel Selection
DRG channels used in an algorithm were selected based on three general approaches. First, channels were selected based on the correlation coefficient between their firing rate and the bladder pressure in the training trial. Channel sets were identified for correlations exceeding each of 0.2, 0.3, 0.4, 0.5, 0.6, and 0.7. The second approach involved using the least absolute shrinkage and selection operator (or LASSO) method,28 which is a linear regression method that minimizes the mean-squared error (MSE) similar to the least-squares method but with an upper bound on the sum of the absolute coefficients (Eq. 2). MSE is minimized for a selected regularization term, λ. This results in some coefficients set to zero and shrinks others towards zero, effectively choosing a model that uses a subset of the input observations. \(f_{k}\) is the training firing rate of length N2 (number of channels) at time bin k, and \(p_{k}\) is the training bladder pressure. \(\beta_{0}\), \(\beta , {\text{and }}\beta_{{k_{2} }}\) are the coefficients. A two-fold cross validation was performed to determine \(\lambda\) and coefficients that minimized MSE. These values were selected to be used in validation (acute) and testing (chronic) sessions. Thus only a subset of DRG channels were used in LASSO selection. The third channel selection method used is all channels.
Linear Regression Algorithm
First, a simple static linear regression (LinReg) was used to determine whether a linear relationship was adequate for modeling the relationship between DRG neural signals and the bladder pressure. During training, a linear filter \(\hat{B}\) was obtained (Eq. 3) using the bladder pressure P train and firing rates F train of the selected channels in the training trial. During validation (acute) or testing (chronic), \(\hat{B}\) was applied to the new firing rates \(F_{{{\text{val}}, {\text{test}}}}\) of the same channels to estimate the bladder pressure \(\hat{P}\) (Eq. 4).
Kalman Filter Algorithm
In order to reduce the effect of noise from the neural inputs, a basic Kalman filter (Kalman) was evaluated. The Kalman filter is a recursive algorithm that combines a physical trajectory model and a neural signal-based observational model using a weighted average method to nominally achieve a better estimation. Below we provide the details regarding development of this model.
To create a Kalman filter model, the data from the training infusion trial was used to calculate the linear filter and noise term (A and W) for the trajectory model and observational model (C and Q). Linear filter A was obtained by regressing the previous bladder state matrix \(P_{k - 1} \varvec{ }\) against the current bladder state matrix \(P_{k}\) (Eq. 5). Each state matrix includes bladder pressure, bladder pressure rate (dP/dk) and a constant term 1 for a series of time points (k = 0, 1, 2,…, N, N = last time interval bin number). This matrix was initialized at [0, 0, 1] at the beginning of each trial. The corresponding noise term (W) was then calculated (Eq. 6).
Next, the linear filter C for the observational model was determined by regressing the pressure state matrix against the firing rate (Eq. 7). The corresponding noise term for the observational model was also calculated (Eq. 8).
During testing, after firing rates were calculated, a trajectory model estimation \(\hat{p}_{{k\left| {k - 1} \right.}} \varvec{ }\) was made (Eq. 9). The error covariance matrix \(E_{k|k - 1}\) (Eq. 10) was calculated to determine how accurate this estimation was. This matrix was used to calculate the Kalman gain \(K_{k}\) (Eq. 11), which was used to obtain a new weighted average estimation \(\hat{p}_{k} \varvec{ }\) that incorporates the firing rate input (Eq. 12). Then a new error covariance matrix \(E_{k} \varvec{ }\) was calculated for the new estimation (Eq. 13) for the next time interval period.
NARMA Algorithm
A more complex NARMA artificial neural network model that took into consideration the nonlinear dynamics between bladder pressure and DRG neural activity was also evaluated. Inputs to the model were firing rates from selected channels and the previous estimated bladder pressure. Two artificial neurons in a hidden layer applied a nonlinear gain (hyperbolic tangent sigmoid function) and a bias to each of the inputs, with outputs summed together to yield a single intermediate output. This intermediate output was then processed through a linear function to give an estimate output (Fig. 3). A general form of the model is given in Eq. 14.
Structure of the NARMA model. The firing rates, f k , at the current time point, k, and the previous time point, k−1, from DRG channels are inputs into the model. The previous estimated bladder pressure, \(\hat{p}\left( {k - 1} \right),\) is also an input to the model. The model outputs the current estimated pressure, \(\hat{p}\left( k \right)\).
In Eq. 14, \(\hat{p}_{k}\) is the output estimated pressure, \(f\) is the input firing rate, and k and k-1 indicate values at the current and prior time points. The model coefficients (\(\alpha_{1}\), \(\beta_{0}\), \(\beta_{1}\)) were identified through training in the NARMA network.
The neural network toolbox in MATLAB was used to create, identify, and validate the models. Each model utilized a Bayesian regularization method to improve generalization and minimize overfitting. Training was stopped when one of two following criteria were met: the algorithm’s performance reached a performance goal of NRMSE < 20% of the recorded bladder pressure or the errors on the validation subset increased consecutively for several epochs.
Performance Measures
The normalized root-mean square error (NRMSE, Eq. 15) and Pearson correlation coefficient (CC, Eq. 16) were calculated to determine how well each model fit the training, validation (acute) and testing (chronic) data sets. We considered an NRMSE under 20% and a CC above 0.8 as our target performance metrics.
For (15), \(\hat{p}_{k}\) and \(p_{k}\) are the estimated and measured bladder pressure, (p max – p min). is the difference between maximum and minimum measured pressure for a trial, and N is the total number of time bins. For (16), \(\hat{P}\) and \(P\) are the estimated and measured pressure stored in vectors, and E is the expected value. Both performance measures are used here as prior decoding studies for bladder and non-bladder applications have relied on either CC2,6,7 or RMSE4,21 to evaluate their results.
Statistical Analysis
In total, three algorithms with 192 possible feature selection method combinations for each algorithm were evaluated in this study (4 firing rate intervals, 3 firing rate smoothing methods, 8 channel selection methods, and on or off of the firing rate normalization). This resulted in 576 total possible combinations. First, we used a mean-value approach to identify the best parameter combinations that yielded the lowest NRMSE and highest CC for each of the three algorithms. Through this approach, parameter sets were identified that had the best mean performance metric across the twelve acute testing data sets, for each of the six total combinations of three algorithms and two performance measures. In a separate analysis, we plotted data residuals on a normal probability plot, which indicated that the data distribution is not normal. Thus, a rank-based non-parametric Kruskal–Wallis test was used to identify parameters that contribute significantly to the estimation accuracy. Any significant parameter was then tested with a post hoc multiple comparison Kruskal Dunn Test, with a Bonferroni correction on the p-values. All statistical analyses were performed in R with the Pairwise Multiple Comparison of Mean Ranks (PMCMR) package. A significance level of 0.05 was used. In the PMCMR package, the Bonferroni correction is performed by multiplying p-values by the appropriate number of comparisons being performed in a given test. These adjusted p-values are reported in the Results below, allowing for a direct comparison to the significance level.
In this manuscript, we present the impact of single variables on each algorithm-performance metric combination. Higher level interactions among variable combinations were also examined, with the outcomes reported in our data repository on the Open Science Framework.23
Chronic Experiment
The primary analyses of this study were performed using data collected in acute, non-survival experiments as detailed above. The optimal algorithm and parameters were selected through training and validation sets from these experiments (NARMA for best NRMSE). To assess the potential utility of this approach, we implemented the best algorithm from acute experiments on data collected from one animal with long-term, chronic DRG implants (two 4 × 8 Blackrock arrays in left S1 and S2 DRG). Surgical implantation and data collection were as described previously.16 Neural recordings and bladder pressure from a bladder fill under dexmedetomidine sedation (0.03 mg/kg IM) were used to create a pressure estimate algorithm that was tested on data collected under similar circumstances 14 days later. Performance measures were calculated as described above.
Results
Bladder Neural Signals
Twelve data sets (a1–a12) were collected from seven acute experiments (Table 1). Raw and analyzed data and MATLAB scripts used in data analysis can be found online.23 There were 4–12 bladder-correlated channels with a correlation coefficient greater than 0.2 in each data set. There was some variability in the number of bladder-correlated channels depending on the firing rate calculation parameters used (Table 2). Most channels identified in the training trials (correlation coefficient > 0.2) were still correlated with bladder pressure in the validation trial (53–68%, depending on feature selection method). However, in some cases there were changes in the number and location of some identified channels (Fig. 4).
Correlation coefficient maps between the measured bladder pressure and firing rate of thresholded DRG neural activity for each electrode channel in S1 and S2 in data set a12 for training (top) and validation (bottom). The firing rates were calculated with Boxcar smoothing at the specified intervals. In this example, the firing rate interval and the time between trials (training to validation) can affect the number of channels with correlation coefficients of interest. Channels with correlation coefficients above 0.2 are indicated by a black square.
Channel Selection
Our results showed that as the correlation coefficient threshold for channel selection increased, the fit increased (Fig. 5). However, the number of trials that have at least one channel with a correlation coefficient greater than or equal to the CC threshold decreases (Table 3). This suggests that a constant threshold for all trials may be impractical, since not all trials have highly correlated channels. Although simply choosing the lowest correlation coefficient threshold would consistently provide the most usable number of channels, a higher correlation coefficient threshold yielded better pressure estimates (Fig. 5). Thus, we utilized a variable channel-selection threshold across data sets, selecting the highest threshold (among 0.2–0.7) that yielded a non-zero number of bladder channels.
The LASSO channel selection method always yielded channels for model use (27 ± 13, mean ± standard deviation) and on average selected at least 3 times more channels than the correlation coefficient channel selection method. This is most likely due to the differences in approach. With LASSO, different combinations of coefficients are evaluated with the set of coefficients that gives the lowest MSE selected. This in turn determines what channels to include in the decoding algorithms. Using the LASSO approach, the selected channels had a wide range of correlation coefficients compared to the approach of selecting channels that are above a certain correlation coefficient threshold.
Algorithm Optimization
A mean-value based approach was used to select the best parameter combinations for each of the three algorithms (Table 4). Across the 12 acute data sets with the best parameter combinations, NARMA provided the lowest mean NRMSE and a Kalman Filter provided the highest mean CC with smallest CC standard deviation (Table 4; Fig. 6). For these parameter combinations, the computational time to make the next bladder pressure estimation during validation are also given in Table 4. Training of each algorithm took no longer than a few seconds (LinReg) to a few minutes (NARMA). Bladder pressure estimations for one dataset using the three algorithms with their NRMSE-best model parameters (selected from Table 4) are shown in Fig. 7.
Use of best model parameters (for NRMSE, in Table 4-upper) for decoding bladder pressure in dataset a4. The top plot shows the measured bladder pressure and the estimated bladder pressure using each algorithim. The NRMSE and CC for each fit is also given. The bottom panel shows the firing rates of the top four channels, with their respective correlation coefficients.
Within NARMA algorithm results, we found that using a correlation coefficient-based channel selection method provided a significant decrease in NRMSE compared to using LASSO (p < 0.0001) or simply including all channels (p < 0.0001). It also yielded a significantly higher CC compared to using LASSO (p < 0.0001) and all channels (p < 0.0001). The firing rate interval was not significant in reducing NRMSE and was marginally significant in increasing CC (p = 0.033), with a 1-s interval slightly superior than a 0.1-s interval (p = 0.049).
Within Kalman filter results, we found that the firing rate interval had a significant role in reducing NRMSE (p = 0.005), with a 0.1-s interval better than 2-s (p = 0.01) and 1-s (p = 0.02) intervals, though the interval was not a significant factor for CC. A correlation coefficient-based channel selection method yielded a significantly lower NRMSE compared to using LASSO (p < 0.0001) or all channels (p < 0.0001). It also provided a significantly greater CC compared to using all channels (p < 0.0001).
Finally, within linear regression results, we have found that firing rate interval did not play a significant role in affecting NRMSE, but was significant in improving CC (p = 0.0001), with a 2-s interval (p < 0.0001) and a 1-s interval (p = 0.019) better than a 0.1-s interval. Similar to NARMA, a correlation coefficient based channel selection method provided a decreased NRMSE compared to using LASSO (p < 0.0001) or all channels (p < 0.0001). It also provided an increased CC compared to using all channels (p value < 0.0001).
Overall, we also found that firing rate smoothing methods alone did not provide any statistical significance in reducing NRMSE or improving CC in any of the three algorithms. The firing rate normalization provided a significant decrease in NRMSE for all three algorithms (NARMA p = 0.0004, Kalman p = 0.013, and Linear p = 0.045), but there was no statistical improvement in CC.
Chronic Data Set
In the chronic experiment, bladder pressure and DRG neural signals were recorded at 34 and 48 days after microelectrode implantation. Use of the best algorithm parameter set for NRMSE (NARMA, Table 4) yielded overall excellent fits on the training and subsequent testing data sets, with NRMSE and CC for testing being 10.7% and 0.61, respectively (Fig. 8). All channels that were highly correlated with bladder pressure on the training date were still identified at the testing date.
Evaluation of best acute-experiment algorithm (NARMA for best NRMSE in Table 4-upper) on data obtained in a chronic experiment. The NRMSE and CC for the training data set (day 34 after implant) were 6.2% and 0.89, respectively. The NRMSE and CC for the testing data set (day 48) were 10.7% and 0.61, respectively. In the training data set (at 170 s) a catheter was bumped which caused a high frequency artifact. This artifact was not removed during the training model fit, and did not affect the testing performance significantly.
Discussion
In this study, we demonstrated the feasibility of using thresholded multi-unit DRG activity to decode bladder pressure (Figs. 7 and 8). We evaluated a broad range of feature selection methods and three algorithms (multivariate linear regression, basic Kalman filter, and nonlinear autoregressive moving average model or NARMA) to evaluate which combinations yielded the best fits for two performance measures—NRMSE and CC (Table 4). Across our twelve acute data sets, NARMA provided the best fit in NRMSE, and channel selection methods had a significant impact on the resulting estimate of bladder pressure (Figs. 4 and 5; Tables 2 and 3).
Towards our ultimate goal of closed-loop neuroprostheses, NRMSE is the more critical of our two performance measures as it minimizes overall error better than CC. Considering this, our findings supported our hypothesis that a NARMA model provides the most accurate estimation of the bladder pressure, with a low NRMSE (Table 4; Figs. 6 and 7). The NARMA model captured important nonlinear dynamics between bladder neuron activity, seen here as thresholded DRG signals, and bladder pressure.27,31 For our secondary performance criteria of CC, a Kalman filter yielded the highest similarity to the results (Table 4). A closed-loop system that does not directly apply absolute pressure value might take advantage of the Kalman filter algorithm (e.g., frequency of non-voiding contractions, relative amplitude of non-voiding contractions, etc.). The multivariate linear regression algorithm was not outstanding in terms of either performance measure.
NARMA models resulted in the best NRMSE fit and a first-order NARMA network with one hidden layer and two artificial neurons was sufficient in capturing dynamics of interest. Further analyses can be done to determine if increasing the order of the model, the number of layers, and/or the number of artificial neurons can improve bladder decoding performance. For example, a prior study utilized two hidden layers in a neural network with 15 neurons per layer to estimate bladder pressure from pudendal nerve electroneurogram (ENG), obtaining greater accuracy than with a linear model.12 However, increasing the complexity of the model can increase overfitting and computational time and may not yield a great improvement compared to current models.
While isolated single units provide more specificity for underlying signals, using threshold-crossing events is more reliable and computationally feasible for online use. In our firing rate calculation for each microelectrode channel, we did not differentiate between any observed large-amplitude single units and small-amplitude threshold crossings. These small-amplitude snippets may be recognized as noise in sorting of single-units, but might still provide useful information. This conclusion is also consistent with previous cortical interfacing studies,7,25 in which there was a consistent conclusion that threshold-crossing events provided a comparable decoding accuracy as single-unit activity.
Only including DRG channels that correlated highly with bladder pressure improved decoding performance. However, as Table 3 shows, there is a tradeoff as the number of channels included in the model decreases. It may not be advantageous to use a constant correlation coefficient threshold for input channel selection. Instead, our data suggests that it is more beneficial to go with the highest correlation threshold that yields a non-zero number of input channels. This approach of using a subset of neural recording channels is comparable to other decoding studies which range from using a single input, such as pudendal nerve ENG,12 to using signals from multiple electrode channels in DRG,2 the spinal cord,24 or the motor cortex,33 and selecting channels based on a combination of the correlation coefficient and subjective judgement.
Statistically, a correlation coefficient-based channel selection method provided the lowest NRMSE in all three algorithms. This result is partially due to this method’s ability to maximize the bladder signal-to-noise ratio compared to using all channels or the LASSO method, by selecting only a few channels that contain a high concentration of relevant sensory information. The LASSO and all channel methods usually selected more channels than the correlation coefficient-based method, thus incorporating more variability in the input channels and decreasing the estimation accuracy. This statistical result is consistent with Table 4.
The firing rate smoothing methods alone did not provide any statistical significance in reducing NRMSE or improving CC in any of the three algorithms, which is consistent with a prior study.8 This effect is in part because both NARMA and Kalman filter algorithms have a recursive mechanism, which has a similar working mechanism as the firing rate smoothing method. Additionally, the thresholded multi-unit activities and relatively large time intervals (> 100 ms) together provide a smoother firing rate compared to what would be obtained for firing rates calculated from single unit activity with smaller time intervals. This statistical outcome is consistent with the varying methods identified across algorithms in Table 4.
Firing rate normalization contributed significantly to reducing the NRMSE in all three algorithms largely due to its ability to reduce the contribution variance among selected channels. As a result, the channels with high firing rates have a more balanced impact on the magnitude of the estimation result compared to channels with low firing rates. However, CC was not expected to be significantly affected because the firing rate only went through a linear transformation. This is consistent with our statistical results, as well as Table 4.
The firing rate interval determines how smooth the firing rate trace is, and how many data points can be used to train from a trial. Larger intervals yield smoother firing rates and potentially improve estimations, especially CC, by providing more channels with correlated firing rates (Tables 2 and 3; Fig. 4). This explains why larger intervals improved CC in Linear Regression. In NARMA and Kalman filter, their recursive mechanisms smoothed out the firing rates, which provides an explanation why larger intervals in this case did not improve estimation further. On the other hand, smaller intervals provide a larger number of data points that can be used for training, which resulted in a reduced NRMSE with the Kalman filter. This tradeoff can be reflected as the inconsistent optimal firing rate interval across different algorithms and performance measures. A drawback of small firing rate intervals is that smaller intervals result in weaker correlations between the firing rate and the bladder pressure (Tables 2 and 3; Fig. 4). In one data set, there were no channels with a correlation greater than 0.2 when using the lowest firing rate interval (Table 3).
Compared to prior studies that used sorted single unit activity to estimate limb states4,29 or bladder pressure,2 our method has a lower computational load due to the simple thresholding method. The accuracy is comparable to prior relevant bladder studies2,12 due to NARMA’s ability to capture the underlying structure of the data sets. Compared to limb state decoding applications, where an artificial neural network might be computationally heavy, bladder decoding is a more suitable application for neural networks because the need for frequent updates is low (every ~ 0.5 s). The step time for new pressure estimates for all models are well below any of the firing rate intervals used here and easily applicable for online decoding (Table 4).
To our knowledge, this work also provides the first proof of concept for decoding bladder pressure from neural recordings using an algorithm based on data collected at an earlier date in a chronic implant (Fig. 8). The feature selection method and algorithm used on the chronic data was based on the best acute study analysis, as discussed above. Although this data set only had 2 weeks between training and testing trials, our prior chronic DRG studies indicated that bladder sensory neurons can be tracked at least 6 weeks after implant using current technology.16 Finally, this preliminary work shows it is possible to translate results from acute studies to chronic studies of bladder pressure.
Our results were obtained from spinally intact felines. Since our goal is to develop a closed-loop neuroprosthesis for bladder control in patients suffering from SCI and other neurogenic bladder dysfunctions, our decoding algorithms need to be assessed in these models. After spinal cord injury, voluntary bladder control is lost and the bladder initially becomes areflexic, later becoming hyperreflexive with re-emergence and remodeling of spinal reflex pathways.1,9 These changes are thought to be partly due to changes in bladder afferent activity. Unmyelinated chemosensitive C-fibers become mechanosensitive and overactive leading to non-micturition contractions at low bladder volumes.10,34 After SCI, there will still be activity from myelinated pelvic afferent fibers that correlate with bladder pressure, but we anticipate increased bladder afferent activity occurring at lower bladder volumes due to C-fibers. We expect that bladder-decoding models will still be effective after chronic SCI, however a model may have low efficacy shortly after injury before neuroplasticity has stabilized. Additional studies are needed to evaluate the consistency of our decoding algorithms at different times after SCI.
Further studies should be performed to test the performance of these algorithms online in real-time and in closed-loop bladder control experiments. In addition, for each of the trials presented here, we trained on only one infusion trial and tested on a later individual trial. Multiple training sets may provide a more robust performance than single-set training, particularly for chronic studies where neural signals may shift over time.16
References
Baptiste, D., M. Elkelini, M. M. Hassouna, and M. G. Fehlings. The dysfunctional bladder following spinal cord injury: from concept to clinic. Curr. Bladder Dysfunct. Rep. 4(4):192–201, 2009. https://doi.org/10.1007/s11884-009-0028-9.
Bruns, T. M., R. A. Gaunt, D. J. Weber. Estimating bladder pressure from sacral dorsal root ganglia recordings. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2011:4239–4242, 2011. https://doi.org/10.1109/IEMBS.2011.6091052.
Bruns, T. M., R. A. Gaunt, and D. J. Weber. Multielectrode array recordings of bladder and perineal primary afferent activity from the sacral dorsal root ganglia. J. Neural. Eng. 8(5):56010, 2011. https://doi.org/10.1088/1741-2560/8/5/056010.
Bruns, T. M., J. B. Wagenaar, M. J. Bauman, R. A. Gaunt, and D. J. Weber. Real-time control of hind limb functional electrical stimulation using feedback from dorsal root ganglia recordings. J. Neural. Eng. 10(2):26020, 2013. https://doi.org/10.1088/1741-2560/10/2/026020.
Bruns, T. M., D. J. Weber, and R. A. Gaunt. Microstimulation of afferents in the sacral dorsal root ganglia can evoke reflex bladder activity. Neurourol. Urodyn. 34(1):65–71, 2015. https://doi.org/10.1002/nau.22514.
Chew, D. J., L. Zhu, E. Delivopoulos, et al. A microchannel neuroprosthesis for bladder control after spinal cord injury in rat. Sci. Transl. Med. 5(210):210ra155, 2013. https://doi.org/10.1126/scitranslmed.3007186.
Christie, B. P., D. M. Tat, Z. T. Irwin, et al. Comparison of spike sorting and thresholding of voltage waveforms for intracortical brain–machine interface performance. J. Neural. Eng. 12(1):16009, 2015. https://doi.org/10.1088/1741-2560/12/1/016009.
Cunningham, J. P., V. Gilja, S. I. Ryu, and K. V. Shenoy. Methods for estimating neural firing rates, and their application to brain-machine interfaces. Neural Netw. 22(9):1235–1246, 2009. https://doi.org/10.1016/j.neunet.2009.02.004.
de Groat, W. C., and N. Yoshimura. Plasticity in reflex pathways to the lower urinary tract following spinal cord injury. Exp. Neurol. 235(1):123–132, 2012. https://doi.org/10.1016/j.expneurol.2011.05.003.
de Groat, W. C., and N. Yoshimura. Changes in afferent activity after spinal cord injury. Neurourol. Urodyn. 29(1):63–76, 2010. https://doi.org/10.1002/nau.20761.
Fry, C. H., F. Daneshgari, K. Thor, et al. Animal models and their use in understanding lower urinary tract dysfunction. Neurourol. Urodyn. 29(4):603–608, 2010. https://doi.org/10.1002/nau.20903.
Geramipour, A., S. Makki, and A. Erfanian. Neural network based forward prediction of bladder pressure using pudendal nerve electrical activity. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2015:4745–4748, 2015. https://doi.org/10.1109/EMBC.2015.7319454.
Jezernik, S., W. M. Grill, and T. Sinkjaer. Detection and inhibition of hyperreflexia-like bladder contractions in the cat by sacral nerve root recording and electrical stimulation. Neurourol. Urodyn. 20(2):215–230, 2001. https://doi.org/10.1002/1520-6777(2001)20:2<215::AID-NAU23>3.0.CO;2-0.
Kao, J. C., P. Nuyujukian, S. Stavisky, S. I. Ryu, S. Ganguli, and K. V. Shenoy. Investigating the role of firing-rate normalization and dimensionality reduction in brain-machine interface robustness. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2013:293-298, 2013. https://doi.org/10.1109/EMBC.2013.6609495.
Karam, R., D. J. Bourbeau, S. Majerus, et al. Real-time classification of bladder events for effective diagnosis and treatment of urinary incontinence. IEEE. Trans. Biomed. Eng. 63(4):721–729, 2016. https://doi.org/10.1109/TBME.2015.2469604.
Khurram, A., S. E. Ross, Z. J. Sperry, et al. Chronic monitoring of lower urinary tract activity via a sacral dorsal root ganglia interface. J. Neural. Eng. 14:36027, 2017. https://doi.org/10.1088/1741-2552/aa6801.
Lin, Y. T., C. Lai, T. S. Kuo, et al. Dual-channel neuromodulation of pudendal nerve with closed-loop control strategy to improve bladder functions. J. Med. Biol. Eng. 34(1):82–89, 2014. https://doi.org/10.5405/jmbe.1247.
Majerus, S. J. A., P. C. Fletter, E. K. Ferry, H. Zhu, K. J. Gustafson, and M. S. Damaser. Suburothelial bladder contraction detection with implanted pressure sensor. PLoS ONE. 12(1):e0168375, 2017. https://doi.org/10.1371/JOURNAL.PONE.0168375.
Melgaard, J., and N. J. M. Rijkhoff. Detecting urinary bladder contractions: methods and devices. J. Sens. Technol. 4:165–176, 2014. https://doi.org/10.4236/jst.2014.44016.
Mendez, A., and M. Sawan. Chronic monitoring of bladder volume: a critical review and assessment of measurement tools. Can. J. Urol. 18(1):5504–5516, 2011.
Mendez, A., M. Sawan, T. Minagawa, and J. J. Wyndaele. Estimation of bladder volume from afferent neural activity. IEEE Trans. Neural Syst. Rehabil. Eng. 21(5):704–715, 2013. https://doi.org/10.1109/TNSRE.2013.2266899.
Nitti, V. W. The prevalence of urinary incontinence. Rev. Urol. 3(Suppl 1):S2–S6, 2001.
Ouyang, Z., S. E. Ross, and T. M. Bruns. Decoding algorithms and dorsal root ganglia neural recordings for estimating bladder pressure. Open Sci. Framew. https://doi.org/10.17605/OSF.IO/ZFYCH.
Park, J. H., C. E. Kim, J. Shin, et al. Detecting bladder fullness through the ensemble activity patterns of the spinal cord unit population in a somatovisceral convergence environment. J. Neural Eng. 10(5):56009, 2013. https://doi.org/10.1088/1741-2560/10/5/056009.
Perel, S., P. T. Sadtler, E. R. Oby, et al. Single-unit activity, threshold crossings, and local field potentials in motor cortex differentially encode reach kinematics. J Neurophysiol. 114(3):1500–1512, 2015. https://doi.org/10.1152/jn.00293.2014.
Rizk, M., and P. D. Wolf. Optimizing the automatic selection of spike detection thresholds using a multiple of the noise level. Med. Biol. Eng. Comput. 47(9):955–966, 2009. https://doi.org/10.1007/s11517-009-0451-2.
Ross, S. E., Z. J. Sperry, C. M. Mahar, and T. M. Bruns. Hysteretic behavior of bladder afferent neurons in response to changes in bladder pressure. BMC Neurosci. 17:57, 2016. https://doi.org/10.1186/s12868-016-0292-5.
Tibshirani, R. Regression shrinkage and selection via the Lasso. J. R. Stat. Soc. Ser. B. 58(1):267–288, 1996.
Weber, D. J., R. B. Stein, D. G. Everaert, and A. Prochazka. Limb-state feedback from ensembles of simultaneously recorded dorsal root ganglion neurons. J. Neural Eng. 4(3):S168–S180, 2007. https://doi.org/10.1088/1741-2560/4/3/S04.
Wenzel, B. J., J. W. Boggs, K. J. Gustafson, and W. M. Grill. Closed loop electrical control of urinary continence. J. Urol. 175(4):1559–1563, 2006. https://doi.org/10.1016/S0022-5347(05)00657-9.
Winter, D. L. Receptor characteristics and conduction velocities in bladder afferents. J. Psychiatr. Res. 8(3):225–235, 1971. https://doi.org/10.1016/0022-3956(71)90021-5.
Wu, W., Y. Gao, E. Bienenstock, J. P. Donoghue, and M. J. Black. Bayesian population decoding of motor cortical activity using a Kalman filter. Neural Comput. 18(1):80–118, 2006. https://doi.org/10.1162/089976606774841585.
Wu, W., A. Shaikhouni, J. P. Donoghue, and M. J. Black. Closed-loop neural control of cursor motion using a Kalman filter. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2004:4126–4129, 2004. https://doi.org/10.1109/IEMBS.2004.1404151.
Yoshimura, N. Bladder afferent pathway and spinal cord injury: possible mechanisms inducing hyperreflexia of the urinary bladder. Prog. Neurobiol. 57(6):583–606, 1999. https://doi.org/10.1016/S0301-0082(98)00070-7.
Acknowledgments
The authors would like to thank Kaile Bennett, Eric Kennedy, Zachariah Sperry, and other members of the Peripheral Neural Engineering and Urodynamics Lab for assistance with surgeries, data collection, and analysis. Research reported in this publication was supported by the Craig H. Neilsen Foundation (Grant # 314980) and by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under Award Number U18EB021760. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Craig H. Neilsen Foundation or the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor Leonidas D Iasemidis oversaw the review of this article.
Shani E. Ross and Zhonghua Ouyang are the Co-first authors.
Rights and permissions
About this article
Cite this article
Ross, S.E., Ouyang, Z., Rajagopalan, S. et al. Evaluation of Decoding Algorithms for Estimating Bladder Pressure from Dorsal Root Ganglia Neural Recordings. Ann Biomed Eng 46, 233–246 (2018). https://doi.org/10.1007/s10439-017-1966-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10439-017-1966-6