Abstract
Initiation and propagation of cell signaling depend on productive interactions among signaling proteins at the plasma membrane. These diffusion-limited interactions can be influenced by features of the membrane that introduce barriers, such as cytoskeletal corrals, or microdomains that transiently confine both transmembrane receptors and membrane-tethered peripheral proteins. Membrane topographical features can lead to clustering of receptors and other membrane components, even under very dynamic conditions. This review considers the experimental and mathematical evidence that protein clustering impacts cell signaling in complex ways. Simulation approaches used to consider these stochastic processes are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cell signaling, used for both intracellular and intercellular communication, is essential for the healthy physiological functioning of multi-cellular organisms. Ligand binding to a transmembrane receptor triggers an intracellular signaling cascade that results in altered cell behavior. The proper integration of different environmental signals is critically important to many biological processes, including cell survival, differentiation, proliferation, and migration.10,39,42,49,85,89 Aberrations in signal transduction have been implicated in numerous pathologies, from allergy and asthma to many different cancers.10,15,29,37,39,49,75,81,89 Signal transduction pathways have therefore been studied extensively, and many drugs developed to target them.10,22,29,49,75,81
Knowledge of the structure of the plasma membrane and of signaling processes continues to improve, due to advances in experimental techniques and imaging technologies.46,84,87 There is considerable evidence for the concept that the cell membrane is compartmentalized into microdomains, such as protein islands88 and lipid rafts.56 Receptor clustering in small or large aggregates (illustrated schematically in Fig. 1) at discrete locations has been noted in many cell types,1,6,31,39,66,73,89 prompting intense interest in roles for membrane microdomains in signal propagation and preliminary mathematical studies to understand both formation of clusters and their role in cell signaling.8,17,18,21,35,36,48,63,76,77 There is general agreement that the composition of these microdomains is quite heterogeneous and, further, that their stability is influenced by the dynamic interactions of the cortical cytoskeleton with membrane proteins and lipids. The cytoskeleton also limits diffusion of membrane constituents by forming “picket fences” and “corrals.”43,72 The role of these membrane features in promoting or limiting protein–protein interactions remains controversial, since there is strong potential to both enhance and inhibit signaling.3,17,55,58 To help resolve these issues, several groups are developing spatially realistic mathematical simulations of receptor motion, aggregation/clustering, and activation in the cell membrane.
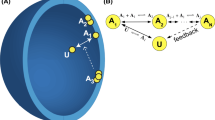
Schematic representation of microdomains and receptor clustering. Left: Cartoon representation of features that can subcompartmentalize the plasma membrane, including rafts or islands, and the cortical cytoskeletal network. These features are highly dynamic, permitting rapid exchange by diffusion. Right: Representation of the distribution of receptors (yellow, blue symbols) in and out of domains (pink regions) formed by these features. Arrows point to various states, including monomers, dimers, and aggregates. Receptors that are transiently trapped in domains are locally crowded (arrow, top right) and appear as clusters by imaging technologies. This molecular crowding can be more pronounced upon ligand stimulation, due in part to formation of dimers and larger aggregates with decreased diffusive mobility. This review considers the experimental and computational evidence that molecular crowding influences receptor dimerization/aggregation and recruitment of signaling proteins
It is important to note that parameters for these mathematical models rely on powerful new experimental techniques. High resolution microscopy techniques, such as transmission electron microscopy (TEM) and photoactivation light microscopy (PALM), have been applied to map the spatial distribution of signaling molecules in fixed cells.47,88 These snapshot images of protein distributions can be supplemented with powerful new live cell imaging approaches, including fluorescence resonance energy transfer (FRET), fluorescence lifetime correlation spectroscopy (FLCS) and single particle tracking (SPT) experiments.46 These techniques can generate key information regarding the kinetics of protein–protein interactions, including rates of dimerization, size of receptor aggregates, and changes in diffusion properties.50 These rich data sets support the development of more accurate and detailed mathematical models, that in turn improve understanding of biological results.
Key Concepts and Definitions Relevant to the Consideration of Protein Clustering in the Plasma Membrane
In this brief review, we focus attention on the mathematical simulation of protein clustering in the plasma membrane, an initial step in many signaling pathways. The protein species considered may be a surface receptor that is triggered by binding to an extracellular ligand or an intracellular signaling partner, such as an adaptor protein or enzyme that propagates signaling through the cell interior. We define clustering as the non-random spatial distribution of a membrane species, which can be observed and experimentally validated through modern technologies. “Snap-shot” images of membrane proteins often capture some level of clustering even before the onset of ligand binding to receptors or active signaling.89 We hypothesize that these basal levels of clustering arise from brief, non-productive interactions among proteins as they encounter one another while diffusing in the plasma membrane or when proteins are transiently co-confined in a raft, island or corral (Fig. 1). Thus clustering in this sense is not synonymous with oligomerization, which reflects the direct and measurable interaction between membrane components. It is important to point out that stable oligomers cannot be distinguished from unstable clusters in imaging techniques using fixed cells, such as TEM and PALM. However, new imaging protocols can now accurately measure the dynamics of protein–protein interactions at the molecular scale.46 A recent example from the Spatiotemporal Modeling Center is the simultaneous SPT of pairs of EGFR molecules, each labeled by virtue of binding to different colors of quantum dot probes; only with two QD-bound receptors were both coincident and exhibited correlated motion, could they pass the stringent criteria for oligomerization.50 The concept of clustering becomes particularly important as we consider the data suggesting that the spatial proximity of proteins can promote protein–protein interactions, including oligomerization, by increasing the likelihood of productive collisions.
Choosing the Right Modeling Approach
Mathematical models constructed to date to study signal transduction pathways are of varied complexity. They can be classified conveniently as deterministic methods, in which inherent temporal and spatial fluctuations in diffusion and reaction rates are ignored, and stochastic methods, which attempt to capture these fluctuations (Fig. 2). The simplest modeling approach is to assume that the system of interest is well mixed, without any spatial concentration gradients, and describe the reactions by a system of ordinary differential equations (ODEs). The utility of ODE modeling is enhanced by systematic sensitivity analysis, which examines automatically changes in model behavior due to parameter variation.60,61 Such a deterministic, well-mixed approach continues to be widely used,77 and has produced useful results.7,61 However, these approaches do not take into account either spatial inhomogeneities or stochastic fluctuations, which can be significant when the number of molecules in the region of interest is small. At a slightly higher level of complexity, some spatial description is provided by dividing the region of interest into separate well-mixed compartments. Additional ODEs are needed to describe inter-compartmental species translocation reactions, thus mimicking spatial movement.
Classes of mathematical models for molecular processes in cells and the scales at which they are applicable to signaling processes. A possible quantitative guide is the size of the largest element that can be treated as spatially homogeneous (horizontal axis) and the typical number of molecules of one species in the element (vertical axis). The suggested spatial resolution is determined by the size of the biological element of interest and current computational capabilities. Spatially resolved models are resource-intensive, and are therefore generally applied to small subsystems. Cell-level models of large signaling networks are typically well mixed; spatial Monte Carlo studies rarely scale beyond a few hundred nanometers. A promising approach for multi-scale applications is a combination of compartment-based models at the large scales and fully spatial simulations focused on a few important processes within small structural elements of the membrane. Temporal fluctuations arise largely from the discrete and stochastic nature of the underlying molecular processes. The relative magnitude of temporal fluctuations (ΔN) decreases as the number of particles increases. The discrete nature of the particle number can thus be ignored when N is significantly greater than 1. That is, deviations from the expected average behavior can be neglected when the expected magnitude of the fluctuations is small compared to N
These well-mixed, ODE-based continuum pathway models41 were expanded to include spatial inhomogeneity9,71 by solving partial differential equations (PDEs) that include molecular diffusion effects. Stochastic methods that assume spatially well mixed systems have also been developed to account for temporal fluctuations.27,45 Stochastic PDEs include both spatial information and temporal fluctuations. The most detailed, and thus most complex, models are fully spatial, stochastic methods that track the movement of individual molecules.4,11,17,18,30,35,36,63,78 However, the computational burden increases rapidly with increasing complexity of the modeling approach. Figure 2 summarizes the various modeling approaches and their range of applicability.
Mathematical simulation of events in the plasma membrane faces unique challenges. Membrane proteins are constantly undergoing random motion in the plane of the membrane, where the diffusion rate is influenced by the environment, such as hindrance by microdomains, and thus varies both spatially and temporally. Optimally, the spatial location of every protein needs to be predicted, in order to capture clustering imposed by membrane topography and to predict the outcomes of both transient and prolonged protein–protein binding events. Fully spatial, stochastic methods offer capabilities that can capture accurately the dynamics of these events, but can be associated with prohibitively high computation cost. Novel hybrid approaches show promise for solving some of these computational challenges.
Finally, this section would not be complete without introducing the unique power of rule-based approaches.20,33 Here, molecular interactions in signaling networks are treated as systems of encoded rules. Molecules are represented as structural objects that have modular domains and associated states representing conformations or covalent modifications of these domains. Importantly the input files and model specification blocks are compatible with multiple types of computational approaches, including coupled ODEs that result in deterministic solutions of reaction kinetics as well as stochastic methods.
Applications in Specific Signaling Pathways
Sections below briefly summarize mathematical models that have been developed to study signal transduction pathways, with emphasis on methods developed by our group and others to capture the influence of clustering and other spatial aspects. We focus on three representative signal transduction pathways (EGFR, Ras/MAPK, and GPCR) where protein clustering has been implicated, and on the modeling approaches used to approach this unique set of challenges.
Our Group’s Focus: Spatial Aspects of Signaling Through the Epidermal Growth Factor Receptor
A member of the ErbB family of plasma membrane receptors, EGFR is critically important to many biological processes, including embryonic development and carcinogenesis.10,39,89 Upon binding any one of several ligands, including EGF, the ErbB receptors homo- or hetero-dimerize. Dimerization is followed by transphosphorylation of tyrosine residues in receptor cytoplasmic tails, which enables recruitment of cytosolic signaling proteins. The reader is referred to Figs. 2 and 3 in the article by Telesco and Radhakrishnan74 within this same issue, for diagrams of EGFR/ErbB1 dimerization, phosphorylation, and adaptor protein recruitment. Subsequently, these complexes activate many different signaling cascades, including the Ras-MAPK pathway discussed in the next section.
There exists considerable experimental evidence for preexisting clusters of resting EGFR (Fig. 3) and for dynamical changes after addition of ligand.1,6,39,66,73,89 We have built simulation platforms at different levels of complexity, in order to evaluate the impact of EGFR clustering in the plasma membrane.
Experimental results and mathematical model predictions of EGFR clustering. (a) Experimental evidence for EGFR clustering in absence of ligand. Electron micrograph of gold particle-labeled EGF receptors in resting A341 cells (~2 million EGFR/cell), reveals a non-random distribution and provides evidence for receptor co-confinement. (b) Spatial domain used in lattice-free Monte Carlo simulation.35 The spatial domain simulated by the off-lattice Monte Carlo procedure was a square of area 2 μm2, representative of a small region in the plasma membrane. This region was modified to include many islands or preferred domains (the gray rectangles within the membrane patch), to simulate the receptor-trapping microdomains seen in (a). Movement of receptors into and out of the simulated microdomains over a time period of 30 s is indicated by the thin colored tracings. Receptor trapping in the microdomains was reproduced mathematically by stipulating that receptors had a greater probability of entering these regions than of leaving them. (c) Simulation predictions of receptor clustering in absence of ligand. The predicted particle positions after 30 s of simulation time are indicated by the black dots. The Hopkins statistical test (inset) was used to test the randomness of receptor distribution. The right shift of the distribution (compared to the random or normal distribution shown in red) confirms the clustered nature of the receptors. The predicted receptor distribution compares well with the experimental observation in (a). (d) Simulations using a coupled spatial/nonspatial stochastic algorithm (CSNSA) support the conclusion that EGFR clustering promotes activation of the adaptor SOS. ODE models confirm this conclusion, using a fast diffusion coefficient to override contributions from membrane spatial organization (from Hsieh et al.35 and Costa et al.18)
Approaches and Methodology
Our first attempt to develop a spatial model of the EGFR pathway was a simple compartmental model that accounted for receptor density differences observed in the plasma membrane, with some regions having high-receptor density and others displaying low-receptor density.52 The focus of this study was to explore whether the added computational complexity associated with spatial modeling was justified. Our initial goal was to determine if the non-uniform receptor distribution in the cell membrane could account for the experimentally observed, concave-up Scatchard plot for binding of EGF ligand to its receptor. We simply optimized the distribution of receptors into high- and low-density regions, and were able to determine the parameter space that allowed for a concave-up Scatchard plot. This first attempt at compartmentalized spatial modeling showed that accounting for the spatial organization of receptors was highly valuable, and should be pursued, to enable both qualitative and quantitative understanding of cell signaling involving (at least) the EGFR.
This study convinced us of the utility of spatial modeling of membrane-bound receptors and of its importance in understanding cell signaling. We have now accumulated extensive experience in developing spatially realistic simulations of the cell membrane and also addressed the initiation of signaling.13,17,18,35,36,51–54 Next, we summarize our development of lattice-based and lattice-free (or off-lattice) methods, as well as our use of hybrid approaches.
Lattice-Based Monte Carlo (MC) Approaches
In lattice-based models, molecules are located at discrete grid points in the spatial domain and diffusion is restricted to movement to an unoccupied neighboring point. Lattice-based MC simulations have become very popular in the physics, chemistry, materials, and engineering communities, as they provide spatio-temporal information at significantly reduced computational cost, compared to off-lattice simulations.5,14,16,28,90 The MC method is a coarse graining of molecular dynamics (MD) simulations,5 because MD is impractical for rare event dynamics, such as hopping between deep minima of a potential energy surface. The MC method stochastically solves an underlying master equation using pseudo-random numbers, by constructing the probability with which the various states of the system have to be weighted according to a Markov process. MC simulations can provide continuous time information. Gillespie26,27 established the foundations of time-dependency for chemical reactions in a spatially homogeneous system. His approach is easily applicable to arbitrary complex computational systems, and is often referred to as the kinetic or dynamic MC method. Despite important algorithmic implementations (e.g., dependency graphs,25 lists of neighbors, binary-tree search, etc.), MC simulations are seriously plagued by (1) the presence of fast reactions that occur in the large biochemical networks seen in biology and (2) the execution of one event at a time.
Our Spatial Kinetic Monte Carlo (SKMC) method52,53 utilizes a modified null-event, lattice-based MC algorithm.18,54 The spatial domain, representing a small region of the plasma membrane, is a two-dimensional square lattice of side ℓ, divided into a large number of much smaller square bins of side a (≪ℓ). The SKMC algorithm consists of first randomly selecting an occupied lattice site, and then choosing either a successful (reaction or diffusion) or unsuccessful (null) event, based on calculated probabilities. If a successful event is chosen, it is executed. The transition rate \( \Upgamma_{i \to j}^{d} \), for diffusion of species from any site i (i.e., lattice point i) to a nearest-neighboring site j is defined as
where \( \Upgamma^{d} = 4D/a^{2} \) and D is the diffusion coefficient of the species located at site i. The term B i denotes the set of four possible nearest-neighboring sites to which diffusion can occur in two dimensions from site i. Because species are allowed to diffuse only to an unoccupied site, we define an occupancy function σ j for each of the four nearest-neighboring sites, in order to simplify the procedure for computing the transition rate for diffusion. For any site k (=i or j), σ k is set equal to 1 if the site is occupied, or to 0 if the site is unoccupied. The transition rate for a chemical reaction at site i, \( \Upgamma_{i}^{r} \), depends on the reaction type and is directly related to the standard reaction rate.
The probability \( p_{i}^{x} \) of an event x (=r reaction or d diffusion) at site i is computed by using the relation
where \( \Upgamma_{\max } \) is a normalization constant, defined as
where the multiplicative factor of 4 accounts for events occurring in the four directions of the two-dimensional square lattice. Finally, the time step \( \Updelta t \) used to advance the simulation time is computed as \( \Updelta t = 1/\Upgamma_{\max } \).
Rule-Based, Non-lattice Simulator
Our non-lattice, stochastic simulator is an alternative approach.35,36 In the lattice-free method, particles are not confined to discrete points in space but are randomly repositioned upon undergoing a diffusion event. Receptors and other proteins in the 2D membrane and 3D cytosolic space are represented by sphere-like particles with radii determined from experimental data and their coarse-grained molecular models. At each time step, species diffusion is simulated as Brownian motion (Fig. 3). In addition, species have the potential to react with spatially nearby species. This simulator was designed for flexible model development and deployment by a modularized and rule-based approach. It tracks the individual reactions of multistate molecules and accommodates complex situations.
Hybrid Approaches
We continue to improve our basic SKMC algorithm, leading to increased efficiency and speed of the simulations. One significant advance was the coupling of our lattice-based SKMC simulations on the cell membrane to well-mixed stochastic simulations within the cytosol.18 In Costa et al.,18 we describe the development of an algorithm that couples a spatial stochastic model of membrane receptors with a nonspatial stochastic model of cytosolic reactions. Our novel hybrid algorithm provided a computationally efficient method to evaluate the effects of spatial heterogeneity on the coupling of receptors to cytosolic signaling partners. For well-mixed systems results obtained using a compartmental ODE method compared well with those generated with our hybrid model. Thus, for sufficiently high receptor copy number, the far simpler ODE model may be adequate. However, for spatially inhomogenous systems where the receptors numbers are low, the hybrid method was significantly superior to the ODE model.
EGFR Density, Through Clustering or Overexpression, Influences Signaling Output
We have applied these methods to study the early molecular mechanisms involved in EGFR signaling. For example, we applied the lattice-based spatial stochastic model of the plasma membrane to examine the influence of cytoskeletal corral openings on EGFR clustering.17 Clustering was shown to depend on both receptor concentration and picket fence density. For high picket fence densities, clustering increased with increasing receptor concentration in the range examined. Conversely, low receptor concentrations combined with small corral sizes inhibited clustering; at normal to high receptor concentration, maximal clustering occurred at an intermediate corral size (~100 nm). These results indicate that both the number of clusters and the average cluster size are likely to be complex functions of receptor density and microdomain size. It follows that compartmentalization of the plasma membrane could either inhibit or enhance signaling, concepts that require further exploration.
The non-lattice, rules-based simulator allowed us to explore the effect of EGFR overexpression and its relation to carcinogenesis.35 We postulated that increased receptor density in membrane microdomains or protein islands might lead to more frequent interactions between non-ligand bound receptors and, further, that large numbers of these short-lived interactions might explain EGFR signaling known to occur even in the absence of ligand.6 One important aspect was consideration of EGFR extracellular domain conformation, based upon structural studies showing that the resting EGFR is predominantly in a “closed” conformation. Binding of ligand is proposed to stabilize the extended conformation and expose the dimerization arm. In our simulations, we assumed that the resting EGFR “fluxes” between the open and closed states, but spends 99% of its time in the closed state. This property translates to a low probability that two diffusing monomers will collide under conditions where both expose their dimerization arms and are therefore competent to form a complex. The 2D simulation space included membrane microdomains that transiently trapped receptors (as in Fig. 3), setting up clusters undergoing dynamic exchange. Remarkably, at levels of receptors typical of most normal cells, co-confinement in membrane microdomains lowered the threshold for ligand-independent receptor dimerization but resulted in very modest signaling output. When the simulation space was populated with densities typically seen in tumors with EGFR gene amplification, which can express millions of EGFR per cell, the percent of activated receptors could exceed 10% with our parameter values. Clustering had little effect in these cases, since the overall density on the membrane was already very high.
We have used both lattice and non-lattice models to consider how spatial aspects might affect the recruitment of signaling molecules to the phosphorylated EGFR tail.18,36 In Hsieh et al.,36 we also considered the combinatorial complexities associated with the facts that EGFR has multiple phosphorylation sites and, further, that each phosphotyrosine site is capable of binding multiple partners. We used coarse-grained molecular docking simulations to show that steric hinderance can impose important constraints on the composition of adaptor proteins capable of docking simultaneously on the EGFR tail. Modeling predictions in Hsieh et al.36 were quantitatively consistent with experimental data for the kinetics of both EGFR phosphorylation and recruitment of adaptor proteins. Importantly, both papers provide mathematical support for the conclusion that clustering of receptors can amplify signaling by promoting sequential binding of adaptor proteins. These results provide confidence in our models, and have led to ongoing studies of other growth factor receptors that initiate signaling through dimerization, particularly VEGFR, as well the heterodimerizing members of the ErbB family. This field continues to advance, as demonstrated by the hybrid approaches of Radhakrishnan and colleagues74 that consider ErbB structural and diffusion properties using increasingly complex models. Additional aspects of cell surface topography, such as the induction of membrane curvature by endocytic adaptor proteins, are new concepts that will provide important insight into the control of signal transduction through the biophysical principles of membranes.
Work by Others: The Case of Signaling via Ras/MAPK Pathways
The Ras superfamily consists of over 100 small GTP-binding proteins (or GTPases), which respond to various extracellular stimuli to regulate important signal transduction pathways.81,85 These proteins, which have low intrinsic GTPase activity, “switch” between active GTP-bound and inactive GDP-bound conformations. The processes mediated by GTPases include cell division, differentiation, apoptosis, and migration, cytoskeletal reorganization, and intracellular protein trafficking.75 Abnormalities in these pathways are seen in various pathologies, including obesity, diabetes, inflammatory diseases, cardiovascular disease, neurological disease, and cancer.15,75,81 Therefore the pharmacological targeting of GTPases and/or their signaling pathways is an active field.81
The Ras/Raf/MEK/ERK mitogen-activated protein kinase (MAPK) pathway has been investigated extensively, both in the clinic and the laboratory, and by mathematical modeling.7,22,23,32,34,40,41,57,61,68,69,76,77,86 Activation of a number of receptors, including EGFR, leads to guanine nucleotide exchange (dissociation of GDP, gain of GTP) by membrane-tethered Ras, thereby activating it. The activated Ras in turn activates Raf (Ras-associated factor), the first kinase in the cascade. Subsequently, Raf activates MEK (MAPK/extracellular signal-regulated kinase), which then activates ERK (extracellular signal-regulated kinase). The translocation of phosphorylated ERK to the nucleus and activation of transcription factors mediates many cellular activities.
Numerous mathematical models have been developed to study this pathway.7,23,32,34,40,41,57,61,68,69,76,77,86 Much of this work uses compartmental models and ODEs to follow the temporal evolution of activated ERK, and does not consider clustering in the plasma membrane. However, Tian et al.76,77 have mathematically evaluated various spatial aspects of Ras signaling, including clustering in the plasma membrane. This group utilized a hybrid approach to simulate reactions in the cell membrane and those in the cytosol, enabling them to separate the contribution of the plasma membrane structure to the signal. They combined the well-mixed stochastic model of Gillespie26,27 to simulate reactions in the membrane with an ODE model for the cytosolic reactions. They assumed that the number of RasGTP clusters was proportional to the EGF concentration, and these clusters served as platforms for recruiting Raf to the plasma membrane for activation. The lifetime of RasGTP clusters was assumed to be normally distributed over a measured value. Plasma membrane reactions, in addition to binding and activation of Raf by RasGTP clusters, included recruitment by activated Raf of the KSR–MEK–ERK complex from the cytosol and activation of MEK by activated Raf and of ERK (MAPK) by activated MEK. KSR (kinase suppressor of Ras) is a scaffold protein that facilitates MAPK activation by providing binding sites for assembly of the signaling complex. The recruitment of both Raf and the KSR–MEK–ERK complex was modeled as occurring through random collisions with the plasma membrane. With dissolution of a nanocluster, all recruited proteins diffused back to the cytosol, where the activated MEK and ERK continued their roles. Using this model in conjunction with biological experiments, Tian et al.76 concluded that RasGTP clustering is essential for signal transduction. Moreover, the RasGTP clusters operate as sensitive switches in that they produce approximately the same levels of normalized activated ERK over a wide range of ligand concentration. One possible explanation for this behavior is the establishment of locally high concentrations of recruited proteins and thus the spatial restriction of active ERK production to RasGTP nanoclusters, whose generation and lifetime are themselves strictly regulated.76 Tian et al.76 also concluded that the production of RasGTP nanoclusters in direct proportion to ligand concentration can ensure high fidelity of signal transduction.
Subsequently, Tian et al.77 incorporated models for following the temporal evolution of RasGTP clusters in the cell membrane. In particular, they studied K-Ras clustering and how it is influenced by the protein Galectin-3 (Gal3). Previous experimental work had shown that Gal3 is a scaffolding protein recruited to the plasma membrane, where it is necessary for the formation of Ras nanoclusters.70 Their mathematical model77 considered the two species, membrane-bound RasGTP and Gal3, initially in the cytosol. Once Gal3 is recruited by RasGTP, the RasGTP-Gal3 complexes are assumed to diffuse randomly in the plasma membrane and react with one another to form complexes of various sizes.
To simplify the calculation procedure, Tian et al.77 allowed for a maximum cluster size of ten. The various combinations of possible complexes resulted in a total of 27 species and 136 reactions in the plasma membrane. In agreement with our earlier observation, they concluded that spatial stochastic modeling of such a large system poses a considerable computational burden. Therefore they developed an ODE system to follow the temporal evolution of complexes of size 1–10, using a spatial stochastic model to only deduce collision rates among the complexes.35,36 This deterministic system was solved with a Runge–Kutta method suitable for stiff ODEs.60 The collision rates were obtained by initially placing RasGTP randomly in a square-shaped representation of the plasma membrane. Recruitment of Gal3 produces the Ras–Gal complex. These molecules were allowed to diffuse randomly, and a collision was said to occur when the distance between two molecules was less than the sum of their radii. The collisions produced various combinations of Ras–Gal complexes. When a nanocluster, defined as a cluster consisting of five or more RasGTP molecules, formed it was assumed to become immobile in the plasma membrane. During the calculation procedure the total numbers of collisions giving rise to all cluster types were tracked. At the end of the computational time period, the collision rate constants were computed from the total numbers of collisions. Kinetic rate constants for the ODE model were then derived from the collision rate constants, by using a genetic algorithm in conjunction with experimental data. The validity of this deterministic ODE model was checked with results generated with a stochastic simulation algorithm.26 Presumably due to the large numbers of proteins, the stochastic simulations predicted only small fluctuations. This observation supports use of deterministic models when the protein copy number is high, in agreement with our observations.
Using this modeling approach, Tian et al.77 studied clustering of K-Ras-GTP in the plasma membrane arising from interactions with Gal3 for various KRas and Gal3 copy numbers. The simulation time period was sufficiently long for the system to equilibrate. The time to equilibrate was approximately two minutes, an important result because it is in good agreement with the time period required for RasGTP loading in response to stimulation.76 Their results also successfully reproduced the experimental results of Plowman et al.59 that approximately 42% of the RasGTP were in clusters and the average cluster size was approximately 7. Tian et al.77 also generated the equilibrium nanocluster number vs. size histogram. Their results showed that nanoclusters with two to four molecules accounted for only 2.1% of the RasGTP, whereas a cluster size of 5 was the most prevalent. Nanoclusters larger than 5 in size were progressively smaller in number, approximately inversely proportional to the size. The authors speculate that one possible reason for the lowered incorporation of RasGTPGal3 complexes into clusters of size 5 or larger is the remodeling of the lipid environment of the cluster by the stable pentamer. Their results also suggest that cluster formation is only weakly dependent on RasGTP concentration, and is determined by the Gal3 cytosolic concentration. Tian et al.77 concluded that on the basis of their simulations neglecting the formation of clusters with more than 10 RasGTP molecules is reasonable. Notably, this work illustrates the difficulty of spatial modeling of systems with large reaction networks.
Work by Others: G-Protein Coupled Receptors
The GPCRs constitute the largest family of transmembrane receptors, consisting of five subfamilies.2,65 These proteins, whose structure and function were reviewed recently by Rosenbaum et al.,65 are characterized by seven transmembrane spanning α-helical segments.2,24 GPCRs regulate many physiological functions such as vision, gustation, and olfaction.65,82 Neurotransmitters, hormones, and environmental stimuli activate these pathways. GPCRs are also implicated in many human diseases, such as inflammation, retinitis pigmentosa, nephrogenic diabetes insipidus and Kaposi’s sarcoma.24,38,82,83 At present, most pharmaceutical drugs used by humans target GPCRs by serving as agonists or antagonists.21,82
Many aspects of GPCR signaling are well established. In the classical view, binding of ligand to a GPCR induces a conformational change in the receptor. The activated receptor initiates guanine nucleotide exchange (GDP → GTP) in its principal signaling partner, a heterotrimeric (αβγ) G-protein complex. Like ras, heterotrimeric G proteins are tethered to the cytosolic leaflet of the plasma membrane through covalently attached lipids, and assume an active state once bound to GTP. An additional step is required for heterotrimeric G proteins: the separation of the GTP-bound Gα subunit from the Gβγ subunit, which diffuses into the cytosol. The subsequent activation of downstream effector proteins results in various distinct biological reactions.
Recent work has focused on new aspects of GPCR signaling, such as the evidence that at least some GPCRs can form homo- or hetero-dimers.8,24,83 These dimers can interact further to form oligomers.21 Although believed essential for signaling to occur, the dimerization mechanism is well characterized for only a few GPCRs.44 Due to the importance of GPCR signaling in healthy and diseased states, GPCR interactions, along with membrane organization, and their impact on signaling must be well characterized. Mathematical modeling is therefore being used increasingly to help unravel the intricacies of this pathway. A useful review of mathematical models that have been developed to study GPCR signaling is given by Linderman.48
Brinkerhoff et al.8 used triangular lattice-based MC models to simulate receptor dimerization and activation in a two-dimensional plane, examining how dimerization creates clusters of receptors. Their model demonstrates the applicability of MC methods to systems with discrete reactions that are diffusion limited.8 Randomly selected particles undergo either one of two possibilities at each time step: displacement in a random direction by a distance governed by the diffusion coefficient or a chemical reaction. Reaction possibilities considered were receptor dimerization, binding of ligand by receptor, receptor activation of G protein and receptor phosphorylation. This group’s simulations suggest that clustering arises through both dimerization and cross talk between receptors as they approach one another closely and are able to share an effector. They also concluded that the resulting clustering enhances signaling.
Fallahi-Sichani and Linderman21 investigated lipid raft impact on GPCR signaling with a combination of MC (stochastic) and deterministic models. A lattice-based, kinetic MC model was used to establish the effects of low-diffusivity rafts on receptor dimerization and cluster dynamics. The stochasticity of the model allowed for receptor distributions to be examined, leading to parameter estimations for exploring the effects on downstream signaling using an ODE model. The fraction of plasma membrane covered by microdomains (rafts), which was varied from 2 to 30%, had a significant impact on output. At 2% coverage, microdomains amplified the overall response, but at higher coverage the signal was attenuated. They concluded that dimerization and lipid raft trapping cooperatively control the extent and dynamics of GPCR signaling.
Tolle and Le Novere78 developed an off-lattice, Brownian diffusion-based stochastic model, which they used to determine how alpha-amino-3-hydroxyl-5-methyl-4-isoxazolepropionic acid receptor (AMPAR) diffusion in the dendritic spine affects synaptic signaling, specifically long-term potentiation (LTP).79 LTP, an increase in synaptic strength, is a well-studied form of synaptic plasticity, the ability to change the strength of a signal.67,79 Tolle and Le Novere’s79 model accounts for the dendritic spine membrane, membrane receptors, and scaffolding proteins known to bind to membrane receptors. The spatial domain representing the plasma membrane of the synaptic spine was modeled as a square of surface area corresponding to the measured volume of the spine. This square was separated into two different compartments or domains, in order to account for the two physiologically different portions of the plasma membrane: the post-synaptic density (PSD) and the extra-synaptic membrane (ESM). The PSD is a protein-rich region where AMPARs are concentrated,67,79 while the rest of the membrane is classified as the ESM.79 The transmembrane receptor movement within the ESM was modeled with Brownian-type diffusion, while confined motion was used to model the restricted diffusion within the PSD. Simulation results indicate that randomly placed receptors quickly localize to the PSD, which Tolle and Le Novere79 suggest explains the quick onset of LTP.
Concluding Remarks
This review specifically considers the mathematical modeling of protein clustering on the plasma membrane and the evidence that signal transduction can be enhanced by locally high concentrations of proteins that increase the probability of protein–protein interactions. This feature is especially important when the numbers of particles are small. When proteins are overexpressed, as in EGFR amplification in certain cancers, the impact of clustering may not be as significant.35 The role of membrane microdomains in signaling may be quite complex, since both inhibitory and stimulatory effects have been observed experimentally and theoretically.3,17,55,58
Mathematical modeling, in conjunction with biological experiments, is providing new insights into the mechanisms that govern protein clustering in membranes and the resulting impact on signaling. Increasing experimental detail is being matched by increasingly complex models that account for previously ignored biological subtleties.12,19,30,45,62,64,80 An important goal is to predict the functional responses of whole cells and cell-tissue systems, based upon integration of spatial and temporally encoded signals from surface receptors. Achieving this goal will necessitate the development of efficient and accurate multi-scale simulation capabilities. A daunting challenge to mathematical modeling of cell signaling continues to be the scaling up of computationally intense methods developed for studying molecular behavior, to enable predictive modeling at progressively more complex levels, from the cellular to the systemic.
References
Abulrob, A., et al. Nanoscale imaging of epidermal growth factor receptor clustering: effects of inhibitors. J. Biol. Chem. 285(5):3145–3156, 2010.
Alberts, B. Molecular Biology of the Cell (5th ed.). New York: Garland Science, 2008. 1 v. (various pagings)
Allen, J. A., R. A. Halverson-Tamboli, and M. M. Rasenick. Lipid raft microdomains and neurotransmitter signalling. Nat. Rev. Neurosci. 8(2):128–140, 2007.
Andrews, S. S., and D. Bray. Stochastic simulation of chemical reactions with spatial resolution and single molecule detail. Phys. Biol. 1(3–4):137–151, 2004.
Auerbach, S. M. Theory and simulation of jump dynamics, diffusion and phase equilibrium in nanopores. Int. Rev. Phys. Chem. 19(2):155–198, 2000.
Bader, A. N., et al. Homo-FRET imaging enables quantification of protein cluster sizes with subcellular resolution. Biophys. J. 97(9):2613–2622, 2009.
Brightman, F. A., and D. A. Fell. Differential feedback regulation of the MAPK cascade underlies the quantitative differences in EGF and NGF signalling in PC12 cells. FEBS Lett. 482(3):169–174, 2000.
Brinkerhoff, C. J., P. J. Woolf, and J. J. Linderman. Monte Carlo simulations of receptor dynamics: insights into cell signaling. J. Mol. Histol. 35(7):667–677, 2004.
Brown, G. C., and B. N. Kholodenko. Spatial gradients of cellular phospho-proteins. FEBS Lett. 457(3):452–454, 1999.
Bublil, E. M., and Y. Yarden. The EGF receptor family: spearheading a merger of signaling and therapeutics. Curr. Opin. Cell Biol. 19(2):124–134, 2007.
Burrage, K., et al. Modelling and simulation techniques for membrane biology. Brief. Bioinform. 8(4):234–244, 2007.
Chakraborty, A. K., M. L. Dustin, and A. S. Shaw. In silico models for cellular and molecular immunology: successes, promises and challenges. Nat. Immunol. 4(10):933–936, 2003.
Chatterjee, A., et al. Time accelerated Monte Carlo simulations of biological networks using the binomial tau-leap method. Bioinformatics 21(9):2136–2137, 2005.
Chuan Kang, H., and W. Weinberg. Modeling the kinetics of heterogeneous catalysis. Chem. Rev. 95:667–676, 1995.
Colicelli, J. Human RAS superfamily proteins and related GTPases. Sci. STKE 2004(250):RE13, 2004.
Coppens, M. O., A. T. Bell, and A. K. Chakraborty. Dynamic Monte-Carlo and mean-field study of the effect of strong adsorption sites on self-diffusion in zeolites. Chem. Eng. Sci. 54:3455–3463, 1999.
Costa, M. N., K. Radhakrishnan, and J. S. Edwards. Monte Carlo simulations of plasma membrane corral-induced EGFR clustering. J. Biotechnol. 151(3):261–270, 2009.
Costa, M. N., et al. Coupled stochastic spatial and non-spatial simulations of ErbB1 signaling pathways demonstrate the importance of spatial organization in signal transduction. PLoS ONE 4(7):e6316, 2009.
Erban, R., and S. J. Chapman. Stochastic modelling of reaction–diffusion processes: algorithms for bimolecular reactions. Phys. Biol. 6(4):046001, 2009.
Faeder, J., M. Blinov, and W. Hlavacek. Rules-based modeling of biochemical systems with BioNetGen. Methods Mol. Biol. 500:113–168, 2009.
Fallahi-Sichani, M., and J. J. Linderman. Lipid raft-mediated regulation of G-protein coupled receptor signaling by ligands which influence receptor dimerization: a computational study. PLoS ONE 4(8):e6604, 2009.
Friday, B. B., and A. A. Adjei. Advances in targeting the Ras/Raf/MEK/Erk mitogen-activated protein kinase cascade with MEK inhibitors for cancer therapy. Clin. Cancer Res. 14(2):342–346, 2008.
Fujioka, A., et al. Dynamics of the Ras/ERK MAPK cascade as monitored by fluorescent probes. J. Biol. Chem. 281(13):8917–8926, 2006.
Fuxe, K., and T. Kenakin. The changing world of G protein-coupled receptors. J. Recept. Signal Transduct. Res. 30(5):271, 2010.
Gibson, M. A., and J. Bruck. Efficient exact stochastic simulation of chemical systems with many species and many channels. J. Phys. Chem. 104:1876–1889, 2000.
Gillespie, D. T. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 81(25):2340–2361, 1977.
Gillespie, D. T. Stochastic simulation of chemical kinetics. Annu. Rev. Phys. Chem. 58:35–55, 2007.
Gilmer, G. Computer models of crystal growth. Science 208:355–363, 1980.
Govindan, R. A review of epidermal growth factor receptor/HER2 inhibitors in the treatment of patients with non-small-cell lung cancer. Clin. Lung Cancer 11(1):8–12, 2010.
Grima, R., and S. Schnell. Modelling reaction kinetics inside cells. Essays Biochem. 45:41–56, 2008.
Hartman, N. C., and J. T. Groves. Signaling clusters in the cell membrane. Curr. Opin. Cell Biol. 23(4):370–376, 2011.
Hatakeyama, M., et al. A computational model on the modulation of mitogen-activated protein kinase (MAPK) and Akt pathways in heregulin-induced ErbB signalling. Biochem. J. 373(Pt 2):451–463, 2003.
Hlavacek, W., et al. Rules for modeling signal-transduction systems. Sci. STKE 2006:re6, 2006.
Hornberg, J. J., et al. Control of MAPK signalling: from complexity to what really matters. Oncogene 24(36):5533–5542, 2005.
Hsieh, M. Y., et al. Stochastic simulations of ErbB homo and heterodimerisation: potential impacts of receptor conformational state and spatial segregation. IET Syst. Biol. 2(5):256–272, 2008.
Hsieh, M. Y., et al. Spatio-temporal modeling of signaling protein recruitment to EGFR. BMC Syst. Biol. 4:57, 2010.
Hynes, N. E., and G. MacDonald. ErbB receptors and signaling pathways in cancer. Curr. Opin. Cell Biol. 21(2):177–184, 2009.
Insel, P. A., et al. Impact of GPCRs in clinical medicine: monogenic diseases, genetic variants and drug targets. Biochim. Biophys. Acta 1768(4):994–1005, 2007.
Keating, E., A. Nohe, and N. O. Petersen. Studies of distribution, location and dynamic properties of EGFR on the cell surface measured by image correlation spectroscopy. Eur. Biophys. J. 37(4):469–481, 2008.
Kholodenko, B. N., J. F. Hancock, and W. Kolch. Signalling ballet in space and time. Nat. Rev. Mol. Cell Biol. 11(6):414–426, 2010.
Kholodenko, B. N., et al. Quantification of short term signaling by the epidermal growth factor receptor. J. Biol. Chem. 274(42):30169–30181, 1999.
Kitaura, J., et al. Evidence that IgE molecules mediate a spectrum of effects on mast cell survival and activation via aggregation of the FcepsilonRI. Proc. Natl. Acad. Sci. USA 100(22):12911–12916, 2003.
Kusumi, A., et al. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 34:351–378, 2005.
Lambert, N. A. GPCR dimers fall apart. Sci. Signal. 3(115):pe12, 2010.
Li, H., et al. Algorithms and software for stochastic simulation of biochemical reacting systems. Biotechnol. Prog. 24(1):56–61, 2008.
Lidke, D. S., and B. S. Wilson. Caught in the act: quantifying protein behaviour in living cells. Trends Cell Biol. 19(11):566–574, 2009.
Lillemeier, B. F., et al. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat. Immunol. 11(1):90–96, 2010.
Linderman, J. J. Modeling of G-protein-coupled receptor signaling pathways. J. Biol. Chem. 284(9):5427–5431, 2009.
Lo, H. W. Nuclear mode of the EGFR signaling network: biology, prognostic value, and therapeutic implications. Discov. Med. 10(50):44–51, 2010.
Low-Nam, S. T., et al. ErbB1 dimerization is promoted by domain co-confinement and stabilized by ligand binding. Nat. Struct. Mol. Biol. 18(11):1244–1249, 2011.
Mayawala, K., C. A. Gelmi, and J. S. Edwards. MAPK cascade possesses decoupled controllability of signal amplification and duration. Biophys. J. 87(5):L01–L02, 2004.
Mayawala, K., D. G. Vlachos, and J. S. Edwards. Heterogeneities in EGF receptor density at the cell surface can lead to concave up scatchard plot of EGF binding. FEBS Lett. 579(14):3043–3047, 2005.
Mayawala, K., D. G. Vlachos, and J. S. Edwards. Computational modeling reveals molecular details of epidermal growth factor binding. BMC Cell Biol. 6:41, 2005.
Mayawala, K., D. G. Vlachos, and J. S. Edwards. Spatial modeling of dimerization reaction dynamics in the plasma membrane: Monte Carlo vs. continuum differential equations. Biophys. Chem. 121(3):194–208, 2006.
Miura, Y., K. Hanada, and T. L. Jones. G(s) signaling is intact after disruption of lipid rafts. Biochemistry 40(50):15418–15423, 2001.
Nagy, P., et al. Lipid rafts and the local density of ErbB proteins influence the biological role of homo- and heteroassociations of ErbB2. J. Cell Sci. 115(Pt 22):4251–4262, 2002.
Orton, R. J., et al. Computational modelling of the receptor-tyrosine-kinase-activated MAPK pathway. Biochem. J. 392(Pt 2):249–261, 2005.
Pike, L. J. Lipid rafts: bringing order to chaos. J. Lipid Res. 44(4):655–667, 2003.
Plowman, S. J., et al. H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc. Natl. Acad. Sci. USA 102(43):15500–15505, 2005.
Radhakrishnan, K. Combustion kinetics and sensitivity analysis. In: Numerical Approaches to Combustion Modeling, edited by E. S. Oran, and J. P. Boris. Washington, DC: AIAA, 1991, pp. 83–128.
Radhakrishnan, K., et al. Sensitivity analysis predicts that the ERK–pMEK interaction regulates ERK nuclear translocation. IET Syst. Biol. 3(5):329–341, 2009.
Radhakrishnan, K., et al. Quantitative understanding of cell signaling: the importance of membrane organization. Curr. Opin. Biotechnol. 21(5):677–682, 2010.
Reddy, A. S., S. Chilukuri, and S. Raychaudhuri. The network of receptors characterize B cell receptor micro- and macroclustering in a Monte Carlo model. J. Phys. Chem. B 114(1):487–494, 2010.
Resat, H., L. Petzold, and M. F. Pettigrew. Kinetic modeling of biological systems. Methods Mol. Biol. 541:311–335, 2009.
Rosenbaum, D. M., S. G. Rasmussen, and B. K. Kobilka. The structure and function of G-protein-coupled receptors. Nature 459(7245):356–363, 2009.
Saffarian, S., et al. Oligomerization of the EGF receptor investigated by live cell fluorescence intensity distribution analysis. Biophys. J. 93(3):1021–1031, 2007.
Santamaria, F., et al. Quantifying the effects of elastic collisions and non-covalent binding on glutamate receptor trafficking in the post-synaptic density. PLoS Comput. Biol. 6(5):e1000780, 2010.
Sasagawa, S., et al. Prediction and validation of the distinct dynamics of transient and sustained ERK activation. Nat. Cell Biol. 7(4):365–373, 2005.
Schoeberl, B., et al. Computational modeling of the dynamics of the MAP kinase cascade activated by surface and internalized EGF receptors. Nat. Biotechnol. 20(4):370–375, 2002.
Shalom-Feuerstein, R., et al. K-ras nanoclustering is subverted by overexpression of the scaffold protein galectin-3. Cancer Res. 68(16):6608–6616, 2008.
Slepchenko, B. M., et al. Quantitative cell biology with the virtual cell. Trends Cell Biol. 13(11):570–576, 2003.
Suzuki, K., et al. Rapid hop diffusion of a G-protein-coupled receptor in the plasma membrane as revealed by single-molecule techniques. Biophys. J. 88(5):3659–3680, 2005.
Szabo, A., et al. Quantitative characterization of the large-scale association of ErbB1 and ErbB2 by flow cytometric homo-FRET measurements. Biophys. J. 95(4):2086–2096, 2008.
Telesco, S. E., and R. Radhakrishnan. Structural systems biology and multiscale signaling models. Ann. Biomed. Eng., 2012. doi:10.1007/s10439-012-0576-6
ten Klooster, J. P., and P. L. Hordijk. Targeting and localized signalling by small GTPases. Biol. Cell 99(1):1–12, 2007.
Tian, T., et al. Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat. Cell Biol. 9(8):905–914, 2007.
Tian, T., et al. Mathematical modeling of K-Ras nanocluster formation on the plasma membrane. Biophys. J. 99(2):534–543, 2010.
Tolle, D. P., and N. Le Novere. Meredys, a multi-compartment reaction–diffusion simulator using multistate realistic molecular complexes. BMC Syst. Biol. 4:24, 2010.
Tolle, D. P., and N. Le Novere. Brownian diffusion of AMPA receptors is sufficient to explain fast onset of LTP. BMC Syst. Biol. 4:25, 2010.
Turner, T. E., S. Schnell, and K. Burrage. Stochastic approaches for modelling in vivo reactions. Comput. Biol. Chem. 28(3):165–178, 2004.
Vigil, D., et al. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat. Rev. Cancer 10(12):842–857, 2010.
Vilardaga, J. P., et al. G-protein-coupled receptor heteromer dynamics. J. Cell Sci. 123(Pt 24):4215–4220, 2010.
Waller, A., et al. Receptor binding kinetics and cellular responses of six N-formyl peptide agonists in human neutrophils. Biochemistry 43(25):8204–8216, 2004.
Wells, N. P., et al. Time-resolved three-dimensional molecular tracking in live cells. Nano Lett. 10(11):4732–4737, 2010.
Wennerberg, K., K. L. Rossman, and C. J. Der. The Ras superfamily at a glance. J. Cell Sci. 118(Pt 5):843–846, 2005.
Wiley, H. S., S. Y. Shvartsman, and D. A. Lauffenburger. Computational modeling of the EGF-receptor system: a paradigm for systems biology. Trends Cell Biol. 13(1):43–50, 2003.
Wilson, B. S., J. M. Oliver, and D. S. Lidke. Spatio-temporal signaling in mast cells. Adv. Exp. Med. Biol. 716:91–106, 2010.
Wilson, B. S., et al. Exploring membrane domains using native membrane sheets and transmission electron microscopy. Methods Mol. Biol. 398:245–261, 2007.
Yang, S., et al. Mapping ErbB receptors on breast cancer cell membranes during signal transduction. J. Cell Sci. 120(Pt 16):2763–2773, 2007.
Zhdanov, V. P., and B. Kasemo. Kinetic phase transitions in simple reactions on solid surfaces. Surf. Sci. Rep. 20:111–189, 1994.
Acknowledgments
This work was supported by NIH R01CA119323 (to BW), NIH P50GM085273 (the New Mexico Spatiotemporal Modeling Center), and NIH K25CA131558 (to AH).
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor Michael R. King oversaw the review of this article.
Rights and permissions
About this article
Cite this article
Radhakrishnan, K., Halász, Á., McCabe, M.M. et al. Mathematical Simulation of Membrane Protein Clustering for Efficient Signal Transduction. Ann Biomed Eng 40, 2307–2318 (2012). https://doi.org/10.1007/s10439-012-0599-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10439-012-0599-z