Abstract
We developed a cryo-imaging system, which alternates between sectioning (10–40 μm) and imaging bright field and fluorescence block-face image volumes with micron-scale-resolution. For applications requiring single-cell detection of fluorescently labeled cells anywhere in a mouse, we are developing software for reduction of out-of-plane fluorescence. In mouse experiments, we imaged GFP-labeled cancer and stem cells, and cell-sized fluorescent microspheres. To remove out-of-plane fluorescence, we used a simplified model of light-tissue interaction whereby the next-image was scaled, blurred, and subtracted from the current image. We estimated scaling and blurring parameters by minimizing an objective function on subtracted images. Tissue-specific attenuation parameters [μ T: heart (267 ± 47.6 cm−1), liver (218 ± 27.1 cm−1), brain (161 ± 27.4 cm−1)] were found to be within the range of estimates in the literature. “Next-image” processing removed out-of-plane fluorescence equally well across multiple tissues (brain, kidney, liver, etc.), and analysis of 200 microsphere images gave 97 ± 2% reduction of out-of-plane fluorescence. Next-image processing greatly improved axial-resolution, enabled high quality 3D volume renderings, and improved automated enumeration of single cells by up to 24%. The method has been used to identify metastatic cancer sites, determine homing of stem cells to injury sites, and show microsphere distribution correlated with blood flow patterns.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Cryo-imaging2,8,15,19,26,29 provides single cell resolution and sensitivity over an entire specimen, which is not possible with in vivo, small animal imaging systems such as CT, MRI, PET, SPECT, intra-vital imaging or bioluminescence. Cryo-imaging consists of a modified, bright-field/fluorescence microscope; a robotic imaging system positioner; a customized, whole mouse motorized cryomicrotome; control system; and analysis/visualization software. By alternately sectioning and imaging the specimen, the system acquires brightfield color and fluorescent image volumes, providing micron-scale, detailed views of anatomy and cells labeled with fluorescent reporter genes or exogenous fluorophores. The other whole-mouse modalities named above do not offer the combination of resolution and contrast mechanisms found in cryo-imaging. Optical imaging modalities such as intra-vital imaging6 do not offer the field of view (FOV) or depth of field of cryo-imaging. Image quality with traditional optical microscopy techniques (i.e. confocal microscopy) degrade with depth due to the accumulating effect of absorbance and scatter of light by tissue and are limited to imaging structures near the tissue surface (≈100 μm).5,13,30 Unlike other optical imaging modalities, cryo-imaging does not suffer from image degradation at depth, but rather only images structures at the surface by physically sectioning the tissue. This allows cryo-imaging to image an entire mouse with single cell sensitivity and resolution over the entire volume.
By imaging with high resolution and sensitivity, it is possible to identify fluorescently labeled single cells or cell clusters within a mouse. Once cells are identified, cell locations can be mapped relative to the tissue anatomy in the high contrast, 3D cryo-image color volumes. The combination of single cell detection and high resolution anatomical data would benefit many potential applications across many areas of biomedical research, including cancer biology/therapy, stem cell biology/therapy, mouse phenotyping, imaging agent development, drug delivery systems, etc.
Out of plane fluorescence presents a challenge to single cell detection in cryo-imaging. Typically, in epifluorescence imaging, an objective lens with a high numerical aperture lens is used, which in turn has a small focal depth, limiting the out-of-plane fluorescence.27 With our cryo-imaging system, we require a long working distance (≈70 mm) so as to limit the exposure of the objective lens to the cold temperatures (≈−25 °C) deep in the cryostat chamber and to guard against possible debris from cutting. We also require a large field of view to minimize the number of images needed to image the entire specimen block-face. To satisfy field of view and depth of focus constraints, we often use a 0.11 NA objective lens giving a 477 μm depth of focus and leading to inclusion of out-of-plane fluorescence. Out-of-plane fluorescence gives a subsurface haze in 2D images and an elongation artifact along the z-axis that appears as a “comet tail” in 3D. The “comet tail” artifact can connect separate structures, leading to quantification errors, particularly when counting fluorescently labeled cells, such as stem cells.
In conventional microscopy, blurring by the optical system often exceeds that due to scattering in tissue, and this is exploited in 3D computational optical sectioning, where images are acquired at different focal planes throughout the sample. Deconvolution algorithms accounting for the microscope optical 3D PSF create high quality 3D image stacks.24 The earliest algorithms for computational optical sectioning were nearest-neighbor algorithms which did not require measurement of the PSF. Typically, images acquired above and below are digitally blurred and subtracted from the image of interest to remove out of plane blurring. Such nearest neighbor algorithms motivate our “next-image” algorithm for the removal of subsurface scatter, with one important difference. Since cryo-images are collected using a stereo microscope with a large depth of focus (≈477 μm), the optical PSF does not noticeably blur features over the range at which out-of-plane fluorescence is visible (≈120 μm). Blurring in cryo-images is primarily due to scatter within the tissue.
In this paper, we develop and evaluate “next-image” processing for reducing out-of-plane fluorescence in cryo-imaging. In experiments, we image homogeneous phantoms and whole mice following intra-cardiac injection of fluorescently labeled cells or microspheres. Because light scattering and absorption can vary between tissues, a key feature of our experiments is to determine potential dependence of next-image parameters on tissue type. In sections that follow, we develop the algorithm, describe our fluorescence correction software, parameter estimation software, process cryo-image volumes, compare 3D reconstructions of original and processed data, and quantify the ability to count cells with and without processing.
Next-Image Algorithm for Reduction of Out-of-Plane Fluorescence
Theory
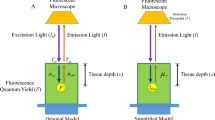
In the formation of a block-face image, photons from the light source, I 0, reach the specimen and a percentage is transmitted into the tissue, T at, dependent upon the block-face index of refraction. The photons that enter the specimen are absorbed (μ A,ex) and scattered \( (\mu _{{{\text{S,ex}}}}^{\prime} = (1 - g)\mu _{{{\text{S,ex}}}} ) \) according to Beer’s law, dependent upon the attenuation coefficient of the specimen, \( \mu _{{{\text{Ex}}}} = \mu _{{{\text{S,ex}}}}^{\prime} + \mu _{{{\text{A,ex}}}} \) (events/cm).1 A percentage of photons that reach fluorophores embedded in the specimen are absorbed by the fluorophore and result in the fluorescent emission of photons, which is dependent upon the quantum efficiency of the fluorophore. Photons are emitted from the fluorophore in all directions at a different wavelength from the excitation photons. The emitted photons that travel back toward the imaging system, F i , are scattered \( (\mu _{{{\text{S,em}}}}^{\prime} = (1 - g)\mu _{{{\text{S,em}}}} ) \) and absorbed (μ A,em), \( \mu _{{{\text{Em}}}} = \mu _{{{\text{S,em}}}}^{\prime} + \mu _{{{\text{A,em}}}} \) (events/cm), and a percentage are transmitted at the tissue–air interface, T ta, and reach the detector. This simplification is used to describe the fluorescent intensity in cryo-images at a given distance in tissue as shown in Eq. (1).
Equations describing a series of cryo-images are given below where increasing i indicates subsequent tissue slicing down through the block, F i is the in-plane fluorescence and F k, k>i is out-of-plane fluorescence in cryo-image i and in-plane fluorescence in image k.
where μT = μEm + μEx.
To perform next-image processing, the next cryo-image is attenuated and subtracted from the current image. Substitution from above gives
where the right hand side is a measurement we desire free of subsurface fluorescence. This calculation was insufficient to remove out-of-plane fluorescence as it did not take into account scattering. Reinisch et al.21 discussed that in a highly scattering media, such as tissue, and at a similar scale, scatter can be modeled by a Gaussian. We modify Eq. (3) to include Gaussian blurring with kernel h, giving the equation below.
Assuming constant slice thickness and μ T, we can simplify to give our working equation where k = exp(−μ T s).
We understand that there are inherent limitations and assumptions built into the above equations, which will be addressed later.
Estimation of Parameters
We developed a computer program to estimate next-image parameters, k and σ. We assume small fluorescent structures such as fluorescent microspheres or cells. The user manually selects a region of interest (ROI) containing a single fluorescent structure in a given tissue. The program then automatically propagates the user specified ROI to include each 2D cryo-image in which the fluorescent object is >C times the background signal. The user defined C should give the best tradeoff between maximizing object detection and minimizing falsely classifying background.
Our software automatically estimates next-image parameters, k and σ. For an image region containing out-of-plane fluorescence, the “next” image region is blurred and scaled by k and σ, respectively, and subtracted from the “current” image region. The Levenberg–Marquardt algorithm is used to minimize an objective function, F obj, subject to constraints, 0 < k < 1 and 0 < σ < 100 μm. The objective function, F obj, is given by
where R s(i, k) is the subtracted image region, μ Rs is the mean value of the subtracted image region, and Σ provides for the summation of intensity values across the region. The objective function is minimized when pixel intensities in the ROI are reduced to the approximate value of the mean, which is a good estimate of the subtracted background. To help justify Eq. (6), we note its similarity to the variance calculated over the subtracted image region. It will penalize any deviations from a constant mean value. In preliminary experiments, we found Eq. (6) superior to variance. Optimization is stopped when the objective function changes less than a tolerance value.
For each tissue of interest, parameters were determined from many fluorescent cells or microspheres; we used at least 20 microspheres or cells, and 2–4 images per microsphere or cell, giving a minimum of 60 2D ROIs upon which parameters were estimated. We found it necessary to reject some (≈5%) of parameter sets, {k, σ}, as outliers. This occurred whenever there was a very low signal to noise ratio in the subtracted image, typically leading to inaccurate estimation of σ. Rather than using parameter estimates, we rejected outliers if the mean of the absolute value of the intensity in the subtracted image region was not reduced to within 1.5 times the background signal of the original image. Outliers were discarded and the mean of remaining values was used as the k and σ value for the corresponding tissue type.
Removal of Out-of-Plane Fluorescence
Given next-image parameters, k and σ, we now describe how to remove subsurface fluorescence in cryo-images. We start with bright field and fluorescence cryo-images, which have been carefully aligned along z (Fig. 2b). Using the bright field images containing the tissue of interest, the user segments either a volume of interest in the tissue or the entire tissue. Within this segmented region, previously determined k and σ values are used with Eq. (5) to remove out-of-plane fluorescence from all cryo-images in the volume. Pseudocode for the next-image processing algorithm implemented in our parameter estimation and fluorescence removal software has been provided in Table 1.
Experimental Methods
Instrument
The cryo-imaging system (Fig. 1) consists of a modified large section cryo-microtome (Model 8250, Vibratome Inc, St. Louis, MO), XYZ robotic positioner carrying an imaging system which consists of a stereo microscope (SZX12, Olympus, Japan), GFP fluorescence filters (Exciter: HQ470/40x, Dichroic: Q495LP, Emitter: HQ500LP, Chroma, Rockingham, VT), low light digital camera (Retiga Exi, QImaging Inc., Canada), brightfield light source, and epi-illumination fluorescent light source (XCite 120PC, EXFO, Canada). The cryo-imaging system is controlled by a control computer running Labview (National Instruments, Austin, TX). The stereo microscope uses multiple objectives (0.036 NA and 0.11 NA) and zoom settings (7–90×), and the FOV can be varied to cover an entire mouse or down to a small organ with a maximum in-plane resolution of 3.1 μm. To enable very high resolution imaging over a large FOV, the XYZ robotic positioner moves the imaging system over the entire specimen, creating a micron-scale tiled image acquisition. Once collected, images were aligned using Amira (Mercury Computer Systems Inc., Chelmsford, MA) to correct for small (micron-scale) misalignments and corrected by hand where necessary. Our fluorescence correction and parameter estimation software was written in Matlab (Mathworks, Natick, MA). All processing and alignment was performed on a PC running Windows XP with a 3 GHZ Intel Pentium 4 processor and 2 GB memory.
Animal Protocol
Animals are covered under various IACUC-approved projects. Prior to being euthanized mice were anesthetized with pentobarbital through a tail vein injection. The mice were then euthanized in a method approved by Case Animal Resource Center (ARC), which consists of euthanizing the animal with pentobarbital. Following sacrifice the mouse was covered with the embedding compound optimal cutting temperature compound (OCT) (Tissue Tek, Ted Pella, Inc., Redding, CA), ensuring that the carcass is completely covered and no air bubbles will be formed in the next step of embedding. The mouse was then embedded in OCT inside an aluminum foil mould. The entire mould is snap-frozen in liquid nitrogen for twenty minutes to reduce ice crystal formation. Following this the mould assembly was removed from the liquid nitrogen bath and placed inside the cryomicrotome chamber to equalize the specimen temperature to that of the cryomicrotome. After 3 h the mould was removed and the frozen specimen mounted on the microtome stage and the slice thickness was set to 40 μm. Mouse specimens were prepared as described and imaged at 7× magnification, with a pixel-size of 15.6 μm and a focal depth of 477 μm, unless otherwise specified.
Animal Experiments
To investigate processing in a controlled fashion with an almost constant bright fluorescence signal, we conducted experiments using fluorescent microspheres. Prior to sacrifice, a mouse was anesthetized with pentobarbital and a solution of saline and 15 μm green fluorescent microspheres (FluoSpheres, Invitrogen, Carlsbad, CA) was injected into the left ventricle of the heart, and time was given for the microspheres to circulate prior to sacrifice. Data sheets on the microspheres indicate that size variation is 15.4 ± 1.6 μm and fluorescence was found to vary by <4% between beads. In addition, to test alignment accuracy, graphite fiducials were inserted into the OCT surrounding the mouse.
In a parabiosis experiment, a GFP mouse was surgically joined with a wild type littermate at 4 weeks of age; 4 weeks later the tibia of the negative mouse was fractured; and four more weeks later the conjoined mice were sacrificed. Imaging was at 40× magnification, giving a pixel size of 2.9 μm.
Approximately 5 million fluorescently labeled murine Lewis lung carcinoma (LLC) cells were injected as a metastatic cancer model via tail vein injection into an adult mouse. The cells were allowed to deposit and 7 days post injection the animal was sacrificed and cryo-imaged at 25× magnification, with a 0.11 NA objective, and an image resolution of 6.21 μm.
In all experiments, the excitation peak of GFP is at 475 nm and the emission peak is at 509 nm.
Phantom Experiments
An imaging phantom was created by embedding 15 μm diameter green fluorescent microspheres (FluoSpheres, Invitrogen, Carlsbad, CA) within OCT. The phantom was then snap frozen in liquid nitrogen, mounted to a stage inside the imaging cryo-microtome and imaged. Slice thickness was set at 5 μm and image magnification at 27×, with pixel size of 4.0 μm.
To test the stability of our fluorescence imaging system, we imaged a fluorescent microsphere test slide (FocalCheck, Invitrogen, Carlsbad, CA) over the course of an hour at 25× magnification.
Results
We performed several tests to ensure our system met necessary engineering requirements for removal of out-of-plane fluorescence. First, the fluorescence imaging system was tested for fluctuations in steady state imaging conditions, by imaging a fluorescent test slide consisting of stably fluorescent microspheres. Fluorescent image intensity was measured over a 1-h period and found to be very consistent as the average intensity across a single microsphere varied by ≈1%. The small variations in image intensity affected the entire image, and therefore is most likely due to fluctuations in light source intensity. Fluctuations appeared random when plotted as a function of time. Second, to ensure that fluorescent intensity was directly proportional to fluorescent concentration, the relationship between fluorescent emission and excitation was tested on a fluorescent test slide and found to be linearly related. The stably fluorescent microsphere test slide was imaged at varying levels of excitation light intensity by adjusting the light source intensity. A decrease of 1/4, 1/2, and 3/4 in light source intensity was found to correspond to a decrease by approximately 1/4, 1/2, and 3/4 respectively of the measured fluorescent intensity. Third, the accuracy of image registration was verified to be accurate to less than one pixel. To test registration accuracy, cryo-images of a mouse (Fig. 2a) with a graphite fiducial embedded next to the mouse were aligned using only image information from the mouse. The center point of the fiducial was determined in each aligned image and a line was fit to the center point with respect to sectioning depth. The fiducial was found to be accurately reconstructed as a straight line with a Euclidian distance error less than one pixel (Fig. 2b).
Example cryo-image and evaluation of image alignment. A single 2D cryo-image of an entire mouse is shown (a). Different tissues are clearly distinguishable in the brightfield color image (a). A graphite fiducial was embedded in OCT alongside a mouse and cryo-imaged. Cryo-images were aligned with the graphite fiducial masked out of the images so it did not influence the alignment. The center of mass of the fiducial was determined in each cryo-image and fit to a straight line. The euclidian distance errors from a straight line were <1. The graphite fiducial was surface rendered and overlaid on a 2D brightfield cryo-image of the mouse
The peak intensity of multiple microspheres in the imaging phantom was plotted as a function of depth from the surface to evaluate the exponential attenuation model (Fig. 3). Because the fluorescence intensity across multiple microspheres varies by <4% and is very stable with time, we lump measurements from multiple microspheres without normalization. In a cryo-image volume of 130 images, the peak intensity, mean value, and standard deviation of 5 microsphere over 17 images was recorded along with the depth of the microsphere from the surface. The microsphere depth was determined as the z distance from the current image to the last image in which the microsphere was visible. Data were well fit to an exponential fit (f(x) = exp(−μ*x) + offset, where x is the depth from the surface in microns, offset is the background, and f(x) is the intensity) (Fig. 3). The correlation coefficient between the data and the fitted exponential with μT = 187 cm−1 was 0.9973. Not only does the quality of the fit support the model, it indicates a consistent slice thickness and μT.
Exponential fit of intensity as a function of depth from the surface. Microspheres were embedded in OCT and imaged with a slice thickness of 5 μm. Five microspheres were used over 17 images without normalization to determine the mean value, which is shown, along with the standard deviation of the intensity at each depth from the surface. The standard deviation was found to be between .87 and 6.48% of the mean value at each distance location
Prior to next-image processing, fluorescent microspheres appeared as “comet tails” when viewed from the “side” in a 3D view (Fig. 4a). Subsurface fluorescence is seen at distances >100 μm. Out-of-plane fluorescence is more apparent in the imaging phantom than in tissue due to its relatively low absorption and scattering and its lack of autofluorescence background. As a result, the phantom is more difficult to successfully process, and it presents good data for evaluation and testing. Next-image processing was performed using μT = 194 cm−1 and σ = 7.2 μm as determined using the parameter estimation software. The estimated value of μT is very similar to the coefficient (μT = 187 cm−1) of the exponential fit in Fig. 3. Analysis of a single microsphere visible in two consecutive images was processed (Figs. 4c and 4d) with (Fig. 4f) and without (Fig. 4e) blurring. With blurring (σ = 7.2 μm), out-of-plane fluorescence was completely removed. Without blurring, an outer, halo ring of subsurface fluorescence was present. In the next-image processed data 97 ± 2% of out-of-plane fluorescent intensity was removed (Fig. 4b), quantified as follows. In a ROI, pixels above a threshold value, T, were labeled as out-of-plane fluorescence. The threshold value T was set equal to the sum of the mean value and standard deviation of the background (T = μBKG + σBKG). Visual inspection was performed to assure that no background pixels were included. The same ROI was analyzed following next-image processing, and the sum of the intensity of those pixels greater than T was calculated. This was a more conservative calculation than obtaining average intensity values with and without processing. The small amount of remaining out-of-plane fluorescence was barely visible and located along a few pixels in the same xy location (Fig. 4b). Post processing the microspheres appeared as objects with a 16 μm diameter, within the range supplied by the manufacturer of 15.4 ± 1.6 μm.
Removal of out-of-plane fluorescence from cryo-images of fluorescent microspheres. 2D maximum intensity projection (MIP) side view of a cryo-image stack of fluorescent microspheres embedded in OCT before (a) and after next image processing (b). In (b) a microsphere is visible that was obscured by out-of-plane fluorescence in the original cryo-image. From the same data set two consecutive images (c, d) were processed with σ = 0 μm resulting in a “halo effect” around the microsphere (e, f) that is removed when σ = 7.2 μm was used (g). The image processed without a blurring function (f) has a max value of 15 and a mean value of ≈4, while the image processed with a blurring function (g) has a max value of 3 and a mean value of ≈1.5. Grayscale windowing is identical in (a) and (b), (c), (d) and (e), and (f) and (g). Grayscale windowing was adjusted for different figure sets dependent upon intensity and features of interest
A next-image parameter library was created from the whole-mouse microsphere data (Fig. 5a). Microspheres were found in almost all organs (Figs. 5b and 6) except the lungs because microspheres injected on the arterial side could not pass through tissue capillaries before entering the lungs. Microspheres concentrated in the brain, kidney, liver, and areas containing large skeletal muscle. These areas receive a large percentage of the blood flow in the mouse, 14%, 22%, 27%, and 15% respectively.11 After processing, individual microspheres are visible, often with multiple microspheres, ≈5–13, along a microvessel. Groups of microspheres are especially visible in the brain (Figs. 6c and 6e). By rotating the volume rendering it is possible to view the individual microspheres (Fig. 6f) that appear as a line segment from a different angle (Fig. 6e). The concentration of microspheres in other organs, such as the kidney (Figs. 5b and 5c), was lower and aggregates of microspheres were smaller. The kidney was found to contain 5295 connected components, approximately half as many as the brain. The concentration of microspheres in tissues is commonly used to measure regional blood flow3 and the difference in microspheres implies that blood flow to the brain is approximately twice the blood flow to a single kidney, closely matching reported values of brain blood flow being 1.6 times the blood flow to a kidney.11 Regional differences in blood flow can be seen in the kidney (Figs. 5b and 5c), where large numbers of microspheres were trapped by the network of capillaries in the nephrons of the medulla and outer cortex.
3D whole mouse volume rendering of fluorescent microspheres. Microspheres were too small to see when viewing the entire mouse and are instead shown as larger white spheres of equal size (a). Some spheres appear dimmer or larger due to transmission loss caused by the brightfield volume rendering and perspective effects (a). A white sphere was placed to show a location in which multiple microspheres were present in a cube 100 μm in length. A 3D surface rendering of the kidney and associated vasculature are shown (c). A 2D brightfield cryo-image of the kidney is shown overlaid with the corresponding next-image processed fluorescent image (b)
Volume rendering of brightfield and fluorescent cryo-images of fluorescent microspheres in a mouse brain. A surface rendering of the segmented brain is shown along with a brightfield volume rendering of the mouse (a). A slab of the fluorescent volume (100 images) was volume rendered and displayed on top of a brightfield image (b, c). Renderings of the microspheres from the original data (b) appear brighter than an identical rendering of the next-image processed data (c) due to the presence of out-of-plane fluorescence. Next-image processing was performed with μT = 161 cm−1 and σ = 11.2 μm. The distortion due to out-of-plane fluorescence is apparent in a zoomed in region of the brain showing only the volume rendered beads (d–f). Streaks due to out-of-plane fluorescence are visible in the unprocessed data (d) that are clearly removed in the processed data (e, f). Because the microspheres were trapped in the microvasculature of the brain multiple microspheres often deposited next to one another and appear as a line segment outlining the vessel (e). By rotating the volume rendering it is possible to view the individual microspheres (f) that appear as a line segment from a different angle (e)
In all tissues of interest, microspheres were present for the determination of next-image parameters. Typically, microspheres were visible for 80–160 μm in depth. The parameter library is given in Table 2. As described in “Discussion”, mean μT values fell within ranges of reported μT values obtained with much different methods. Reported μT values were typically spread over a large range due to different techniques used to determine tissue properties, different wavelengths of light used, different tissue preparation methods, and tissue harvested from different species.
We analyzed next-image parameters for dependence upon depth and tissue type (Fig. 7). Although no consistent trend is seen with depth, data values from different tissues show a clear separation. A t-test was performed on the measured μT and σ values independently between each tissue with α = .05. With respect to μT all tissues except the heart and fat tissue; liver, muscle and kidney; and muscle, kidney, and the adrenal gland were found to be statistically different. For all tissues that were not statistically different with respect to μT, the σ values were found to be statistically different.
3D plot of measured attenuation coefficients, σ values, and microsphere depth for various tissues. Values for σ and μT were determined using our fluorescence correction software in various tissues (see figure legends). These parameters were determined using images of beads at different distances from the surface. For both σ and μT there was no discernable relationship between bead depth and the measured value
To further analyze the significance of tissue type on next-image parameters, we processed images from different tissues using correct parameters (Figs. 8b, 8e, and 8h) and those from a different tissue (Figs. 8c, 8f, and 8i). Images processed with incorrect parameters showed artifacts. If k was too large then negative values were present in the processed images (Figs. 8c and 8f). If k was too small then out-of-plane fluorescence was incompletely removed (Fig. 8i). Upon visual inspection it was found that images processed with correct parameters were artifact free.
Comparison of processed cryo-images using different parameter sets. Unprocessed 2D cryo images from the heart, fat, and brain (a, d, g) were processed using parameters specific to the tissue (b, e, h) and parameters from different tissue (c, f, i). The tissue type is shown in the original images (a, d, g) in the upper right corner. Processed images (b, c, e, f, h, i) are marked in the lower right corner with the parameters used in next-image processing. Grayscale windowing is the same across each row. Images of the heart (a–c) and fat (d–f) appear brighter than images of the brain (g–i). This is because images of the heart (c) and brain (f) processed using incorrect parameters contained negative values and correspondingly shifted the grayscale windowing
Next-image parameters successfully removed out-of-plane fluorescence and improved microsphere and cellular quantification (Figs. 6, 9, and 10). Prior to processing, individual cancer cells in the adrenal gland (Fig. 9a) and microspheres in the brain (Fig. 6a) were difficult to distinguish and quantify due to the “comet trail” of out-of-plane fluorescence which tended to “join” individual cells. Next-image processing was performed in a region of interest for both tissues using μT = 161 and 206 cm−1 and σ = 11.2 and 5.8 μm for the brain and adrenal gland respectively (Figs. 6, 9c, and 10b). Next-image parameters in the brain and adrenal gland were determined using our parameter estimation software. Fluorescent structures were determined to be single microspheres through visual analysis and integrated intensity. The integrated intensity of single microspheres was found to vary by <34%. The variation is due to changes in attenuation depending upon the depth of the microsphere from the surface. A 15 μm diameter microsphere with slice thickness of 40 μm can vary in depth by up to 25 μm, corresponding to a change in attenuation of 33.0% (assuming mu = 160 cm−1). Connected component analysis (CCA) of unprocessed cryo-images found 2835 and 8856 clusters in the adrenal gland and brain respectively. The same analysis of next-image processed data found 3739 and 10317 clusters in the adrenal gland and brain respectively. The increase in the number of clusters of approximately 24 and 14% in the adrenal gland and brain respectively, demonstrates that cells and microspheres were incorrectly joined into single clusters in the original data.
Consecutive cryo-image of GFP positive LLC cells in the adrenal gland (a, b). Two consecutive cryo-images are shown, with image (b) taken after a single 40 μm section was removed from the block face shown in image (a). Significant out-of-plane fluorescence is visible at the arrows in (a). Results of next image processing of (a) is shown in (c). Out-of-plane fluorescence was removed and a few in-plane individual cells remain. Cells vary in size and intensity depending on GFP expression and cell size
Comparison of unprocessed and processed cryo-image volumes of cancer cells in the adrenal gland. Approximately 5 million cancer cells were injected into a mouse and brightfield and fluorescent cryo-images were collected. The adrenal gland was manually segmented from the brightfield cryo-images and surface rendered in purple. Vasculature was also manually segmented from the brightfield cryo-images and surface rendered and is shown rendered in red. The 3D renderings of the adrenal gland and vasculature are sitting atop the corresponding cryo-imaging bright-field image slice of the adrenal gland and surrounding tissue. GFP positive LLC cells were segmented through connected component analysis (CCA) and surface rendered (green) in both the original (a) and next-image processed data (b). Figure (b) is a volume rendering of the unprocessed fluorescence data. Figure (a) is the same figure using “next image” processed fluorescence data
Next-image processing successfully removed out-of-plane fluorescence, greatly improving the fluorescence contrast of the in-plane GFP-positive cells, while reducing autofluorescence from the bone and surrounding tissues (Figs. 11 and 12). Bone is highly autofluorescent, due primarily to the presence of collagen.20 The femur of a wild type mouse that shared circulation with a surgically joined GFP positive transgenic littermate was imaged in fluorescence and brightfield. Fluorescent stem cells, 12–20 μm, were visible across multiple images, along with deeper autofluorescent bone (Figs. 11a and 11b). Next-image processing was performed with parameters, μT = 214 cm−1 and σ = 9 μm, from the mouse perfused with microspheres.
Next-image processing of cryo-images containing individual stem cells. A cryo-image of fluorescently labeled cells at a fracture site in a mouse before (a) and after a single 40 μm section was taken (b). Images were taken of a parabiosis mouse model in which a GFP positive transgenic mouse and a GFP negative littermate had surgically joined circulatory system. Performing next image processing with μ = 214 cm−1 and σ = 9 μm on (a) out-of-plane fluorescence was removed (c). Out-of-plane fluorescence was successfully removed leaving in-plane fluorescence from two surface GFP positive cells
Increased contrast in next-image processed cryo-images of GFP positive skeletal muscle. Brightfield and fluorescent cryo-images were collected of the GFP positive littermate in the parabiosis experiment. A brightfield cryo-image (a) of the GFP positive mouse skeletal is shown along with the corresponding fluorescent image (b). Out-of-plane fluorescence is visible along the periphery of the muscle (*) and inside of the vasculature (arrow). Processing removed out-of-plane fluorescent (c) resulting in significant improvement in image contrast as well as removing the halo effect (*) and out-of-plane fluorescence visible inside of the vasculature (arrow)
Next-image processing removed out-of-plane fluorescence in large textured structures, improving contrast and resolution in GFP positive mouse skeletal muscle. Following next-image processing the orientation of individual muscle fibers (Figs. 12c and 12d) which were blurred in the original cryo-images due to out-of-plane fluorescence (Fig. 12b), were visible. In the original cryo-images out-of-plane fluorescence is visible along the periphery of the muscle and the inside of muscle vasculature. Processing with parameters taken from the library determined for skeletal muscle, μT = 214 cm−1 and σ = 9.0 μm, removed out-of-plane fluorescent resulting in significant improvement in image contrast, as well as removing a “halo” surrounding the muscle (Fig. 12c). In the processed images, muscle fibers are more apparent due to increased contrast caused by the removal of out-of-plane fluorescence. A magnified view shows single muscle fibers and fiber orientation with diameter ≈20 μm (Fig. 12d). However, not all muscle fibers were distinguishable from their neighbors. This is most likely due to the relatively large slice thickness of 40 μm in comparison to the average muscle fiber diameter range of 10–80 μm.11
Discussion
Next-image processing removed out-of-plane fluorescence, improving resolution, contrast, 3D visualization, and quantification of single cells and cell clusters. Using our parameter estimation and fluorescence correction software, we successfully determined tissue specific processing parameters and removed subsurface haze (Figs. 6, 8, 10, 11, and 12). 3D reconstructions of processed cryo-images show that the “comet tail” artifact had been removed in imaging phantoms (Fig. 4) and in whole organs (Figs. 6 and 10). Comet tails caused separate microspheres to appear as one larger object (Fig. 4). Processing removes overlaps, improving the quantification of fluorescent objects (Figs. 6 and 10). Analysis of 2D cryo-images (Figs. 8, 9, 11, and 12) showed that out-of-plane fluorescence had been significantly reduced, increasing contrast sufficiently in 2D images of the skeletal muscle of a GFP mouse so that the orientation of muscle fibers was visible (Fig. 12). Next-image processing of cryo-images of fluorescent cells in the parabiosis experiment removed large autofluorescent background haze caused by the bone, further improving single cell contrast (Fig. 11).
Optical properties of tissue depend upon wavelength. The EGFP (enhanced green fluorescent protein) labeled cells and microspheres have the same excitation and emission spectra, allowing us to use parameters from microsphere experiments on cell data. We recognize that experiments utilizing fluorophores with different excitation and/or emission wavelengths may have different optical properties, requiring a different set of next-image parameters for a given tissue. Similarly, a different slice thickness will change the amount of attenuation and scatter between images. While using Beer’s law one can estimate the change in k for a given slice thickness, the precise relationship between blur (σ) and slice thickness is not simply determined. Therefore a change in slice thickness will necessitate the estimation of σ for the given slice thickness.
Next-image processing is necessary for 3D quantitative analysis of fluorescently labeled cells and microspheres. Next-image processing correctly separated microspheres (Figs. 4a and 4b) incorrectly joined due to out-of-plane fluorescence in unprocessed data. Following next-image processing, single cell identification was possible in the adrenal gland (Figs. 9c and 10b) and single microsphere identification was possible anywhere in a mouse (Figs. 6d and 6e). CCA was used to perform cell counting in the adrenal gland before and after processing, with 24% more cell clusters found in the processed data set. Similarly, CCA was used to count the number of microspheres in the brain before (8856 clusters) and after processing (10317 clusters), with an increase of 14% in the number of clusters found after processing.
Equations developed for next-image processing (Eqs. 1, 2, 5) simplify light-tissue interaction. Our model assumes that scatter, absorption, and the index of refraction are homogenous throughout an organ. Attenuation and scatter are simply modeled by exponential attenuation and Gaussian blurring respectively. Despite the assumptions and simplifications next-image processing works quite well. The affect of many of the assumptions was minimal, i.e. variations in μT within an organ would result in a change to k within the background noise level. Our model of scatter through Gaussian convolution, was validated through experimental evidence (Figs. 4c–4g), which was consistent with published reports in the literature.23 There are other approaches for modeling image formation, such as the diffusion equation in radiative transfer7,12,14,22,28 or evaluating solutions using Monte Carlo simulations9,10,16,22,28 that more exactly model the interaction of light in tissue. Such methods would not be amenable for a fast image processing approach for hundreds of GB of data and tens of thousands of images. By using an exponential model of attenuation we effectively limited processing time to the time taken to load and save the images. As a result, we opted for the simplified model.
Instrumental methods are available to reject out-of-plane fluorescence. In a confocal scanning microscope, out-of-focus light is strongly attenuated by the confocal detector pinhole.17 Another method is the use of structured light, which projects a single spatial-frequency grid pattern onto the specimen.17,18 Another less common method is to use polarized light, where light from the surface will retain polarization while light from a depth undergoes scattering events and looses polarization.25 Yet another possibility is computational optical sectioning in which a stack of focal sections is recorded and the contribution of out-of-focus signal to a given section from structures in other sections is computationally removed.4 Potentially some of these methods could be applied in concert with cryo-imaging. However, design constraints for whole mouse cryo-imaging (high field of view, large working distance, low NA, etc.) greatly limit the applicability of confocal or computational optical sectioning approaches.
Computational optical sectioning suggests an alternative to our next-image processing of cryo-images to reduce effects of light propagation through tissue. To at least a first approximation, a cryo-image volume of a point fluorescence source would yield a cryo-image PSF, not to be confused with an optical PSF, which has minimal impact in our case. Iterative deconvolution using the cryo-imaging PSF could be an attractive option, at least in some cases requiring high quality processing. However, the computation time can be excessive. For example, three different commercial deconvolution software packages, Huygens Deconvolution Software (Scientific Volume Imaging, The Netherlands), AutoDeblur (Media Cybernetics, Bethesda, Maryland), and Amira, were used to perform deconvolution on a small cryo-image stack (300 sections). The fastest deconvolution software (AutoDeblur) required ≈60 min, while the slowest software package (Amira) required ≈90 min to process 300 images. In comparison, next-image processing runs extremely fast, requiring only 37 s to process the same data set. Next-image processing allows us to efficiently process whole mouse datasets (typically consisting of ≈24 FOVs, totaling ≈30,000 brightfield and fluorescent images) in a minimal amount of time.
Measured tissue specific parameters were found to be necessary and independent of the object used to measure the parameters. Estimated parameters, μT and σ, were independent of the depth of the structure used to estimate the parameters. Parameters from different tissues were separable in 3D feature space (Fig. 7), and using incorrect parameters in different tissues introduced artifacts (Fig. 8), verifying the use of tissue specific parameters. Images processed with the correct parameters (Figs. 8b, 8e, and 8h), did not contain negative values (black holes as seen in Figs. 8c and 8f), caused by k being too large. The brain has a lower attenuation coefficient, which leads to a larger k value (k = exp[−μT * slice thickness]), and correspondingly removes in-plane fluorescence, as well as out-of-plane fluorescence in the heart (Fig. 8c) and fat (Fig. 8f) images, creating negative values. When k was too small, out-of-plane fluorescence was not entirely removed (Fig. 8i, arrow).
For all tissue types analyzed the estimated attenuation values fell within the range of reported values in the literature. Reported μT values in the literature were typically spread over a large range due to the use of different techniques to determine tissue properties, including different wavelengths of light, tissue preparation methods, and tissue harvested from different species.4 The standard deviation of estimated μT and σ values ranged from 12 to 18% and 14 to 24% of the mean value respectively. The relatively large standard deviation was most likely due to differences in the actual scattering and absorption coefficient across a tissue, as variations in tissue properties within an organ lead to changes in optical properties. However, despite the large standard deviations the measured μT values were found to be statistically different for all but the most similar tissues. In all cases in which μT values were not statistically different, the σ values were found to be statistically different. Table 2 shows a correlation between larger σ values for tissues with a larger reported scattering coefficient. For example tissues with higher reported scattering coefficients and lower absorption coefficients, fatty tissue (μS = 270 cm−1, μA = .49 cm−1) and brain (μS = 90.2 cm−1, μA = 2.02 cm−1), had a larger measured σ value.4 Similar tissues such as skeletal muscle and cardiac muscle, heart, had fairly similar measured μT values, 267 ± 47.6 cm−1 and 214 ± 41.5 cm−1, and nearly equal σ values, 9.1 ± 2.1 μm and 9.0 ± 2.1 μm. The presence of blood surrounding the cardiac muscle most likely accounts for the increase in measured μT values without changing μS values between the similar tissues. Blood is highly absorbing with reported μT values of 514–1416 cm−1 and weakly scattering with reported μS values of 1.68–4.87 cm−1.4
We believe that cryo-imaging is a promising technology for imaging fluorescent structures and shows particular promise for imaging fluorescently labeled cells, specifically stem cells in regenerative medicine and metastatic cancer. Cryo-imaging is unique compared to in vivo and microscopic techniques, in that it allows micron scale resolution and information-rich contrast mechanisms over very large 3D fields of view.
References
W. T. Baxter, S. F. Mironov, A. V. Zaitsev, J. Jalife, and A. M. Pertsov, “Visualizing excitation waves inside cardiac muscle using transillumination. Biophys. J. 80, 516-530 (2001). doi:10.1016/S0006-3495(01)76034-1
A. J. Bellezza, J. F. Reynaud, B. A. Hirons, and C. F. Burgoyne, “Digital three-dimensional (3D) reconstruction of the connective tissues of the monkey optic nerve head. Invest. Ophthalmol. Vis. Sci. 43, U1159 (2002).
S. L. Bernard, J. R. Ewen, C. H. Barlow, J. J. Kelly, S. McKinney, D. A. Frazer, and R. W. Glenny, “High spatial resolution measurements of organ blood flow in small laboratory animals. Am. J. Physiol. Heart Circ. Physiol. 279, H2043-H2052 (2000).
W. F. Cheong, S. A. Prahl, and A. J. Welch, A review of the optical-properties of biological tissues. IEEE J. Quantum Electron. 26, 2166-2185 (1990). doi:10.1109/3.64354
T. Collier, A. Lacy, R. Richards-Kortum, A. Malpica, and M. Follen, “Near real-time confocal microscopy of amelanotic tissue: detection of dysplasia in ex vivo cervical tissue. Acad. Radiol. 9, 504-512 (2002). doi:10.1016/S1076-6332(03)80326-4
J. Condeelis and J. E. Segall, “Intravital imaging of cell movement in tumours. Nat. Rev. Cancer 3, 921-930 (2003). doi:10.1038/nrc1231
C. Donner and H. W. Jensen, “Light diffusion in multi-layered translucent materials. ACM Trans. Graph. 24, 1032-1039 (2005). doi:10.1145/1073204.1073308
M. Donoser, M. Wiltsche, and H. Bischof, “A New Automated Microtomy Concept for 3D Paper Structure Analysis. Tsukuba, Japan, 2005), pp. 76-79.
S. T. Flock, M. S. Patterson, B. C. Wilson, and D. R. Wyman, Monte-Carlo modeling of light-propagation in highly scattering tissues. 1. Model predictions and comparison with diffusion-theory. IEEE Trans. Biomed. Eng. 36, 1162-1168 (1989). doi:10.1109/TBME.1989.1173624
S. T. Flock, B. C. Wilson, and M. S. Patterson, Monte-Carlo modeling of light-propagation in highly scattering tissues. 2. Comparison with measurements in phantoms. IEEE Trans. Biomed. Eng. 36, 1169-1173 (1989). doi:10.1109/10.42107
A. C. Guyton and J. E. Hall, Textbook of Medical Physiology, (W.B. Saunders Company, Philadelphia, 1996).
R. C. Haskell, B. J. Tromberg, L. O. Svaasand, T. T. Tsay, T. C. Feng, and M. S. Mcadams, “Boundary-conditions for the diffusion equation in radiative-transfer. Biophys. J. 66, A378 (1994).
F. Helmchen and W. Denk, “Deep tissue two-photon microscopy. Nat. Methods 2, 932-940 (2005). doi:10.1038/nmeth818
A. Ishimaru, “Diffusion of light in turbid material. Appl. Opt. 28, 2210-2215 (1989). doi:10.1364/AO.28.002210
A. Kenzie-Graham, E. F. Lee, I. D. Dinov, M. Bota, D. W. Shattuck, S. Ruffins, H. Yuan, F. Konstantinidis, A. Pitiot, Y. Ding, G. G. Hu, R. E. Jacobs, and A. W. Toga, “A multimodal, multidimensional atlas of the C57BL/6J mouse brain. J. Anat. 204, 93-102 (2004). doi:10.1111/j.1469-7580.2004.00264.x
G. B. Ma, J. F. Delorme, P. Gallant, and D. A. Boas, “Comparison of simplified Monte Carlo simulation and diffusion approximation for the fluorescence signal from phantoms with typical mouse tissue optical properties. Appl. Opt. 46, 1686-1692 (2007). doi:10.1364/AO.46.001686
J. Mitic, T. Anhut, M. Meier, M. Ducros, A. Serov, and T. Lasser, “Optical sectioning in wide-field microscopy obtained by dynamic structured light illumination and detection based on a smart pixel detector array. Opt. Lett. 28, 698-700 (2003). doi:10.1364/OL.28.000698
M. A. A. Neil, R. Juskaitis, and T. Wilson, “Method of obtaining optical sectioning by using structured light in a conventional microscope. Opt. Lett. 22, 1905-1907 (1997). doi:10.1364/OL.22.001905
J. Nissanov, L. Bertrand, and O. Tretiak, “Cryosectioning distortion reduction using tape support. Microsc. Res. Tech. 53, 239-240 (2001). doi:10.1002/jemt.1089
A. I. Prentice, “Autofluorescence of bone tissues. J. Clin. Pathol. 20, 717-719 (1967). doi:10.1136/jcp.20.5.717
L. Reinisch, “Scatter-limited phototherapy: A model for laser treatment of skin. Lasers Surg. Med. 30, 381-388 (2002). doi:10.1002/lsm.10046
B. W. Rice, M. D. Cable, and M. B. Nelson, “In vivo imaging of light-emitting probes. J. Biomed. Opt. 6, 432-440 (2001). doi:10.1117/1.1413210
M. P. Rolf, R. ter Wee, T. G. van Leeuwen, J. A. E. Spaan, and G. J. Streekstra, “Diameter measurement from images of fluorescent cylinders embedded in tissue. Med. Biol. Eng. Comput. 46, 589-596 (2008). doi:10.1007/s11517-008-0328-9
P. Sarder and A. Nehorai, “Deconvolution methods for 3-D fluorescence microscopy images. IEEE Signal Process. Mag. 23, 32-45 (2006). doi:10.1109/MSP.2006.1628876
J. M. Schmitt, A. H. Gandjbakhche, and R. F. Bonner, Use of polarized-light to discriminate short-path photons in a multiply scattering medium. Appl. Opt. 31, 6535-6546 (1992). doi:10.1364/AO.31.006535
J. A. E. Spaan, R. ter Wee, J. W. G. E. van Teeffelen, G. Streekstra, M. Siebes, C. Kolyva, H. Vink, D. S. Fokkema, and E. VanBavel, “Visualisation of intramural coronary vasculature by an imaging cryomicrotome suggests compartmentalisation of myocardial perfusion areas. Med. Biol. Eng. Comput. 43, 431-435 (2005). doi:10.1007/BF02344722
W. Wallace, L. H. Schaefer, and J. R. Swedlow, “A workingperson’s guide to deconvolution in light microscopy. Biotechniques 31, 1076–1078 + (2001).
L. H. Wang and S. L. Jacques, Hybrid model of Monte-Carlo simulation and diffusion-theory for light reflectance by turbid media. J. Opt. Soc. Am. A 10, 1746-1752 (1993). doi:10.1364/JOSAA.10.001746
W. J. Weninger and T. Mohun, “Phenotyping transgenic embryos: a rapid 3-D screening method based on episcopic fluorescence image capturing. Nat. Genet. 30, 59-65 (2002). doi:10.1038/ng785
A. A. Young, I. J. Legrice, M. A. Young, and B. H. Smaill, “Extended confocal microscopy of myocardial laminae and collagen network. J. Microsc. 192, 139-150 (1998). doi:10.1046/j.1365-2818.1998.00414.x
Acknowledgments
The authors wish to thank many collaborators for being early adopters of the cryo-imaging technology and providing samples. Collaborators include: M. Penn (Dept of Cell Biology, Cleveland Clinic), H. von Recum (Dept of Biomedical Engineering, Case Western Reserve University), and G. Muschler (Orthopedic Surgery, Cleveland Clinic). This investigation was conducted in a facility constructed with support from Research Facilities Improvement Program Grant Number C06 RR12463-01 from the National Center for Research Resources, National Institutes of Health. This research is supported by an Ohio Wright Center of Innovation and Biomedical Research and Technology Transfer award, “The Biomedical Structure, Functional and Molecular Imaging Enterprise,” and NIH R41CA124270. Grant Steyer was partially supported from the predoctoral training grant, T32EB007509, Interdisciplinary Biomedical Imaging Training Program. Dr. Wilson has an interest in BioInVision, Inc.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Steyer, G.J., Roy, D., Salvado, O. et al. Removal of Out-of-Plane Fluorescence for Single Cell Visualization and Quantification in Cryo-Imaging. Ann Biomed Eng 37, 1613–1628 (2009). https://doi.org/10.1007/s10439-009-9726-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10439-009-9726-x