Abstract
Magnetic Resonance Imaging (MRI) longitudinal studies conducted to assess changes in tibia bone quality impose strict requirements on the reproducibility of the prescribed region acquired. Registration, the process of aligning two images, is commonly performed on the images after acquisition. However, techniques to improve image registration precision by adjusting scanning parameters prospectively, prior to image acquisition, would be preferred. We have adapted an automatic prospective mutual information based registration algorithm to a MRI longitudinal study of trabecular bone of the tibia and compared it to a post-scan manual registration. Qualitatively, image alignment due to the prospective registration is shown in 2D subtraction images and 3D surface renderings. Quantitatively, the registration performance is demonstrated by calculating the sum of the squares of the subtraction images. Results show that the sum of the squares is lower for the follow up images with prospective registration by an average of 19.37% ± 0.07 compared to follow up images with post-scan manual registration. Our study found no significant difference between the trabecular bone structure parameters calculated from the post-scan manual registration and the prospective registration images (p > 0.05). All coefficient of variation values for all trabecular bone structure parameters were within a 2–4.5% range which are within values previously reported in the literature. Results suggest that this algorithm is robust enough to be used in different musculoskeletal imaging applications including the hip as well as the tibia.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Osteoporosis is a metabolic disorder that results in bone with decreased mechanical strength and increased fracture risk. It primarily targets trabecular bone, which is spongy bone found in skeletal sites such as the vertebrae and the proximal and distal parts of the appendicular skeleton, with both thinning and loss of structure.16 Trabecular bone micro-architecture is of particular importance to bone strength.7 Studies have confirmed that MRI can be used to detect differences in trabecular bone due to age, bone mineral density, and osteoporotic status.21,26 Longitudinal studies conducted to assess changes in bone quality in the tibia impose strict requirements on the reproducibility of data acquired.12,22 The same region must be consistently analyzed between baseline and follow-up image acquisition. To this end, registration is usually performed manually at the MRI scanner based on visually locating anatomic landmarks such as the endplate prior to image acquisition. Additional registration is still needed post-acquisition by manually matching slices.
Techniques to improve image registration precision by prospective registration, adjusting scanning parameters prior to image acquisition, have been reported recently in literature.6,14 A prospective registration technique for proton magnetic resonance spectroscopy of brain longitudinal examinations to track disease progression13 has been developed. This technique utilizes a mutual information registration algorithm25 to register images in a baseline and follow-up exam. The output of the registration algorithm, three translations and three Euler angles, is used to redefine the region to be imaged and thus to acquire a follow-up oblique imaging volume identical to the baseline volume. This pre-registration has provided improved region overlaps as well as generally decreased short-term measurement variability and improved workflow. Much work has been done to optimize and validate prospective registration in MR brain images4,9,15 including methods to prospectively register brain images to an atlas29 and to register brain spectroscopy images.13,14 However, no such techniques have been applied to musculoskeletal imaging. All methods to register musculoskeletal images, such as the radius and tibia, for bone structure analysis have been performed post-acquisition.19,22
When imaging trabecular bone in osteoporosis, the regional variations in structure of bone are inherent and follow-up images registered to the baseline images would have profound impact on the quantitative evaluation of trabecular bone architecture. A study performed by Gomberg et al. 12 investigating the error sources in MRI-based trabecular bone structural parameters found the two main sources of variation to be patient motion and failure to match the ROI. They found that even if the ROI is offset by one slice, BV/TV can vary by a median error of 1%. An implementation of prospective registration to musculoskeletal MRI longitudinal studies would be of significant importance for characterizing trabecular bone. Prospective registration would reduce the need for an additional manual post-processing step that requires substantial expertise. It would therefore reduce the post-processing time and subjectivity while maintaining the precision of trabecular bone measurements such as apparent bone volume fraction (App.BV/TV), apparent trabecular separation (App.Tb.Sp), apparent trabecular thickness (App.Tb.Th) and apparent trabecular number (App.Tb.N).
Methods
Registration Algorithm
Image registration involves aligning two images (either 2D or 3D) by adjusting the parameters of a transformation which maps one image to the other. The parameters are adjusted until a metric function is optimized. We used the same technique as presented by Hancu et al. 13 by implementing a mutual information metric to rigidly register baseline and follow-up low resolution images. Mutual information is a measure of the information one image provides with respect to another. The use of mutual information for medical image registration applications was introduced in 1997 by both Viola and Wells25 and Collignon et al. 8 For two images, the mutual information is computed from the joint probability distribution of the images’ intensity or gray-level values. When two images are perfectly aligned, they should provide maximal information about each other and the joint probability distribution would yield a high mutual information value.23 The information contributed by each of the two images, denoted image A and image B, is entropy which measures the dispersion of a probability distribution. The entropy measure of an image is defined as
Here, p A is the marginal probability distribution, the likelihood of finding pixels of an intensity throughout the imaging volume.3 The joint entropy of the two imaging volumes A and B is defined as
Therefore, the mutual information of two images can be defined as the degree of dependence between image A and image B by the Kullback–Leibler17 distance between the joint distribution, p AB (a,b), and the distribution associated with the case of complete independence, p A (a)·p B (b):
For the prospective registration algorithm, image A was the baseline image and image B was the transformed follow-up image. The transformation involves a rotation matrix, characterized by three Euler angles, and a translation vector composed of three translation parameters. The rotation matrix and the translation vector define the movement of a point from the follow-up to the baseline image. A conjugate gradient descent method1 was implemented to search for the six parameters that define the rotation matrix and translation vector which optimize the mutual information, MI(A,B).
Magnetic Resonance Imaging
The right distal tibia of five healthy volunteers (average age of 26 ± 3 years old) were scanned with their informed consent in accordance with the regulations of the Committee of Human Research at the University of California, San Francisco. All scans for each subject were performed on the same day. Subjects were removed from the scanner and repositioned between baseline and follow-up scans. To ensure a consistent clinical position between scans, a leg holder and pads were used in scanning which helped to limit rotation to small angles (<10°). The longitudinal landmark line was aligned with the subject’s lower leg and the transverse landmark line was aligned at the medial malleolus of the tibia.
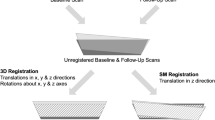
All MR images were acquired axially on a 3-T Signa Scanner (GE Healthcare, Milwaukee, WI, USA) with a modified multi-acquisition SSFP sequence (Steady State Free Procession) applying a maximum intensity projection of two images (MI-SSFP).2 A Nova Medical (Wilmington, MA, USA) four-coil surface phased array receiver coil was used. The scanning procedure is depicted in Fig. 1. After a three-plane localizer, two baseline scans were obtained. The first baseline scan was a low spatial resolution scan in the axial plane with a 256 × 256 matrix, 8 cm FOV, 0.5 mm slice thickness, 64 slices, 60° flip angle, 17/6.5 ms TR/TE, and a scan time of approximately 4 min. The second baseline scan, intended for quantitative analysis and comparison, was a high spatial resolution with a 512 × 384 matrix, 8 cm FOV, 0.5 mm slice thickness, 64 slices, 60° flip angle, 17/6.5 ms TR/TE, and approximately 16 min of scan time (Fig. 2).
The volunteers were then removed from the scanner and repositioned for the follow up scans. After a three plane localizer, four follow-up scans were obtained. The first two follow-up scans used the same protocols as the first two baseline scans, one low spatial resolution follow-up scan for registration (∼4 min scan time) and a high spatial resolution scan for quantitative trabecular bone structure analysis and comparison (∼16 min scan time). The low-resolution axial baseline and follow-up scans were then registered using the mutual information based rigid registration scheme described previously. The registration was performed while the patient remained in the scanner and took less than 1 min, including the time to upload the baseline and follow-up volumes.
The final two follow-up scans required a modification of the MI-SSFP sequence to allow for the input of the six registration parameters (three translations and three rotations) output by the mutual information registration algorithm. Oblique scans were acquired with the same parameters as the first two follow-up scans except for input parameters from the registration. The prescription of these oblique scans had the same tibia coverage and slice orientation as the baseline axial images.
Registration Algorithm Validation
To validate the registration algorithm, the output of the registration algorithm was compared to known values. One of the low-resolution images of one of the baseline scans was rotated and translated by a known amount selected with a random number generator. The rotation was limited to ±8° and the translation was limited to ±10 mm. The baseline volume and the transformed volume were then registered and the registration error was computed as the difference between the known translation and the output of the registration algorithm. This procedure was repeated 50 times.
Registration Performance Evaluation
Visual inspection of 3D surface renderings and subtraction images aided in evaluating the success of the prospective registration with tibial images. Using an in-house developed software based on MATLAB (The MathWorks, Inc., Natick, MA), the inner cortical shell of the tibiae were segmented semi-automatically with a Bezier-spline and edge detection based method on a slice by slice basis. Segmented contours were then stacked to create a 3D surface. This was performed on the baseline and follow-up low spatial resolution images which were then visualized together in a 3D surface rendering. Subtraction images were created by subtracting the low spatial resolution follow-up volumes from the low spatial resolution baseline volumes. Subtraction images were created for both follow-up image with the prospective registration and the follow-up image without registration. To quantify the improvement in image alignment seen in the subtraction images, the sum of the squares was calculated for each slice in the volume and then averaged across the volume.
Analysis of Trabecular Bone Microarchitecture
Tibial trabecular bone structure analysis was performed using software developed at our institution using IDL (Interactive Data language, Research Systems, Inc., Boulder, CO). Due to the use of surface coils, a correction for the spatial variation in the coil detection sensitivity was required for accurate image analysis. The images were coil-corrected with a low-pass-filter based coil sensitivity correction.22 A volumetric region of interest (ROI) was manually defined using a graphics cursor. Moving proximally from the slice where the growth plate ends (endplate), the volumetric ROI consisted of 20 axial slices of the volume and included only trabecular bone and bone marrow. Each volumetric ROI requires between 10 and 20 min (30–60 s per slice) to accurately define. The ROI for the high-resolution baseline image and follow-up image without registration were then registered manually by visually matching corresponding slices. The same ROI was used on the follow-up with prospective registration as for the baseline image and did not require the additional ROI definition and manual ROI registration. The resulting ROI was divided into two 10-slice thick regions due to inherent changes in trabecular bone structure with distance from the end plate. After the ROI had been defined and aligned, an image intensity histogram based thresholding technique was used to binarize the ROI into trabecular bone and marrow phases.22 Previously described methods20 were then used to compute the apparent trabecular structural parameters: App.BV/TV, App.Tb.Sp., App.Tb.Th., and App.Tb.N. To determine the effects of using the prospective registration on trabecular parameters, the data was analyzed using a repeated-measures analysis of variance procedure.10 This statistical procedure was chosen to help distinguish between the variability between the experimental subjects, the variability due to different post-processing methods, and the variability of the measurements within the same subject. The reproducibility of the technique was verified by calculating the short term coefficient of variation.11
Results
Table 1 presents the results of the registration validation by showing the registration errors (mean ± standard deviation) as determined by subtracting known transformations from registration outputs. The average error in rotations was ∼0.2° and in translations was ∼1.1 mm which are within reasonable accuracy for tibia registration.
By image subtraction and 3D surface rendering of the segmented tibiae, the improvement in image alignment can be assessed. Figure 3 shows representative results of the prospective registration. The improvement from the registration can be seen by looking at the subtraction images (Figs. 3c and 3f) and corresponding tibia segmentations (Figs. 3d and 3g). Displayed next to the subtraction images are the corresponding low-resolution follow-up images without registration (Fig. 3b) and with prospective registration (Fig. 3e). The baseline low-resolution scan is also shown for comparison (Fig. 3a). It can be seen in the results that the second follow up scan is more closely oriented with the baseline scans. For example, in Fig. 3c the edges of the cortical bone are misaligned with higher intensity in the subtraction image, and clear separation of the red and green tibial renderings in Fig. 3d is visible. In Fig. 3f, the high intensity differences within the tibial edge are reduced, and there is considerably more overlap in the red and green tibial renderings in Fig. 3g.
Visual comparison of prospective registration versus follow-up without registration for low spatial resolution images (0.313 × 0.313 × 0.5 mm3) of the tibia. (a) Axial slice of baseline image (b) Axial slice of follow-up image without registration (c) Subtraction of a and b (d) Rendering of non-registered tibiae (green = low resolution baseline, red = low resolution follow-up) (e) Axial slice of follow-up image with prospective registration (f) Subtraction of a and e (g) Rendering of prospectively registered tibiae (green = low resolution baseline, red = low resolution follow-up)
To quantify the dispersion seen in the subtraction images, the sum of the squares was calculated for each slice. The graph in Fig. 4 shows the average of the sum of the squares across all slices for the volume for each of the five subjects. Sum of the squares was lower for the follow up images with prospective registration by an average of 19.37% ± 0.07.
Our study did not observe differences between the trabecular bone structure parameters calculated from the post-scan manual registration and the prospective registration images. Figure 5 shows the trabecular bone parameters calculated for one of the tibiae. There is very little variation in the parameters between images. The results of a repeated-measures analysis of variance indicate there is no significant difference between the trabecular bone parameters calculated from the prospective registration images and those calculated from the post-scan manual registration (p > 0.05). Additionally, our study also found little difference in the coefficient of variation when evaluating the post-scan registration and the prospective registration for the four parameters in the two different bone regions. All values were within a 2–4.5% (Fig. 6) range, which are within values previously reported.22
The raw data for the trabecular bone parameters for one of the tibiae in both regions is shown. Very little variation in the parameters was found between images which demonstrates that the trabecular bone parameters found with the prospective registration are just as accurate as those found using the manual post-scan registration technique
Discussion
In this work we have demonstrated the feasibility of using a mutual information based method to prospectively register longitudinal MR images of tibia scans. We have developed a unique MI-SSFP sequence which allows for the input of the registration results to scan oblique registered high spatial resolution tibia images.
The coefficient of variation of the trabecular bone structure parameters is within the same range for both registration types and the repeated-measures analysis of variance indicates that there is no significant difference in trabecular bone parameters between baseline and follow-up images for both registration types. These findings demonstrate that the trabecular bone structure parameters found with the prospective registration are just as accurate as those found using the established post-scan manual registration technique.22
The use of automatic prospective registration ensures that the ROI is placed on the same slice for both the baseline and follow-up. Automatic prospective registration also has benefits by eliminating the need for subjectively finding the longitudinal reference location (endplate location) for all follow-up scans. In addition, the use of prospective registration at clinical sites will ensure that the same region is scanned in the baseline and follow-up. Often there is a discrepancy in the region scanned and the images cannot be utilized for quantitative comparison.
Automatic prospective registration allows for a time savings and an easy analysis of multiple ROIs, as the regions defined on the baseline scans can also be applied directly to the registered follow-up images. Automatic prospective registration adds 4 min to the baseline scan time and 5 min (4 min of scan time and 1 min for the registration) to the follow-up scan time. However it eliminates the need for an additional ROI to be manually generated for the follow-up images, which allows for a 10–20 min savings in post-processing time. For longitudinal studies, where hundreds of patients are being scanned and analyzed, the time savings in post-processing could be substantial.
The prospective registration algorithm implemented in this study was the same algorithm utilized by Hancu et al. 13 It is optimized for magnetic resonance spectroscopy studies with a mutual information metric, a rigid body transformation, and a conjugate gradient descent optimizer. Although the results from implementing this prospective registration technique are just as accurate as the results from the current post-scan manual registration technique, the algorithm may produce better results if optimized for musculoskeletal imaging. Since the registration is intra-modality, an intensity-based metric such as a cross-correlation metric,24 a least squares metric,28 or a voxel-intensity-based method27 may be more appropriate. Additionally, musculoskeletal images contain joints surrounded by soft tissue that deform depending on the subject position. This deformable soft tissue makes rigid registration more challenging for musculoskeletal images compared to brain images. The registration can be improved by cropping the image so that the entropy is only calculated in a region with minimal soft tissue. This would minimize the effects of the non-rigid movement of the soft tissue on the registration algorithm.
Automatic retrospective registration, registration after the images have been acquired, may be an alternative to prospective registration. However, prospective registration requires no interpolation of the data, which is required in automatic retrospective registration. All interpolation methods smooth images to some degree and images with sharp-edge details, such as high-resolution trabecular bone images, are much more effected.18 Prospective registration also ensures that the correct region is being acquired at scan time. Retrospective registration will fail if there is a large difference in the regions scanned and therefore little overlap between the baseline and follow-up images.
Prospective registration may have a bigger impact on hip images since the reproducible positioning of the subjects is not as easy as when imaging the tibia. Carpenter et al. 5 performed a study investigating the reproducibility of bone structure parameters in the proximal femur. They suggested that the high coefficients of variation, ranging from 6.5% to 13.5% may be partially due to patient repositioning. Additionally, the images of the proximal femur were acquired with a slice thickness of 1mm, double the slice thickness in this study, which may contribute to additional partial volume effects. More experiments need to be conducted to investigate if prospective registration can minimize partial volume and patient repositioning effects when imaging the proximal femur.
This study proves that it is possible to implement prospective registration to a musculoskeletal application. Prospective registration ensures that the same region is analyzed in both the baseline and follow-up images, saves post-processing time, preserves the reproducibility of the trabecular bone parameters, and requires no interpolation. The results suggest that it may be robust enough to be used in different musculoskeletal imaging applications including the hip.
References
Atkinson K. An Introduction to Numerical Analysis. Chichester: Wiley; 1989
Bangerter N. K., Hargreaves B. A., Vasanawala S. S., Pauly J. M., Gold G. E., Nishimura D. G. Analysis of multiple-acquisition SSFP. Magn. Reson. Med. 2004;51(5):1038–1047
Bankman, I. N. Handbook of Medical Imaging: Processing and Analysis San Diego. CA: Academic, pp. 542–543, 2000
Benner, T., J. J. Wisco, and A. J. van der Kouwe, et al. Comparison of manual and automatic section positioning of brain MR images. Radiology 239:246–254, 2006
Carpenter, D., R. Krug, S. Banerjee, and S. Majumdar. Analyzing Trabecular Bone Structure in the Proximal Femur with High-Resolution Parallel Magnetic Resonance Imaging. International Bone Densitometry Workshop, Kyoto, Japan 2006
Chu, W.-J., C. Pan, J. W. Pan, and H. P. Hethereinton. Reproducibility of 1H Spectroscopic Imaging of the Human Hippocampus. In: Proceedings of the 12th International Society of Magnetic Resonance Medicine 2004
Ciarelli M. J., Goldstein S. A., Kuhn J. L., Cody D. D., Brown M. B. Evaluation of orthogonal mechanical properties and density of human trabecular bone from the major metaphyseal regions with materials testing and computed tomography. J. Orthop. Res. 1991;9(5):674–682
Collignon A., Maes F., Vandermeulen D., Marchal G., Suetens P. Multimodality image registration by maximization of mutual information. IEEE Trans. Med. Imaging 1997;16(2):187–198
Gedat E., Braun J., Sack I., Bernarding J. Prospective registration of human head magnetic resonance images for reproducible slice positioning using localizer images. J. Magn. Reson. Imaging 2004;20(4):581–587
Glantz S. A. Primer of Bio-Statistics. United States of America: McGraw-Hill Companies, Inc.; 1997
Gluer C. C., Blake G., Lu Y., Blunt B. A., Jergas M., Genant H. K. Accurate assessment of precision errors: how to measure the reproducibility of bone densitometry techniques. Osteoporos. Int. 1995;5(4):262–270
Gomberg B. R., Wehrli F. W., Vasilic B., et al. Reproducibility and error sources of micro-MRI-based trabecular bone structural parameters of the distal radius and tibia. Bone 2004;35(1):266–276
Hancu I., Blezek D. J., Dumoulin M. C. Automatic repositioning of single voxels in longitudinal 1H MRS studies. NMR Biomed. 2005;18(6):352–361
Hartmann S. L., Dawant B. M., Parks M. H., Schlack H., Martin P. R. Image-guided MR spectroscopy volume of interest localization for longitudinal studies. Comput. Med. Imaging Graph 1998;22(6):453–461
Itti L., Chang L., Ernst T. Automatic scan prescription for brain MRI. Magn. Reson. Med. 2001;45(3):486–494
Kleerekoper M., Villanueva A. R., Stanciu J., Rao D. S., Parfitt A. M. The role of three-dimensional trabecular microstructure in the pathogenesis of vertebral compression fractures. Calcif. Tissue Int. 1985;37(6):594–597
Kullback S., Leibler R. A. On information and sufficiency. Ann. Math. Stat. 1951;22(1):79–86
Lehmann T. M., Gonner C., Spitzer K. Survey: interpolation methods in medical image processing. IEEE Trans. Med. Imaging 1999;18(11):1049–1075
Magland, J., B. Vasilic, W. Lin, and F. W. Wehrli. Automatic 3D Registration of Trabecular Bone Images Using a Collection of Regional 2D Registrations. In: Proceedings of the 14th International Society of Magnetic Resonance Medicine 2006
Majumdar S., Genant H. K. Assessment of trabecular structure using high resolution magnetic resonance imaging. Stud. Health Technol. Inform. 1997;40:81–96
Majumdar S., Genant H. K., Grampp S., et al. Correlation of trabecular bone structure with age, bone mineral density, and osteoporotic status: in vivo studies in the distal radius using high resolution magnetic resonance imaging. J. Bone Miner Res. 1997;12(1):111–118
Newitt D. C., Van Rietbergen B., Majumdar S. Processing and analysis of in vivo high-resolution MR images of trabecular bone for longitudinal studies: reproducibility of structural measures and micro-finite element analysis derived mechanical properties. Osteoporos. Int. 2002;13:278–287
Pluim J. P., Maintz J. B., Viergever M. A. Mutual-information-based registration of medical images: a survey. IEEE Trans. Med. Imaging 2003;22(8):986–1004
Rizzo, G., P. Pasquali, and M. C. Gilardi, et al. Multimodality biomedical image integration: use of a cross-correlation technique.; pp. 219–220, 1991
Viola P., Wells W. M. Alignment by maximization of mutual information. Int. J. Comp. Vis. 1997;24(2):137–154
Wehrli F. W., Hwang S. N., Ma J., Song H. K., Ford J. C., Haddad J. G. Cancellous bone volume and structure in the forearm: noninvasive assessment with MR microimaging and image processing. Radiology 1998;206:347–357
Woods R. P., Cherry S. R., Mazziotta J. C. Rapid automated algorithm for aligning and reslicing PET images. J. Comput. Assist. Tomogr. 1992;16:620–633
Woods R. P., Grafton S. T., Holmes C. J., Cherry S. R., Mazziotta J. C. Automated image registration: I. General methods and intrasubject, intramodality validation. J. Comput. Assist. Tomogr. 1998;22:141–154
van der Kouwe A. J., Benner T., Fischl B., et al. On-line automatic slice positioning for brain MR imaging. Neuroimage 2005;27(1):220–230
Acknowledgments
This work is funded by NIH grant award program number ROI-AR49701. The authors thank David Newitt and Ben Hyun for their insight on trabecular bone analysis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Blumenfeld, J., Carballido-Gamio, J., Krug, R. et al. Automatic Prospective Registration of High-Resolution Trabecular Bone Images of the Tibia. Ann Biomed Eng 35, 1924–1931 (2007). https://doi.org/10.1007/s10439-007-9365-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10439-007-9365-z