Abstract
Summary
Longitudinal studies of bone using high-resolution medical imaging may result in non-physiological measurements of longitudinal changes. In this study, we determined that three-dimensional image processing techniques best capture realistic longitudinal changes in bone density and should therefore be used with high-resolution imaging when studying bone changes over time.
Introduction
The purpose of this study was to determine which longitudinal analysis technique (no registration (NR), slice-match (SM) registration, or three-dimensional registration (3DR)) produced the most realistic longitudinal changes in a 3-year study of bone density and structure using high-resolution peripheral quantitative computed tomography (HR-pQCT).
Methods
We assessed HR-pQCT scans of the distal radius and tibia for men and women (N = 40) aged 55–70 years at baseline and 6, 12, 24, and 36 months. To evaluate which longitudinal analysis technique (NR, SM, or 3DR) best captured physiologically reasonable 3-year changes, we calculated the standard deviation of the absolute rate of change in each bone parameter. The data were compared between longitudinal analysis techniques using repeated measures ANOVA and post hoc analysis.
Results
As expected, both SM and 3DR better captured physiological longitudinal changes than NR. At the tibia, there were no differences between SM and 3DR; however, at the radius where precision was lower, 3DR produced better results for total bone mineral density.
Conclusions
At least SM or 3DR should be implemented in longitudinal studies using HR-pQCT. 3DR is preferable, particularly at the radius, to ensure that physiological changes in bone density are observed.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Recent developments in high-resolution imaging are providing important insight into microarchitecture, mineralization, and biomechanics of bone tissue. Specifically, high-resolution peripheral quantitative computed tomography (HR-pQCT) is a non-invasive and low-radiation imaging method used to assess bone microarchitecture and volumetric bone mineral density at the peripheral sites of the radius and tibia with an isotropic voxel size of 82 μm or 61 μm, depending on the scanner generation. HR-pQCT has been used to compile normative databases providing age- and sex-specific reference data [1,2,3,4] and has been used in risk assessment of distal radius fractures [5,6,7]. When HR-pQCT measurements are taken at multiple time points, longitudinal changes in bone have been sensitively assessed for osteoporosis therapies [8,9,10] and bone loss prevention therapies [11]; however, these longitudinal changes are subject to precision errors.
Although bone is a highly adaptive tissue that responds to changes in hormones, physical loading, diet, and disease condition, the normal trajectory of bone adaptation follows a stable pattern that does not oscillate dramatically over time. Nevertheless, longitudinal HR-pQCT measures can sometimes exhibit large oscillations in bone parameters that are clearly not physiological, but rather due to precision errors related primarily to motion artifact and patient positioning. To reliably track longitudinal changes over time, it is important to investigate the same spatial regions of bone, regardless of limb positioning during measurements. Minimizing positioning error between baseline and follow-up is a challenge, and errors occur even for experienced operators. These errors are manifested in non-physiological patterns of longitudinal change.
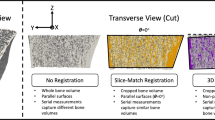
To address the challenges with repositioning error, image registration techniques have been proposed [12, 13] (Fig. 1). The most commonly used technique, a slice-match (SM) method, has been implemented in the HR-pQCT software (Image Processing Language, v5.16, Scanco Medical AG, Brüttisellen, Switzerland). This method aligns two image stacks by translating along the axis perpendicular to the slices (the longitudinal axis) until the two-dimensional (2D) cross-sectional areas of the stacks are best aligned. This method was shown to improve short-term reproducibility of HR-pQCT measurements [12, 14, 15]. However, SM does not account for rotational misalignment around any axis and may provide inconsistent results when there is angular repositioning error between repeat scans.
Illustration of the differences between slice-match (SM) registration and three-dimensional (3D) registration. 3D registration rotates and translates follow-up images in six degrees of freedom to most accurately align the intensity of each voxel in the images. SM translates the images in the longitudinal direction only (one degree of freedom) to find the best correlation between the total area of the bones in each image. The hashed regions of the bone indicate the volume of interest identified by each registration method
Three-dimensional image registration (3DR) was also shown to improve reproducibility in HR-pQCT [12, 16, 17] and microcomputed tomography (μCT) [13, 18,19,20,21] studies. In contrast to SM, 3DR aligns images by applying a combination of rotations and translations (Fig. 2) determined by an optimization metric that matches voxel intensities between two images [22]. In HR-pQCT, 3DR was shown to improve short-term reproducibility in cortical geometry and microarchitectural parameters compared with SM registration at both skeletal sites [12]. In combined short-term (1-week) and long-term (4-month) reproducibility studies of HR-pQCT measurements, 3DR demonstrated non-significant improvements in total and cortical volumetric BMD compared with SM at both skeletal sites [16]. As opposed to reproducibility studies which measure pairs of images, longitudinal studies often involve several measurements per participant with the intention of studying changes in the bone microarchitecture and density. It is not clear whether a simple registration technique such as SM suffices for longitudinal studies or if more complex approaches such as 3DR are necessary to capture physiological changes.
Process for identifying the volume of interest (VOI) using three-dimensional (3D) registration. Each image was registered to the baseline scan, and a box was generated containing all the information for each registered image. The logical intersection of the boxes from each time point was determined to create a single box defining the VOI. This box was transformed back to each image’s original orientation and used to crop the images to include only the VOI. In the second row, we see the overlap between baseline (red) and follow-up images (green). In the third row and below, bone in green is included in the VOI, whereas bone in gray is excluded from the VOI
The main objective of this study was to determine whether no registration (NR), SM, or 3DR most closely match what would be expected as physiological long-term change in bone density and microarchitecture. To achieve this, we performed a short-term reproducibility analysis comparing NR, SM, and 3DR based on matched pairs to establish baseline reproducibility. To study longitudinal changes, we quantified the variability in bone parameters over a 3-year period with five scans per participant. We hypothesized that by accounting for the translational and rotational misalignment between follow-up measurements, 3DR will produce physiological trends of longitudinal measurements in bone microarchitecture and density that minimize oscillations due to inherent repositioning error.
Methods
Participants
The present study used a subset of participants from the Calgary Vitamin D Study [11]. Participants were men and women aged 55–70 years, with women at least 5 years post-menopause. This cohort was vitamin D sufficient, non-osteoporotic, not currently, or within the last 2 years taking bone active medication, and any chronic diseases were stable [11, 23]. The reproducibility analysis included 38 women and 31 men (64.7 ± 5.1 years) who were randomly selected from the Vitamin D cohort to participate in the reproducibility study. Fifty-one participants completed two scans at both the radius and tibia, and the remaining participants completed two scans at either the radius or tibia, following repositioning between scans, resulting in a total of 60 radius and 60 tibia scans. Certain participants were not eligible for scans at both sites as they had already reached the allowable radiation dose. To study longitudinal changes, 21 women and 19 men (62.0 ± 3.8 years at baseline) were selected from the Calgary Vitamin D Study. The 40 participants were selected from a pool of 311 to reflect the widest variability of change over the 3 years. The study was approved by the Conjoint Health Research Ethics Board at the University of Calgary and Health Canada. All participants gave written consent before participating in the study.
HR-pQCT image acquisition
Density and morphometric parameters at the distal radius and tibia were determined using second generation high-resolution peripheral quantitative computed tomography (XtremeCT, Scanco Medical, Brüttisellen, Switzerland) with an isotropic voxel size of 61 μm and 168 slices over a 10.2-mm length. HR-pQCT scans occurred at baseline and a same-day follow-up for the reproducibility analysis and at baseline, 6, 12, 24, and 36 months for studying longitudinal changes.
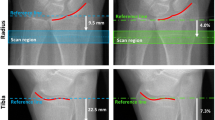
A standard in vivo scanning protocol was used. Scans of the non-dominant radius and left tibia were acquired 9.5 mm and 22.5 mm, respectively, proximal from the reference line. If a previous distal radius or tibia fracture was reported at baseline, the opposite limb was scanned. Scans were graded for motion artifacts following scan acquisition [24] and had motion scores in the radius and tibia of three or better. The reference line was placed independently at baseline scans at the mid-inclination notch for the radius and at the plateau portion of the tibial endplate for the tibia. For both the reproducibility and longitudinal studies, during follow-up scans, technicians followed the standard protocol of placing the reference line as close as possible to the baseline reference line. Two technicians were involved in image acquisition, and their combined precision scores were previously reported as less than 4% for all bone parameters and sites [25].
Standard analysis
A semi-automated contouring method was used to generate masks of the cortical and trabecular compartments independently for each scan in the study (Image Processing Language, v5.16, Scanco Medical AG, Brüttisellen, Switzerland). A whole-bone mask was defined based on the periosteal surface of the bone. A trabecular mask was defined based on the endosteal surface of the bone. A cortical mask was defined as the difference between the whole-bone mask and the trabecular bone mask.
Image registration and VOI selection
Three methods were used to identify the volume of interest (VOI) for density and microarchitecture analysis in the baseline and follow-up images: (1) no registration (NR), (2) slice-match registration (SM), and (3) 3D registration (3DR).
The VOI for NR was identified by the masks generated from the semi-automated contouring technique; no volume of bone was excluded from the analysis.
SM registration was performed using the manufacturer’s software package (Image Processing Language, v5.16, Scanco Medical AG, Brüttisellen, Switzerland). Briefly, the total cross-sectional area (Tt.Ar) was computed on a slice-by-slice basis for each measurement. The algorithm computed an optimal offset between follow-up scans using cross-correlation of Tt.Ar. The images were translated in the longitudinal direction until the correlation metric was optimized (Fig. 1), and the resulting offset was used to define the common VOI present in all scans.
The 3DR algorithm employed an intensity-based, rigid body registration technique [12] (Image Processing Language, v5.16, Scanco Medical AG, Brüttisellen, Switzerland). The whole bone mask (cortical and trabecular regions) was used to identify the region to sample image data, and each follow-up image was registered to the baseline image. To initialize the registration, the centers of mass in the masked regions of the follow-up and baseline images were aligned. A cross-correlation metric and the downhill simplex optimization scheme were used. Results of the 3D image registration were used to define a consistent VOI on all images. To prevent interpolation error that would result from directly rotating the grayscale images, a common mask of the largest common volume, defined as the intersection of all registered time points, was transformed to the image spaces of all follow-up scans (Fig. 2). All morphometric and density parameters were determined in the follow-up image space within the largest common volume defined by the common mask.
To quantify repositioning error with 3DR, we calculated the rotation angle between the longitudinal axes of the baseline and each follow-up scan. For all registration techniques, the percent overlap was defined as the common mask volume divided by the total baseline image volume.
Density and microarchitectural measurements
Density and microarchitecture were measured in the VOIs defined by each registration method. Within each VOI, the standard morphologic analysis was applied to determine total volumetric BMD (Tt.BMD; mg HA/cm3) and trabecular volumetric BMD (Tb.BMD; mg HA/cm3); trabecular bone volume fraction (Tb.BV/TV; %), number (Tb.N; /mm), thickness (Tb.Th; mm), separation (Tb.Sp; mm), and area (Tb.Ar; mm2) [26]; total cross-sectional area (Tt.Ar; mm2); and cortical volumetric BMD (Ct.BMD; mg HA/cm3), thickness (Ct.Th; mm), porosity (Ct.Po; %), and area (Ct.Ar; mm2) [27, 28].
Statistical analysis
To assess short-term reproducibility, the coefficient of variation (CV) and root mean squared coefficient of variation (CVRMS, %) were calculated for each registration technique. The smallest change to be considered statistically significant, least significant change (LSC), was calculated as the product of CVRMS and 2.77 [29, 30] and expressed in parameters units by multiplying LSC (%) by the group mean of each bone variable. We examined differences in reproducibility by comparing CVs across registration techniques using a repeated measures analysis of variations (ANOVA) followed by post hoc analysis (Tukey’s honest significant difference (HSD) test) of pairwise comparisons.
To study the effect of registration techniques on detecting longitudinal changes, we selected a metric to quantify which technique best minimized non-physiological oscillations in longitudinal data. The metric we selected was based on the absolute rate of change in all bone parameters between time points,
where xi is the value of the bone parameter at time point i, ∆ti is the time between measurement i and measurement i − 1, and vj is the absolute rate of change between measurement i and measurement i − 1. We then calculate the standard deviation of the absolute rate of change between measurements,
where σ is standard deviation, \( \overline{v} \) is the mean absolute rate of change, and n is the number of intervals between time points (i.e., n = 4). The registration technique that provides the least oscillations, which we presume is the most representative of real physiological change, will minimize σ.
We calculated σ for each bone parameter, each participant, and each registration technique. The σ values were transformed with a logarithmic (log10) function to ensure a normal distribution. We examined differences between NR, SM, and 3DR in log-transformed σ values for all bone parameters through a repeated measures ANOVA followed by post hoc analysis (Tukey’s HSD test) of pairwise comparisons. All statistical analyses were performed (Stata, v15.0) with statistical significance set at p < 0.05.
Results
Short-term reproducibility
CVRMS (%) and LSC (parameter units) for the reproducibility analysis are presented in Table 1. The common region was lower at the radius (96% for SM, 95% for 3DR) than at the tibia (98% for both SM and 3DR). At the radius, the misalignment between the longitudinal axes of the baseline and follow-up scans ranged from 0.09 to 4.65°, with an average misalignment of 1.15°. At the tibia, the misalignment was generally smaller than the radius and ranged from 0.03 to 3.23° with an average misalignment of 1.02°.
SM and 3DR significantly improved reproducibility compared with NR for all parameters except Ct.Po and Tb.Th at the radius (Table 1). At the tibia, only the reproducibility of Tb.BMD, Ct.Th, and Tb.BV/TV was improved with either SM or 3DR compared with NR. In general, with SM and 3DR, we noticed the greatest improvement in reproducibility for density measures, particularly at the radius where angular alignment between scans was worse. There were generally less improvements using SM or 3DR compared with NR for microarchitectural parameters. Short-term reproducibility did not differ between SM and 3DR for any parameter.
Longitudinal changes in density and microarchitectural parameters
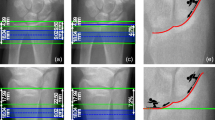
Four representative cases of individuals’ longitudinal changes in Tt.BMD and the corresponding absolute rate of change v are presented in Fig. 3. If the rate of change in Tt.BMD is smooth (i.e., there are small or no oscillations in Tt.BMD), v is relatively constant across time intervals. Participants A and D show large oscillations in Tt.BMD with NR, some oscillations with SM, and the least oscillations with 3DR. This is illustrated in Fig. 3, where v of Tt.BMD is most constant with 3DR. Participant B shows similar oscillations in Tt.BMD between SM and 3DR, while NR demonstrates large oscillations. Participant C has similar oscillations in Tt.BMD for all registration techniques. The oscillation in v is captured by the standard deviation σ of v. Smaller oscillations in Tt.BMD correspond to a lower σ, indicating changes over time that are more likely representative of true physiological adaptation.
Difference in measured Tt.BMD (left column) and the absolute rate of change v of Tt.BMD (right column) for three participants. NR no registration, SM slice match registration, 3DR 3D registration, Tt.BMD total bone mineral density, M00 month 0, baseline, M06 month 6, M12 month 12, M24 month 24, M36 month 36
The group differences in σ of Tt.BMD at the radius and tibia are illustrated in Fig. 4. At the radius, 3DR resulted in significantly lower σ than NR and SM. At the tibia, there are no significant group differences between registration techniques.
Average standard deviation (σ) of the absolute rate of change (v) for Tt.BMD over all participants (N = 40). The error bars represent the standard deviation over all participants of σ. Significance between groups is denoted as single asterisk (p < 0.05) and double asterisks (p < 0.001). NR no registration, SM slice-match registration, 3DR 3D registration
The group differences in σ for all bone parameters are presented in Table 2. At the radius, most parameters demonstrated a significant improvement in σ between NR and SM, as well as between NR and 3DR. At the tibia, there are few improvements in σ using SM or 3DR compared with NR, with the exception of Ct.Th and Tb.Ar, suggesting that registration is not as critical for studying longitudinal trends at the tibia. Nevertheless, at the tibia, σ of Tt.BMD showed a similar trend as at the radius with improvement in 3DR compared with NR (p = 0.054). At the tibia, 3DR only demonstrated improved σ over SM for Tb.BV/TV.
At the radius, the misalignment between the longitudinal axes of the baseline and follow-up scans ranged from 0.28 to 8.77°, with an average misalignment of 2.03°. At the tibia, the misalignment ranged from 0.27 to 7.73° with an average misalignment of 2.74°.
Discussion
This study demonstrates that registration improves short-term reproducibility and the estimate of longitudinal changes in bone density and structure with HR-pQCT. By using 3-year longitudinal data, we found that both SM and 3DR improved our ability to detect realistic longitudinal changes for most parameters compared with NR at both skeletal sites. Equally, at the radius, where repositioning is more challenging compared with the tibia, there were benefits of using 3DR compared with SM, particularly for density estimates.
Previously reported short-term reproducibility scores for HR-pQCT measurements using SM [12, 14,15,16, 31, 32] and 3DR [12, 16] are consistent with the present study. MacNeil and Boyd reported enhanced short- (1-week) and long-term (4-month) reproducibilities in most parameters using 3DR compared with SM; however, none of the improvements was statistically significant [16]. Ellouz and colleagues reported significant improvement in short-term reproducibility using SM and 3DR compared with no registration for Ct.Th and Tt.BMD at the radius and Ct.Th at the tibia [12]. The data we provided are a comprehensive assessment of short-term reproducibility for the second-generation HR-pQCT scanner and provide a basis to assess the LSC.
Similar to previous studies investigating registration methods [12, 16], the reproducibility of tibia measurements was better than radius measurements. This could be due to the tibia being a larger bone than the radius, the greater variance of the radius bone along its length [33], the difficulty of positioning the radius, or the fact that motion artifact is more prevalent at the radius. In general, we observed more improvements in short-term reproducibility with registration at the radius than at the tibia. The reproducibility of tibia measurements with NR is closer to machine reproducibility and therefore does not benefit as dramatically by using registration. Regardless, it is usually more convenient to analyze both skeletal sites using the same methodology. As there is no increase in reproducibility scores with registration at the tibia (i.e., there is no downside to registration), it may be beneficial to implement registration at both sites to accrue the benefits at the radius.
The reproducibility of some microarchitectural parameters benefits less from registration than others. For example, we observed consistently poor reproducibility in Ct.Po for all registration techniques, with no improvement using SM or 3DR. This is reflected in the high LSC values reported for Ct.Po [12, 25, 31], and it suggests that the resolution limits of HR-pQCT may be a more important factor affecting reproducibility than finding common regions of interest by registration techniques. It is possible that some parameters do not benefit from registration because they are a feature of the skeletal site and do not vary greatly with spatial position [33]. For example, if Ct.Po does not vary along the length of the radius or tibia, then finding a common volume through registration cannot improve its reproducibility. On the other hand, Ellouz and colleagues noted significant improvements in Ct.Po using 3DR compared with SM at both the radius and the tibia [12]. The degree of spatial variation may differ depending on the population, which may explain the different findings in Ct.Po for Ellouz and colleagues from our own (i.e., 21–47 years in Ellouz et al. vs 55–70 years in this study). It is possible that with different populations or scan conditions, registration may show benefit in these parameters. It is therefore safer to use either SM or 3DR techniques even for parameters that did not benefit in this study.
To study whether SM and 3DR detect changes in longitudinal data that represent physiological remodeling, we used a metric based on the intra-participant standard deviation, σ, of the absolute rate of change, v, in 3-year longitudinal measurements of bone parameters. Without a ground truth, it is challenging to assess which technique is the most appropriate for measuring longitudinal change. We conceptualized this metric based on the assumption that from one time point to the next, the variation of change should be minimized. In other words, wild oscillations over a 3-year period cannot represent a true physiological adaptation of bone. Figure 3 shows two examples where the oscillations are clearly not physiological (see participants A and D) and another where all three methods of NR, SM, and 3DR produce similar results (see participant C). We expect the large oscillations in participants A and D with NR are the result of measuring very different volumes of bone between time points. The oscillations decrease with SM by reducing translational error in the longitudinal direction, and the oscillations decrease even further with 3DR by reducing translational and rotational errors in six degrees of freedom. Participant B likely had primarily translational misalignment between scans, and participant C likely had small repositioning error in all degrees of freedom.
At the radius, we observed a significant decrease in σ for most bone parameters using both SM and 3DR compared with NR. Comparing SM and 3DR directly, the reproducibility was significantly better for Tt.BMD at the radius, while a non-significant trend was found (p = 0.054) between 3DR and NR for Tt.BMD at the tibia. As total density is an important outcome measure for HR-pQCT, it is reasonable to suggest using 3DR over SM to ensure the reproducibility of that parameter is maximized. It is possible that improvements using 3DR may have been found for microarchitectural measures too, but that the increased motion at the radius compared with the tibia masked our ability to measure those parameters. The sensitivity to motion artifact is less for density measures, particularly Tt.BMD, than for microarchitectural measures.
The benefits of using registration techniques are largely affected by the skills of the medical radiation technologists performing the HR-pQCT scans, and the benefits of registration diminish as the quality of the image acquisition improves. If the error at each time point is purely translational, then SM is equally beneficial as 3DR. However, it is unlikely that repositioning results exclusively in translational errors. We found that in addition to translational errors, there was an angular error about the longitudinal axis at the radius (0.09–8.77°) and tibia (0.03–7.73°), in which case 3DR is beneficial. Experienced medical radiation technologists can minimize the angular and translational positioning error. Although our two imaging technicians had similar precision scores, it is important when designing a study to consider minimizing bias that results from varying skill levels. As such, registration remains a valuable tool to ensure consistency of results.
Our study has limitations that should be noted. First, motion artifact is a source of error during in vivo HR-pQCT image acquisition, and increased participant movements introduce artifactual errors [34] that cannot be corrected through registration techniques. Another limitation is that our sample consisted only of adults between 55 and 70 years meaning our findings may have been different for a younger cohort with high-quality bone or an osteoporotic cohort with extremely poor-quality bone. We reported most of the parameters typically measured using HR-pQCT, but area measurements were only reported for NR and SM. With 3DR, the common volume results in a complex mask which affects areal measurements in slices at the ends of the VOI. If areal measurements are necessary with 3DR, they could be taken on a subsample of complete cross-sectional slices near the middle of the common region. Similarly, we do not report finite element results since it is challenging to conduct a standardized test due to the complex proximal and distal surfaces of the common region from 3DR. The benefit of 3DR over SM may not be evident given these challenges and the additional analysis steps required. However, once the appropriate software is developed, 3DR is as efficient and simple to implement as SM.
In conclusion, 3DR and SM improve short-term reproducibility compared with NR for most HR-pQCT parameters at the radius and tibia. There are no significant differences in short-term reproducibility between SM and 3DR for density or microarchitectural parameters. Equally, SM and 3DR most closely captured physiological-like longitudinal changes in bone parameters at the radius and tibia. 3DR in particular demonstrates improved detection of density measures and should be used in longitudinal studies to aid in studying changes in bone quality over time.
Data availability
The data are not publicly available. Upon reasonable requests to the corresponding author, data may be shared.
References
Macdonald HM, Nishiyama KK, Kang J, Hanley DA, Boyd SK (2011) Age-related patterns of trabecular and cortical bone loss differ between sexes and skeletal sites: a population-based HR-pQCT study. J Bone Miner Res 26:50–62
Khosla S, Riggs BL, Atkinson EJ, Oberg AL, McDaniel LJ, Holets M, Peterson JM, Melton LJI (2006) Effects of sex and age on bone microstructure at the ultradistal radius: a population-based noninvasive in vivo assessment. J Bone Miner Res 21:124–131
Dalzell N, Kaptoge S, Morris N, Berthier A, Koller B, Braak L, van Rietbergen B, Reeve J (2009) Bone micro-architecture and determinants of strength in the radius and tibia: age-related changes in a population-based study of normal adults measured with high-resolution pQCT. Osteoporos Int 20:1683–1694
Burt L, Liang Z, Sajobi TT, Hanley DA, Boyd SK (2016) Sex- and site-specific normative data curves for HR-pQCT. J Bone Miner Res 31:2041–2047
Varga P, Baumbach S, Pahr D, Zysset PK (2009) Validation of an anatomy specific finite element model of Colles’ fracture. J Biomech 42:1726–1731
Christen D, Melton LJI, Zwahlen A, Shreyasee A, Khosla S, Müller R (2013) Improved fracture risk assessment based on nonlinear micro-finite element simulations from HRpQCT images at the distal radius. J Bone Miner Res 28:2601–2608
Samelson EJ, Broe KE, Xu H, Yang L, Boyd S, Biver E, Szulc P, Adachi J, Amin S, Atkinson E, Berger C, Burt L, Chapurlat R, Chevalley T, Ferrari S, Goltzman D, Hanley DA, Hannan MT, Khosla S, Liu CT, Lorentzon M, Mellstrom D, Merle B, Nethander M, Rizzoli R, Sornay-Rendu E, van Rietbergen B, Sundh D, Wong AKO, Ohlsson C, Demissie S, Kiel DP, Bouxsein ML (2019) Cortical and trabecular bone microarchitecture as an independent predictor of incident fracture risk in older women and men in the Bone Microarchitecture International Consortium (BoMIC): a prospective study. Lancet Diabetes Endocrinol 7:34–43
Rizzoli R, Chapurlat RD, Laroche J-M, Krieg MA, Thomas T, Frieling I, Boutroy S, Laib A, Bock O, Felsenberg D (2012) Effects of strontium ranelate and alendronate on bone microstructure in women with osteoporosis. Osteoporos Int 23:305–315
Seeman E, Delmas PD, Hanley DA, Sellmeyer D, Cheung AM, Shane E, Kearns A, Thomas T, Boyd SK, Boutroy S, Bogado C, Majumdar S, Fan M, Libanati C, Zanchetta J (2010) Microarchitectural deterioration of cortical and trabecular bone: differing effects of denosumab and alendronate. J Bone Miner Res 25:1886–1894
Tsai JN, Nishiyama KK, Lin D, Yuan A, Lee H, Bouxsein ML, Leder BZ (2017) Effects of denosumab and teriparatide transitions on the bone microarchitecture and estimated strength: the DATA-Switch HR-pQCT study. J Bone Miner Res 32:2001–2009
Burt LA, Billington EO, Rose MS, Raymond DA, Hanley DA, Boyd SK (2019) Effect of high-dose vitamin D supplementation on volumetric bone density and bone strength. JAMA 322:736–745
Ellouz R, Chapurlat R, van Rietbergen B, Christen P, Pialat JB, Boutroy S (2014) Challenges in longitudinal measurements with HR-pQCT: evaluation of a 3D registration method to improve bone microarchitecture and strength measurement reproducibility. Bone 63:147–157
Boyd SK, Moser S, Kuhn M, Klinck RJ, Krauze PL, Müller R, Gasser JA (2006) Evaluation of three-dimensional image registration methodologies for in vivo micro-computed tomography. Ann Biomed Eng 34:1587–1599
Bonaretti S, Vilayphiou N, Chan CM et al (2017) Operator variability in scan positioning is a major component of HR-pQCT precision error and is reduced by standardized training. Osteoporos Int 28:247–257
Chiba K, Okazaki N, Kurogi A, Isobe Y, Yonekura A, Tomita M, Osaki M (2018) Precision of second-generation high-resolution peripheral quantitative computed tomography: intra- and intertester reproducibilities and factors involved in the reproducibility of cortical porosity. J Clin Densitom 21:295–302
MacNeil JA, Boyd SK (2008) Improved reproducibility of high-resolution peripheral quantitative computed tomography for measurement of bone quality. Med Eng Phys 30:792–799
Shi L, Wang D, Hung VWY, Yeugn BHY, Griffith JF, Chu WCW, Heng PAH, Cheng JCY, Qin L (2010) Fast and accurate 3-D registration of HR-pQCT images. IEEE Trans Inf Technol Biomed 14:1291–1297
Waarsing JH, Day JS, van der Linden JC, Ederveen AG, Spanjers C, De Clerck N, Sasov A, Verhaar JAN, Weinans H (2004) Detecting and tracking local changes in the tibiae of individual rats: a novel method to analyse longitudinal in vivo micro-CT data. Bone 34:163–169
Nishiyama KK, Campbell GM, Klinck RJ, Boyd SK (2010) Reproducibility of bone micro-architecture measurements in rodents by in vivo micro-computed tomography is maximized with three-dimensional image registration. Bone 46:155–161
Lan S, Luo S, Huh BK, Chandra A, Altman AR, Qin L, Liu XS (2013) 3D image registration is critical to ensure accurate detection of longitudinal changes in trabecular bone density, microstructure, and stiffness measurements in rat tibiae by in vivo microcomputed tomography (muCT). Bone 56:83–90
Campbell GM, Tiwari S, Grundmann F, Purcz N, Schem C, Glüer C-C (2014) Three-dimensional image registration improves the long-term precision of in vivo micro-computed tomographic measurements in anabolic and catabolic mouse models. Calcif Tissue Int 94:282–292
Goshtasby A (2005) 2-D And 3-D Image Registration For Medical, Remote Sensing, And Industrial Applications. Hoboken, N.J.: Wiley. pp 63–154
Burt LA, Gaudet S, Kan M, Rose MS, Billington EO, Boyd SK, Hanley DA (2018) Methods and procedures for: a randomized double-blind study investigating dose-dependent longitudinal effects of vitamin D supplementation on bone health. Contemp Clin Trials 67:68–73
Pauchard Y, Liphardt AM, Macdonald HM, Hanley DA, Boyd SK (2012) Quality control for bone quality parameters affected by subject motion in high-resolution peripheral quantitative computed tomography. Bone 50:1304–1310
Manske SL, Davison EM, Burt LA, Raymond DA, Boyd SK (2017) The estimation of second-generation HR-pQCT from first-generation HR-pQCT using in vivo cross-calibration. J Bone Miner Res 32:1514–1524
Manske SL, Zhu Y, Sandino C, Boyd SK (2015) Human trabecular bone microarchitecture can be assessed independently of density with second generation HR-pQCT. Bone 79:213–221
Burghardt AJ, Kazakia GJ, Ramachandran S, Link TM, Majumdar S (2010) Age- and gender-related differences in the geometric properties and biomechanical significance of intracortical porosity in the distal radius and tibia. J Bone Miner Res 25:983–993
Buie HR, Campbell GM, Klinck RJ, MacNeil JA, Boyd SK (2007) Automatic segmentation of cortical and trabecular compartments based on a dual threshold technique for in vivo micro-CT bone analysis. Bone 41:505–515
Glüer C-C (1999) Monitoring skeletal changes by radiological techniques. J Bone Miner Res 14:1952–1962
Bonnick SL, Johnston CC Jr, Kleerekoper M, Lindsay R, Miller P, Sherwood L, Siris E (2001) Importance of precision in bone density measurements. J Clin Densitom 4:105–110
Burghardt AJ, Buie HR, Laib A, Majumdar S, Boyd SK (2010) Reproducibility of direct quantitative measures of cortical bone microarchitecture of the distal radius and tibia by HR-pQCT. Bone 47:519–528
Burghardt AJ, Pialat J-B, Kazakia GJ, Boutroy S, Engelke K, Patsch JM, Valentinitsch A, Liu D, Szabo E, Bogado CE, Zanchetta MB, McKay HA, Shane E, Boyd SK, Bouxsein ML, Chapurlat R, Khosla S, Majumdar S (2013) Multicenter precision of cortical and trabecular bone quality measures assessed by high-resolution peripheral quantitative computed tomography. J Bone Miner Res 28:524–536
Boyd SK (2008) Site-specific variation of bone micro-architecture in the distal radius and tibia. J Clin Densitom 11:424–430
Pauchard Y, Ayres FJ, Boyd SK (2011) Automated quantification of three-dimensional subject motion to monitor image quality in high-resolution peripheral quantitative computed tomography. Phys Med Biol 56:6523–6543
Acknowledgments
The authors thank J.M. Allan, J.A. Allan, S. Gaudet, M. Kan, and B. Love for participant recruitment and A. Cooke, S. Kwong, and D. Raymond for scan acquisition and analysis. The authors also thank the study participants for generously donating their time to support our research.
Funding
This study was funded by Pure North S’Energy Foundation in response to an investigator-initiated research proposal.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The study was approved by the Conjoint Health Research Ethics Board at the University of Calgary and Health Canada. All participants gave written consent before participating in the study.
Conflicts of interest
TDK, CMJdB, LG, DAH, and LAB have nothing to declare. EOB has previously received honoraria from Amgen and Eli Lilly and a research grant from Amgen. SKB has received honorariums from Amgen and Servier and is co-owner of Numerics88 Solutions Inc.
Ethics approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kemp, T., de Bakker, C., Gabel, L. et al. Longitudinal bone microarchitectural changes are best detected using image registration. Osteoporos Int 31, 1995–2005 (2020). https://doi.org/10.1007/s00198-020-05449-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00198-020-05449-2