Abstract
A highly sensitive, precisely specific, environmentally friendly, high-throughput, microwell-plate and microchip-based sandwich assay was developed to detect HIV-1 p24 antigen, a protein biomarker using fluorescent carbon dots. High quantum yield carbon dots were synthesized using citric acid and ethylenediamine as carbon and nitrogen sources by a single-step hydrothermal reaction. The desired amine groups confirmed by FTIR on the carbon dots were coupled to streptavidin by amine–amine coupling reaction using glutaraldehyde. The detection range of the carbon dot based immunoassay (CDIA) was found to be between 20 and 1000 pg/mL in a linear dose-dependent manner. CDIA tested for HIV negative plasma samples showed no false positive results in the detection of HIV-1 p24 antigen. The CDIA was extended to develop a microfluidic carbon dot immunoassay (μCDIA) which exhibited analytical sensitivity in the range of 30–1000 pg/mL. The CDIA and μCDIA can easily be adapted to a lab-on-a-chip platform for use in resource limited settings and can also be multiplexed for the detection of other pathogens like TB and Hepatitis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
In recent years, fluorescence-based immunoassays have become powerful tools in the field of clinical diagnosis, microbial ecology and biochemical studies for the visualization of various biological events such as antigen–antibody binding, enzyme–substrate reactions and in drug discovery (Feng et al. 2003; Shen et al. 2014). In a biosensor, fluorophores are molecular transducers that give out information in the form of light when a particular chemical or biological reaction occurs (Hötzer et al. 2012). Traditionally, organic fluorophores are widely used as fluorescent labels due to their small size, relatively high fluorescence intensity and their commercial availability for various fluorescent immunoassays (Härmä et al. 2011). But organic dyes are vulnerable to photobleaching and always confine their detection capabilities. In recent decades, semiconductor quantum dots (QDs) have become preferred fluorophores because of their tunable emission wavelengths, good photostability, high quantum yield, narrow emission and large Stokes shift (Barroso 2011). QDs such as CdSe (Zhao et al. 2014) and CdTe have been successfully employed to demonstrate various biological processes at the molecular level, and their bio-sensing applications have been studied extensively (Fang et al. 2013; Zhu et al. 2014b). However, preparation, storage and stability methods of these traditional QDs are cumbersome. In addition, the blinking nature and heavy metal toxicity limit their use for biological applications. Therefore, selecting an environmentally friendly material with excellent photoluminescent property as fluorescent label for the sensitive and selective antigen or antibody detection is in ever-increasing demand. Carbon dots (CDs) are newly emerged fluorescent carbon nanoparticles with combined advantages of both semiconducting quantum dots and organic dyes.
CDs are quasi-spherical nanoparticles (<10 nm) with good water solubility (Wang et al. 2014), high quantum yield (Zhu et al. 2013), low toxicity (Yang et al. 2009) and high resistance to photobleaching (Sun et al. 2006). They have been explored for cell imaging (Shi et al. 2015), bio-sensing (Yang et al. 2013), metal ion sensing (Liu et al. 2011) and catalytic applications (Zuo et al. 2015). Arc discharge, laser ablation, thermal cracking of organic compounds, electrochemical oxidation, microwave-mediated synthesis, oxidation of candle soot and hydrothermal routes were used for the synthesis of CDs (Dong et al. 2012; Li et al. 2013; Tang et al. 2012). But hydrothermal reaction is the simplest and most inexpensive method to prepare functionalized CDs in a single step (He et al. 2015).
CDs have been studied for the detection of small biological molecules such as glutathione, tetracyclines and cysteine in clinical samples (Amjadi et al. 2015; Feng et al. 2015; Jana et al. 2015; Shi et al. 2014). Zhu et al. (2014a) used CDs as fluorescent labels in fluorescence immunoassay platform to label the goat anti-human IgG (gIgG) to obtain the CDs–gIgG conjugate, and quantitative measurements of IgG in human sera have been demonstrated in clinical diagnosis for the detection of human immunoglobulin. In this study, the detection is based on fluorescence enhancement caused by the surface polarity of antigen–antibody immunocomplex which is very specific to the proposed system. Wang et al. (2015) demonstrated the combination of CDs and aptamers as a novel fluorescence probe for rapid and sensitive detection of Salmonella typhimurium concentration in solutions of eggshell and tap water with CDs-apt applied as fluorescent probes for bacterium quantitative detection with a detection limit down to 50 cfu/mL. Wu et al. developed a carbon dot (C-Dot)-Linked well-plate-based Immunosorbent Assay for the Detection of Human α-Fetoprotein (AFP) in the concentration range of 0–350 ng/mL of AFP. The fluorescence intensities detected from the CDs on these sandwich immune complexes were positively correlated with the concentrations of AFP antigen (Wu et al. 2015). The only report on the CDs-based immunoassay for detecting p24 antigen was that of the patent filed by Min et al. is relating to a preparation method of a carbon quantum dot test paper strip for detecting a p24 antigen preparing a p24 carbon quantum dot pad and a p24 nitrocellulose membrane and a qualitative analysis (Min et al. 2013). All the existing CDs-based immunoassays referred above are limited either by specific application or sensitivity limits or lack of comprehensive evaluation for immediate application. The present CDIA has the advantage of the use of streptavidin-conjugated CDs which can be adapted easily for the detection of different targets in addition to the present p24 antigen.
p24 antigen is the most abundant viral protein of human immunodeficiency virus (HIV-1) since the capsid contains ~2000 molecules per virus. It is a potential viral marker for the diagnosis of HIV and blood donor screening due to its relatively high levels in blood during early and late stages of infection. There are currently many available diagnostic kits with accuracy of 99.5%, but all of them target antibodies produced by the host which generally takes 3–6 months to appear in the blood (Teeparuksapun et al. 2010). PCR (polymerase chain reaction), a nucleic acid (DNA and RNA) test, is the gold standard for diagnosing HIV but is limited to central laboratories owing to its cost and high complexity. Enzyme-linked immunosorbent assays (ELISAs) are a common primary screening tool in developed countries, but once again their availability in resource-limited setting is restricted due to infrastructure requirements (Haleyur Giri Setty and Hewlett 2014). Previously, we have also studied bio-barcode amplification assay (gold nanoparticles, 0.1–500 pg/mL) and fluorescence immunoassay (europium nanoparticles, 0.5–500 pg/mL) for the sensitive and specific detection of p24 antigen (Tang and Hewlett 2010; Tang et al. 2010). However, the requirement for short oligonucleotides, sophisticated instruments for bio-barcode amplification assay and the cost of rare earth-based europium nanoparticles for fluorescence immunoassay created the need to develop and evaluate new immunoassays.
In the present work, CDs with high quantum yield were synthesized using citric acid and ethylenediamine as carbon and nitrogen sources by a single-step hydrothermal reaction. These CDs have been used as fluorescent labels to develop a highly sensitive, precisely specific, environmentally friendly, high-throughput, carbon dot based immunoassay (CDIA) to detect HIV-1 p24 antigen on both microwell plates and microfluidic chip. With a motivation to provide better health care through point-of-care (POC) tests in resource-limited settings, we translated this bench-top CDIA onto microfluidic chip and also evaluated its sensitivity. Furthermore, the sensitivity of the CDIA and carbon dot based immunoassay μCDIA for p24 antigen in human plasma has been demonstrated through quantitative measurements, resulting in a promising potentially new assay for clinical diagnosis.
2 Experimental section
2.1 Materials
Anhydrous citric acid (99.5%), ethylenediamine and Triton-x-100 were purchased from Sigma. Quinine sulfate dehydrate, sodium borohydride and glutaraldehyde (50%, v/v) were supplied by Himedia. All chemicals used were of analytical grade and used as received. Centrifugal spin columns (10, 30, 100 kDa) were obtained from GE Health Care, UK. Dow Corning Sylgard 184 s polydimethylsiloxane (PDMS) kit was ordered from Ellsworth Adhesives (Germantown, WI, USA). Microfluor 96-well microtiter plates were bought from Thermo Scientific, USA. Phosphate-buffered saline (PBS; 100 mM, pH = 7.2) and Carbonate–bicarbonate buffer (500 mM, pH 9.5) were prepared in the laboratory by following established procedures. Casein Block buffer and wash buffer (0.1% tween-20 in PBS, PBST) was purchased from Thermo Scientific. Double-distilled water was used for all the experiments. HIV-1 p24 antigen (Virogen, USA), C65690M anti-p24 antibody from mouse (Prospec, USA) samples and HIV-1 detector antibody from mouse (biotinylated) were purchased from PerkinElmer (USA). Streptavidin was supplied by Scripps Laboratories, USA.
We obtained all the plasma samples (HIV positive, HBV positive, HCV positive and healthy adults plasma) from Department of Microbiology, Sri Sathya Sai Institute of Higher Medical Sciences, Prasanthigram, India, after approval from the Institutional Ethics Committee (SSSIHL/IEC/PSN/BS/2012/01). For clinical studies, all details of the samples were coded and tested in a blinded manner. All of them were confirmed again using a fourth-generation Microlisa HIV Ag + Ab Elisa kit (J.Mitra & Co Pvt Ltd).
2.2 Synthesis of carbon dots
In a single-step hydrothermal method, amine-functionalized CDs were synthesized from citric acid as carbon source and ethylenediamine as nitrogen source (Zhu et al. 2013). In this procedure, 2.0 mg of citric acid and 2.6 mL of ethylene diamine (1:4 molar ratio) were dissolved in 50 mL of double-distilled water. The solution was then transferred to a 100-mL Teflon-lined autoclave and heated in an oven at 200 °C for 6 h. The reaction mixture was cooled to room temperature, and the solution was collected into a beaker for further purification. This crude extract was first filtered through a 0.2 µm membrane for the removal of any large particles that may be present and the solution was lyophilized. These purified CDs were dissolved with water (10 mg/mL) and stored under dark conditions until further experiments.
2.3 Conjugation of streptavidin to CDs
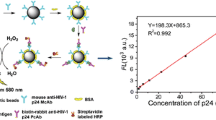
Streptavidin was conjugated to CDs following the procedure reported by Wu et al. (2015). 5 mL of the stock solution (150 mg/mL) of purified CDs was mixed with 1 mL of carbonate–bicarbonate buffer (100 mM, pH 9.5). To this solution, freshly prepared 0.3 mL of 5% (w/v) glutaraldehyde solution (diluted 50% glutaraldehyde (w/v) with water 10 times) was added dropwise with vigorous stirring. The reaction was continued by stirring at room temperature for 1 h. In the next step, 1 mL of streptavidin solution (1 mg/mL in PBS) was added slowly dropwise with stirring. This conjugation mixture was stirred at room temperature for 1 h after which it was transferred to an ice bath. Then, 1.5 mL of freshly prepared sodium borohydride solution (15 mg/mL) was added to the reaction with stirring. This reduction reaction was left overnight at 4 °C. In the next step, streptavidin-conjugated CDs (CD-SA) were isolated by centrifugal spin columns (10 kDa, 30 kDa MWCO) and washed several times with PBS buffer. Finally, the CD-SA solution was diluted to 1 mg/mL with PBS buffer and stored at 4 °C under dark conditions for immunoassay experiments both on microplate and microfluidic chips.
2.4 Measurement of fluorescence polarization
Fluorescence polarization (FP) allows rapid and quantitative analysis of diverse molecular interactions and enzyme activities. The principle of FP derives from the degree of polarization of a fluorophore which is inversely related to its molecular rotation. Therefore, low molecular weight fluorescent species rotate rapidly resulting in low FP values. However, if the fluorescent species are conjugated with large biomolecules such as proteins, antibodies and enzymes, a higher FP value results (Moerke 2009). Fluorescence polarization is expressed as
where I ∥ and I ⊥ represent the horizontally polarized and vertically polarized emission intensities, respectively. G is the correction factor calculated for the sensitivity of the instrument.
FP mode was selected to measure the I ∥ and I ⊥ values for both CDs and streptavidin-conjugated CDs (CD-SA) by exciting them with 340 nm wavelength and measuring their emission at 440 nm wavelength. The corresponding polarization values were calculated from the FP equation. Polarization value is a ratio and does not change with fluorescence intensity; therefore, different concentrations of samples should have the same FP value.
2.5 Characterization
The morphology and size of the CDs was investigated by transmission electron microscope (TEM, JEOL, 2100F electron microscope operated at 200 kV). The TEM sample was prepared by drop casting diluted solution of CDs on to a carbon-coated copper grid and dried under vacuum before the analysis. Surface functionalization was confirmed by Fourier transform infrared (FTIR) spectroscopy (SHIMADZU IR Affinity-1) using KBr pellet method. UV–Vis absorption spectra were obtained using Shimadzu UV-2450 UV–Vis spectrophotometer. The fluorescence emission spectra of nanoparticles in the UV and visible regions were measured by Spectramax M5, multimode microplate reader. The same reader was also used to measure fluorescence for detection of HIV-1 p24 antigens.
2.6 CDIA for the detection of p24 antigen
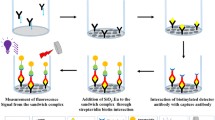
In this study, we have chosen sandwich immunoassay in which p24 antigen was sandwiched between primary and secondary antibodies on polystyrene microtiter wells. The fluorescence signal was collected from the CDs bound to the secondary antibodies. We chose physical adsorption since it is a simple and reproducible method. Hydrophobicity and surface charge of microplates are the main factors for the adsorption of capture antibodies (Qian et al. 2000). The high binding capacity plate has a small number of ionic carboxyl groups resulting in a slightly ionic and hydrophobic surface. The hydrophobic and ionic interactions between the plate and biomolecules facilitate the physical adsorption of antibodies onto the plate (Gibbs and Kennebunk 2001). A capture antibody concentration of 4 µg/mL was prepared by diluting with carbonate–-bicarbonate buffer (100 mM, pH 9.6). 100 µL of this solution was coated on to the black color fluorescence microplates and incubated for 36 h at 4 °C. After incubation, all the wells were washed five times with wash buffer after which 300 µL/well casein blocking buffer was added. Incubation was continued for 30 min at 37 °C to ensure the blocking of nonspecific adsorption sites. Stock antigen p24 solution was diluted with block buffer or plasma sample to prepare different concentrations of antigen. 100 µL of the antigen was added to each well and incubated at 37 °C with shaking for 1 h. After washing all the wells for five times with wash buffer, detector antibody conjugated to biotin (100 µL/well) was added and incubated for 30 min at 37 °C. To this antibody–antigen–antibody complex, 100 µL of CD-SA (0.1 mg/mL) was added to each well and the mixture was incubated again for 30 min at 37 °C with shaking. This was followed by final washing with PBST buffer for five times to avoid nonspecific interactions and reduce background noise. Finally, fluorescent signals from the sandwich immunocomplexes were read with the Spectramax microplate reader (excitation at 340 nm and emission at 440 nm) directly from the microplate/microchip in the end point format. All experiments were performed in triplicate. A schematic representation of the detection of HIV-1 p24 antigen by CD-SA is shown in Fig. 1.
Schematic illustration for the detection of HIV-1 p24 antigen by CD-SA conjugates. CDIA uses capture antibody-coated microtiter wells to capture HIV-1 p24, which is then sandwiched with secondary biotinylated anti-p24 antibody. The CD-SA conjugates recognize and bind the above biotinylated antigen–antibody complex. After extensive washing, the fluorescence signal released from the sandwiched complex is then recorded and quantified with Spectramax M5 multiplate reader
2.7 Fabrication of microfluidic chip for µCDIA
We have fabricated microfluidic chips following the procedure reported by us for the europium nanoparticle immunoassay in our previous study (Liu et al. 2014).The channels were designed by Autodesk AutoCAD, and then, the templates were made by computer-numerical-control (CNC) milling machine. The microfluidic channels were machined on a 100 mm × 100 mm × 1.5 mm acrylic plaque mounted on the CNC stage. PDMS prepolymer was mixed with its curing agent at 10:1 weight ratio and poured on the acrylic plaque to replicate the microchip pattern. PDMS was thermally cured at 65 °C for 1 h, and the resultant elastomeric pad with protruded pattern was subjected to corona discharge activation for 2.5 min. Immediately after treatment, the surface of the patterned PDMS pad was passivated using vapor of (tridecafluoro-1,1,2,2-tetrahydrooctyl)-trichlorosilane in a vacuum desiccator for 2 h. To finish device fabrication, a 1-mm-thick layer of PDMS was partially cured at 65 °C for 18 ~ 20 min and attached to a freshly prepared microchip replicate to seal its open channels. The assembly was cured at 65 °C overnight before use.
The microfluidic chip contains 12 independent reactors with volume of 20 μL each. The microfluidic channels have the channel width: 254 µm; depth: 150 µm, and serpentine-channel bioreactor have the channel width: 1000 µm; depth: 750 µm; and length: 29 mm.
As shown in Fig. 2, our present microfluidic chip contains 12 independent microreactors. The microreactors are serpentine channels, and the total volume of each reactor is 20 µL. We could use micropippete for loading the reagents into the reactors with the help of adapters provided on each reactor.
2.8 µCDIA for the detection of p24 antigen
We extended our CDIA onto a microfluidic chip to develop into point-of-care device for resource-limited settings. To implement CDIA on microfluidic chip, we chose physical adsorption method to immobilize capture antibodies onto PDMS surface. For µCDIA studies, in the first step, 100 μg of capture antibody was dissolved in 1 mL of PBS (100 mM, pH = 7.2) buffer. This 100 μg/mL capture antibody was diluted to 4 μg/mL with PBS buffer. Then, 20 μL of 4 μg/mL capture antibody solution was loaded into each channel with micropipette and incubated at 4 °C for 36 h. This step was followed by replacing the coating antibody solution with PBST as block buffer to avoid the nonspecific adsorption of antigen on the microchannel walls. The PBST solution was removed after 40 min of incubation at 37 °C. HIV-1 p24 antigen sample solutions were loaded into PBST-blocked microchannels, and the device was kept at 37 °C for 15 min. This was followed by washing the channels with PBST buffer to remove loosely bound antigen. In the next step, 20 μL of 10 ng/mL biotinylated secondary antibody (detector antibody) was added to each channel to form antibody–antigen–antibody sandwich immunocomplex and the reaction was allowed to continue for 30 min at 37 °C. After second wash with PBST, CD-SA solution was loaded into each channel and the streptavidin–biotin coupling reaction was allowed to continue at 37 °C for 15 min. After the final wash, the device was placed onto a modified rack of spectraMax M5 for recording the fluorescent signals from the sandwich immunocomplex. In the present study, the reagent pumping into the channel was achieved manually using a rubber pipette filler (Thermo Fisher Scientific) connected to the devices through a section of PTFE tubing (1/32″ID × 1/16″ OD, Cole-Plamer, Vernon Hills, IL, USA).
3 Results and discussion
The CDs were characterized for their optical properties by UV–Vis, photoluminescence (PL) and TEM images and were described in detail in the supplementary information. FP measurements were carried out to investigate the successful functionalization of streptavidin to CDs.
Here, in our work, the binding of streptavidin to CDs has increased its size which slows down its molecular rotation compared to the carbon CDs alone. This has resulted in the higher polarization value for streptavidin-conjugated CDs compared to CDs alone. The FP values of unconjugated and streptavidin-conjugated CDs from the FP equation (Eq. 1) were calculated by measuring the P and S polarization values and are reported in Table 1. As small-sized particles have small FP value and large-sized particles have higher FP value, functionalization could easily be monitored by comparing the FP values. The FP value of the CDs-SA was higher than CDs and confirmed the conjugation of streptavidin to CDs.
3.1 Optimization of fluorescent CDIA for the detection of HIV-1 p24 antigen
p24 antigen is the most abundant viral protein in the early stages of HIV infection even when the antibody concentrations are still low. Therefore, tests were designed to detect p24 antigen at very early stages for better antiretroviral treatment (Pandori et al. 2009). In this study, we adopted p24 antigen assay to study the CDIA performance. We optimized the concentration of CD-SA by measuring the signal-to-blank ratio (S/B) of 100 pg/mL p24 using serial dilutions of CD-SA. All the measurements were repeated three times.
Figure 3a shows that there is no linear relation between the signals and CD-SA concentration. The highest S/B ratio was observed with CD-SA concentration of 2 mg/mL. However, with increase in concentration beyond 2 mg/mL, intensity of the blank increased rapidly when compared to signal which could be due to nonspecific adsorption of the CD-SA. There could be few of the CDs binding to the microplate wells even after extensive washing. Since the fluorescence quantum yield of CDs is very high, the binding of even of a few CDs to the microplate wells would contribute to the extra noise resulting in lower signal-to-noise ratio. We have used this optimized CD-SA concentration to determine the sensitivity of CDIA for the p24 antigen assay. Figure 3b shows a linear dynamic range extending from 20 to 1000 pg/mL. The correlation coefficient was calculated from the linear fit of the data between signal intensity and concentration of antigen as R 2 = 0.9957 which suggests it to be a linear dose-dependent assay. There was clear difference between the blank and signal intensities only up to 20 pg/mL beyond which it was indistinguishable. The fluorescence cut-off value was derived using the sum of the means of the signal intensity of six negative controls plus two times their standard deviation (SD). Samples with signal-to-cut-off (S/CO) ratios equal to or greater than 28.20 were considered positive for HIV-1 p24. Thus, our CDIA could detect p24 antigen reproducibly up to 20 pg/mL. The lower detection limit of the CDIA using CDs as fluorescent probes can be claimed as 20 pg/mL.
In the process of studying various factors for the optimization of CDIA’s analytical performance, we also carried out the effect of concentration of capture antibody coating on the detection of p24 antigen. We coated the microplate with capture antibody concentrations of 1, 2 and 4 µg/mL. As shown in Fig. 4, with the concentrations of 1 and 2 µg/mL, the signal response was weak and no linearity was observed. This might be because at the lower coating concentrations, there may not be enough sites available for the antigen binding when the p24 concentrations are high. But when the plate was coated with a higher concentration of 4 µg/mL, strong signal response was recorded even with lower p24 antigen concentrations. It also exhibited a linear analytical sensitivity from 20 to 1000 pg/mL. This higher dynamic range would be useful for detecting p24 antigen concentrations within days after exposure up to 4 weeks, i.e., prior to seroconversion after which the antigen becomes undetectable. Since there was good S/B ratio even at the lower concentration of p24, in this study, we chose 4 µg/mL as coating concentrations of capture antibody for all our CDIA experiments.
3.2 Analysis of HIV-1 p24 antigen in human plasma samples
To determine the sensitivity of the assay, we first tested 45 plasma samples of healthy individuals and no false positives were observed. In Fig. 5 for all the 45 samples, the scatter plot was plotted for their intensity and all of them appeared below the signal-to-cut-off ratio which confirmed no false positives. Further to evaluate the analytical sensitivity of the assay, known concentrations of p24 antigen were spiked into plasma samples collected from healthy individuals (HIV negative sample). The plasma samples were diluted 100 times for the analysis to avoid interference from other plasma proteins while reducing the sample consumption. Concentrations of 20, 50, 100, 150 and 500 pg/mL purified p24 antigen were added to the HIV negative samples. All experiments were conducted in triplicate, and average signal response was used to calculate the concentration of p24 antigen in the sample from the standard curve (Fig. 3b). From Table 2, the yielded results show that the quantification of p24 antigen by CDIA is comparable to the actual amounts spiked into sample. The higher percent recovery and lower variability of antigen in spiked plasma samples can be attributed to the washing out interferences by diluting plasma samples. Thus, CDIA can quantitatively determine the concentration of HIV-1 p24 antigen in clinical plasma samples.
To determine the specificity of the CDIA, we tested the HIV positive plasma samples with all the optimized conditions as discussed above. Again as a prerequisite, the plasma samples were diluted 100 times prior to the analysis. We tested 25 HIV positive samples (confirmed by both 3rd and 4th generation commercial ELISA kits) using CDIA, and no false negatives were observed. Initially, the three signal intensities of the each sample of the experiments conducted were averaged. Then, mean ratios were calculated by averaging all the 25 samples participated in the experiment. The mean ratio of samples over the cut-off ratio of the negative control for these samples was 124.62 ± 89.54. We also evaluated the assay with eight HBV positive/HIV negative and 10 HCV positive/HIV negative plasma samples for cross-reactivity, and their mean ratios of samples over the cut-off value of the negative controls were 25.63 ± 3.81, 19.42 ± 5.18, respectively. Figure 6a depicts the varying concentrations of fluorescence intensities of CDIA tested on HIV positive samples directly correlates with the presence of p24 in the plasma which points out the progression of the disease in the patient. As shown in Fig. 6b, the CDIA tested on eight HBV and 10 HCV samples showed fluorescence intensities below cut-off ratio. So CDIA showed no false positives when HBV and HCV samples were tested. The CDIA method for detection of p24 antigen had high specificity without interference from other antigens such as HCV and HBV samples. The low cost along with the higher sensitivity would be of an advantage for the adaption of CDIA to resource-limited settings.
a CDIA applied to 25 HIV positive samples and compared with the results from blank and a HIV negative sample. The blue, black and red bars denote the positive sample, blank and the negative samples, respectively. b CDIA applied to eight HBV positive/HIV negative and 10 HCV positive/HIV negative samples which are compared with the result from a blank and a HIV positive sample. The green, blue, black and red bars denote the HBV positive/HIV negative, HCV positive/HIV negative blank and the lowest measured positive samples, respectively
3.3 µCDIA for the detection of HIV-1 p24 antigen
With a motivation to provide better health care through point-of-care (POC) tests in resource-limited settings, we continued our efforts to translate this bench-top CDIA onto microfluidic chip. The immunoassays performed on a microfluidic chip offer several advantages over conventionally used microtiter plates. We evaluated the analytical sensitivity of the µCDIA, and it exhibits a linear dynamic range from 30 to 1000 pg/mL (Fig. 7). The correlation coefficient value R 2 = 0.9737 obtained from the linear fit of the graph suggests it as a linear dose-dependent assay. Below this concentration, we could not distinguish between the signal and background. Therefore, for µCDIA, we considered the detection limit as 30 pg/mL. Table 3 clearly shows the sample/reagent consumption reduced by fivefold except for the block buffer and wash buffer (tenfold). The assay time also shortened by about 34% without compromising on the analytical sensitivity of the assay to a significant degree. Our findings suggest that the µCDIA could be easily adapted for the detection of other infectious diseases associated with HIV such as tuberculosis and influenza. For example, we can choose for tuberculosis, antigens such as ESAT-6, CFP-10 and their corresponding antibodies to develop our µCDIA by employing streptavidin-conjugated CDs to make this microfluidic chip a useful multiplex platform.
When compared with conventional ELISA in terms of cost, µCDIA is a more cost-effective platform as shown in Table 4. The chip which supports 12 reaction zone costs $1 which is much lesser compared to $2.5 which is cost for 12 wells. Also, the significantly lower volumes in case of µCDIA bring down the cost for the reagents in µCDIA compared to conventional ELISA. More importantly, the synthesis of carbon dots by hydrothermal methods lowers the cost significantly when considered in comparison with cost of the enzyme and substrate reagents. When the total cost was compared, the estimated cost for µCDIA is less than 3 dollars which is around 3–4 times lesser than detection using a conventional ELISA platform.
4 Conclusions
The potential of the CDs as fluorophores was explored for the development of a high-throughput CDIA to detect infectious agents such as HIV with enhanced sensitivity. Synthesis of the high quantum yield CDs was made possible through using citric acid and ethylenediamine as carbon and nitrogen sources. It was demonstrated that the CDIA is capable of detecting HIV-1 p24 antigen in a broad range from 20 to 1000 pg/mL, thus giving a 100-fold enhancement of detection over the conventional colorimetric assays. This assay will be useful in the early identification and monitoring of the disease and thus help in the planning of the treatment. This highly sensitive p24 assay can also help improve blood safety by reducing the antibody negative window period in blood donors in resource-limited settings where nucleic acid testing is not practical or feasible. The µCDIA has the complete capability for development into a lab-on-a-chip platform device with high sensitivity to detect the early infections and good specificity because of the disease-specific antigen and antibody pair. This could be adapted for the detection of other antigens such as ESAT-6 and CFP-10 for tuberculosis. This could also improve the test and treat approach, by relatively reducing the time and the reagent consumption for the HIV tests.
References
Amjadi M, Abolghasemi-Fakhri Z, Hallaj T (2015) Carbon dots-silver nanoparticles fluorescence resonance energy transfer system as a novel turn-on fluorescent probe for selective determination of cysteine. J Photochem Photobiol A 309:8–14
Barroso MM (2011) Quantum dots in cell biology. J Histochem Cytochem 59(3):237–251
Dong Y, Chen C, Zheng X, Gao L, Cui Z, Yang H, Guo C, Chi Y, Li CM (2012) One-step and high yield simultaneous preparation of single- and multi-layer graphene quantum dots from CX-72 carbon black. J Mater Chem 22(18):8764–8766
Fang M, Yuan J-P, Peng C-W, Pang D-W, Li Y (2013) Quantum dots-based in situ molecular imaging of dynamic changes of collagen IV during cancer invasion. Biomaterials 34(34):8708–8717
Feng J, Shan G, Maquieira A, Koivunen ME, Guo B, Hammock BD, Kennedy IM (2003) Functionalized europium oxide nanoparticles used as a fluorescent label in an immunoassay for atrazine. Anal Chem 75(19):5282–5286
Feng Y, Zhong D, Miao H, Yang X (2015) Carbon dots derived from rose flowers for tetracycline sensing. Talanta 140:128–133
Gibbs J, Kennebunk M (2001) Immobilization principles-selecting the surface. ELISA Tech Bull 1:1–8
Haleyur Giri Setty MK, Hewlett IK (2014) Point of care technologies for HIV. AIDS Res Treat 2014:20
Härmä H, Toivonen J, Soini JT, Hänninen P, Parak WJ (2011) Time-resolved fluorescence immunoassay for C-reactive protein using colloidal semiconducting nanoparticles. Sensors 11(12):11335
He J, Lei B, Zhang H, Zheng M, Dong H, Zhuang J, Xiao Y, Liu Y (2015) Using hydrogen peroxide to mediate through a one-step hydrothermal method for the fast and green synthesis of N-CDs. RSC Adv 5(116):95744–95749
Hötzer B, Medintz IL, Hildebrandt N (2012) Fluorescence in Nanobiotechnology: sophisticated fluorophores for novel applications. Small 8(15):2297–2326
Jana J, Ganguly M, Pal T (2015) Intriguing cysteine induced improvement of the emissive property of carbon dots with sensing applications. Phys Chem Chem Phys 17(4):2394–2403
Li L, Wu G, Yang G, Peng J, Zhao J, Zhu J-J (2013) Focusing on luminescent graphene quantum dots: current status and future perspectives. Nanoscale 5(10):4015–4039
Liu M, Zhao H, Chen S, Yu H, Zhang Y, Quan X (2011) Label-free fluorescent detection of Cu(ii) ions based on DNA cleavage-dependent graphene-quenched DNAzymes. Chem Commun 47(27):7749–7751
Liu J, Du B, Zhang P, Haleyurgirisetty M, Zhao J, Ragupathy V, Lee S, DeVoe DL, Hewlett IK (2014) Development of a microchip Europium nanoparticle immunoassay for sensitive point-of-care HIV detection. Biosens Bioelectron 61:177–183
Min H, Yi P, Hao L (2013) Preparation method of carbon quantum dot test paper strip for detecting P24 antigen. Google Patents, China
Moerke NJ (2009) Fluorescence polarization (FP) assays for monitoring peptide-protein or nucleic acid-protein binding. Curr Protoc Chem Biol 1:1–15
Pandori MW, Hackett J, Louie B, Vallari A, Dowling T, Liska S, Klausner JD (2009) Assessment of the ability of a fourth-generation immunoassay for human immunodeficiency virus (HIV) antibody and p24 antigen to detect both acute and recent HIV infections in a high-risk setting. J Clin Microbiol 47(8):2639–2642
Qian W, Yao D, Yu F, Xu B, Zhou R, Bao X, Lu Z (2000) Immobilization of antibodies on ultraflat polystyrene surfaces. Clin Chem 46(9):1456–1463
Shen J, Li Y, Gu H, Xia F, Zuo X (2014) Recent development of sandwich assay based on the nanobiotechnologies for proteins, nucleic acids, small molecules, and ions. Chem Rev 114(15):7631–7677
Shi Y, Pan Y, Zhang H, Zhang Z, Li M-J, Yi C, Yang M (2014) A dual-mode nanosensor based on carbon quantum dots and gold nanoparticles for discriminative detection of glutathione in human plasma. Biosens Bioelectron 56:39–45
Shi W, Fan H, Ai S, Zhu L (2015) Preparation of fluorescent graphene quantum dots from humic acid for bioimaging application. New J Chem 39(9):7054–7059
Sun YP, Zhou B, Lin Y, Wang W, Fernando KS, Pathak P et al (2006) Quantum-sized carbon dots for bright and colorful photoluminescence. J Am Chem Soc 128(24):7756–7757
Tang S, Hewlett I (2010) Nanoparticle-based immunoassays for sensitive and early detection of HIV-1 capsid (p24) antigen. J Infect Dis 201(Supplement 1):S59–S64
Tang S, Zhao J, Wang A, Viswanath R, Harma H, Little RF, Yarchoan R, Stramer SL, Nyambi PN, Lee S, Wood O, Wong EY, Wang X, Hewlett IK (2010) Characterization of immune responses to capsid protein p24 of human immunodeficiency virus type 1 and implications for detection. Clin Vaccine Immunol 17(8):1244–1251
Tang L, Ji R, Cao X, Lin J, Jiang H, Li X, Teng KS, Luk CM, Zeng S, Hao J, Lau SP (2012) Deep Ultraviolet photoluminescence of water-soluble self-passivated graphene quantum dots. ACS Nano 6(6):5102–5110
Teeparuksapun K, Hedström M, Wong EY, Tang S, Hewlett IK, Mattiasson B (2010) Ultrasensitive detection of HIV-1 p24 antigen using nanofunctionalized surfaces in a capacitive immunosensor. Anal Chem 82(20):8406–8411
Wang W, Lu YC, Huang H, Feng JJ, Chen JR, Wang AJ (2014) Facile synthesis of water-soluble and biocompatible fluorescent nitrogen-doped carbon dots for cell imaging. Analyst 139(7):1692–1696
Wang R, Xu Y, Zhang T, Jiang Y (2015) Rapid and sensitive detection of Salmonella typhimurium using aptamer-conjugated carbon dots as fluorescence probe. Anal Methods 7(5):1701–1706
Wu Y, Wei P, Pengpumkiat S, Schumacher EA, Remcho VT (2015) Development of a carbon dot (C-Dot)-linked immunosorbent assay for the detection of human α-fetoprotein. Anal Chem 87(16):8510–8516
Yang ST, Wang X, Wang H, Lu F, Luo PG, Cao L, Huang Y (2009) Carbon dots as nontoxic and high-performance fluorescence imaging agents. J Phys Chem C 113(42):18110–18114
Yang Z, Li Z, Xu M, Ma Y, Zhang J, Su Y, Gao F, Wei H, Zhang L (2013) Controllable synthesis of fluorescent carbon dots and their detection application as nanoprobes. Nano Micro Lett 5(4):247–259
Zhao M-X, Li Y, Zeng E-Z, Wang C-J (2014) The application of CdSe quantum dots with multicolor emission as fluorescent probes for cell labeling. Chem Asian J 9(5):1349–1355
Zhu S, Meng Q, Wang L, Zhang J, Song Y, Jin H, Zhang K, Sun H, Wang H, Yang B (2013) Highly photoluminescent carbon dots for multicolor patterning, sensors, and bioimaging. Angew Chem Int Ed 52(14):3953–3957
Zhu L, Cui X, Wu J, Wang Z, Wang P, Hou Y, Yang M (2014a) Fluorescence immunoassay based on carbon dots as labels for the detection of human immunoglobulin G. Anal Methods 6(12):4430–4436
Zhu Y-D, Peng J, Jiang L-P, Zhu J-J (2014b) Fluorescent immunosensor based on CuS nanoparticles for sensitive detection of cancer biomarker. Analyst 139(3):649–655
Zuo J, Jiang T, Zhao X, Xiong X, Xiao S, Zhu Z (2015) Preparation and application of fluorescent carbon dots. J Nanomater 2015:13
Acknowledgements
All authors are grateful to Bhagawan Sri Sathya Sai Baba for his constant inspiration and guidance. L. A. Avinash Chunduri acknowledges UGC Govt. of India for BSR fellowship. We acknowledge with gratitude Prof. S. Sampath (IPC Department, IISc Bangalore, India) for providing TEM facility.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclaimer The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Food and Drug Administration, U.S. Department of Health and Human Services.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chunduri, L.A.A., Haleyurgirisetty, M.K., Patnaik, S. et al. Development of carbon dot based microplate and microfluidic chip immunoassay for rapid and sensitive detection of HIV-1 p24 antigen. Microfluid Nanofluid 20, 167 (2016). https://doi.org/10.1007/s10404-016-1825-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10404-016-1825-z