Abstract
Accurate and affordable rapid diagnostic tests (RDTs) are indispensable but often lacking for many infectious diseases. Specifically, there is a lack of highly sensitive malaria RDTs that can detect low antigen concentration at the onset of infection. Here, we present a strategy to improve the sensitivity of malaria RDTs by using capillary-driven microfluidic chips and combining sandwich immunoassays with electroless silver staining. We used 5 μm fluorescent beads functionalized with capture antibodies (cAbs). These beads are self-assembled by capillary action in recessed “bead lanes”, which cross the main flow path of chips microfabricated in Si and SU-8. The binding of analytes to detection antibodies (dAbs) and secondary antibodies (2ndAbs) conjugated to gold nanoparticles (NPs) allows the formation of a silver film on the beads. Such silver film masks the fluorescent core of the bead inversely proportional to the concentration of antigen in a sample. We illustrate this method using the recombinant malaria antigen Plasmodium falciparum histidine-rich-protein 2 (rPfHRP2) spiked in human serum. This antigen was a recombinant HRP2 protein expressed in Escherichia coli, which is also the standard reference material. The limit of detection (LOD) of our immunoassay was found to be less than 6 ng mL−1 of rPfHRP2 within 20 min, which is approaching the desired sensitivity needed in the Target Product Profile (TPP) for malaria elimination settings. The concept presented here is flexible and may also be utilized for implementing fluorescence immunoassays for the parallel detection of biomarkers on capillary-driven microfluidic chips.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Today many cases of infectious diseases remain untreated even though the diseases are curable. One of the main reasons for this situation is that many patients do not have access to diagnostic services. Furthermore, late diagnosis of infectious diseases such as malaria increases the risk of its spreading and makes treatment more difficult (World Health Organization 2011). To address this challenge, the World Health Organization (WHO) published in April 2018 its first Essential Diagnostics List, a catalogue of essential tests needed to diagnose global priority diseases such as malaria, HIV and tuberculosis, as well as most common conditions such as diabetes (World Health Organization 2018a). For test developers, the release of this list indicates the importance of designing tests such as malaria RDTs that are affordable, safe, and of high quality.
As the declining trend in global malaria burden has stalled, efforts to eliminate this scourge require disruptive innovations to obtain more powerful tools (World Health Organization 2017). Accurate and effective malaria RDTs remain a key intervention leading to more appropriate treatment and achieving the ambitious goal of eliminating malaria by 2030 (The malERA Consultative Group on Diagnoses and Diagnostics 2011). Most current malaria RDTs are based on a lateral flow assay format, which is cheap to produce and easy to use. However, the major disadvantage of this type of test is the inability to detect low antigen concentrations or low parasite densities at early stages of infection (Mcmorrow et al. 2011). A TPP was proposed by the Foundation for Innovative New Diagnostics (FIND) and Program for Appropriate Technology in Health to define the desired LOD at 6 ng mL−1 PfHRP2 for the ideal malaria RDTs detecting HRP2 in elimination settings (Program for Appropriate Technology in Health 2014). There are only few malaria tests that can meet this requirement at present (Das et al. 2018; Jimenez et al. 2017).
There are several examples of microfluidic chips that have been developed with promising analytical performance. We reviewed the current landscape of malaria RDTs and how recent advancement in technology can help in detecting the last few parasites in malaria elimination settings (Pham et al. 2018a). Like current malaria RDTs, many of them use a nitrocellulose membrane, or so-called paper-based microfluidics, to facilitate the detection of malaria. Yager et al. demonstrated a lateral flow immunoassay incorporated on a paper-patterned ‘card’. On this card, all reagents were stored in dry state. By folding the card, a sequential chemical analysis could be triggered, resulting in the detection of malaria PfHRP2 (Lafleur et al. 2012; Stevens et al. 2008). Cooper et al. developed a paper-based LAMP device that is capable of detecting and differentiating Plasmodium falciparum, Plasmodium vivax and being pan-specific for the other Plasmodium species directly from finger-pricked samples within 45 min (Xu et al. 2016). It certainly would be beneficial to further develop these technologies using less reagents and improving sensitivities and precision.
Furthermore, it is challenging for RDT developers to evaluate the performance of prototypes due to a lack of standardised reference materials. To avoid this caveat, it is important to have access to established reference materials at early stages of test development. In a joint project of the WHO Global Malaria Programme, FIND and the Centers for Disease Control and Prevention (CDC), the performance of commercially available malaria RDTs has been systematically evaluated and compared using high quality of panels of recombinant antigens (Foundation for Innovative New Diagnostics 2017). Such a recombinant antigen is used in this study to evaluate the performance of the prototype.
To address the above-mentioned shortcomings of current malaria RDTs, we developed an assay for detecting malaria PfHRP2 achieving clinically relevant LOD. This builds on our previous work on immuno-gold silver staining (IGSS) assays and capillary-driven microfluidic chips where species and anti-species Abs were used to perform a sandwich immunoassay on beads. These beads are fluorescent, functionalized with cAbs, and self-assembled by capillary action in microstructures of the chips. The accumulation of antigens and dAbs conjugated to gold NPs on the functionalized beads results in the electroless deposition of a silver film, which leads to a corresponding attenuation of the fluorescence signal from the core of beads (Pham et al. 2018b). Here, we functionalized the fluorescent microbeads with cAbs directed against PfHRP2 (denoted as α-PfHRP2 or cAb). In presence of PfHRP2, a complex between this antigen and dAbs conjugated with gold NPs is formed and captured on the surface of the microbeads. Then, gold NPs catalyse the formation of a silver film on the microbeads with a continuous flow of reagents for silver staining. The attenuation of fluorescence emitted from the core of the beads strongly correlates to the concentration of rPfHRP2 spiked in human serum samples, leading to a highly sensitive detection of malaria PfHRP2 infection.
2 Materials and methods
2.1 Antibodies and reagents
All reagents were purchased from Sigma-Aldrich unless otherwise stated. Deionised water was obtained from a Simplicity 185 system (Millipore, Billerica, MA). PBS was prepared by dissolving one PBS tablet in 200 mL of deionised water. Solutions of 1% and 5% w/v BSA in PBS were prepared by dissolving BSA in PBS solution. Solutions of PBST 0.1% and 0.01% were prepared by adding appropriate volume of Tween 20 to PBS solutions. A monoclonal mouse α-PfHRP2 IgG 2b (G18K) (100 μg mL−1, ThermoFisher Scientific) was used as cAb. A monoclonal mouse α-PfHRP2 IgG 2a (H36F) (1.0 μg mL−1, ThermoFisher Scientific) was used as dAb. A monoclonal rabbit α-mouse IgG 2a (1.0 μg mL−1, Abcam) was used as 2ndAb. Polyclonal donkey anti-rabbit IgGs conjugated with 6 nm gold nanoparticles and polyclonal donkey anti-rabbit IgGs conjugated with enzyme horse radish peroxide (HRP) were from Abcam. 3,3′-5,5′-tetramethylbenzidine in water (BM Blue POD substrate) was used as a substrate for enzyme-based immunoassays. A 1:1 v/v mixture of silver A and Silver B from SE 100–1 KT silver enhancer kit was prepared prior to the signal amplification step. Transparent Costar 96-well microtiter plates were from Sigma-Aldrich.
Fluorescent streptavidin PMMA beads (PolyAn GmbH, Red 25, 5.9 μm diameter) were used as substrates to integrate cAb into microfluidic chips. An antibody concentration kit (Abcam) was used to enrich the concentration of cAbs 10 times for the biotinylation step. A biotin fast conjugation kit (type B) (Abcam) was used to biotinylate cAbs. Recombinant HRP2 protein (rPfHRP2) expressed in Escherichia coli (Microcoat Biotechnologie GmbH, Germany) based on the HRP2 sequence of the W2 P. falciparum strain was selected as target analyte. A serial dilution of target analyte was prepared in 1% BSA solutions in PBS and human serum (Biowest) for assays on microtiter plates and in microfluidic chips, respectively. Prior to use, human serum was filtered using 0.2 μm diameter blood filter (Millipore).
2.2 Fabrication of microfluidic chips and integration of beads
Microfluidic chips were fabricated on 4-in. Si wafers (Si-Mat, Kaufering, Germany). A wafer layout comprising 28 chips and 2 layers was written to 5 × 5 in.2 glass/Cr photomasks (Nanofilm, USA) using a direct laser writing tool (DWL 2000, Heilderberg Instruments). A mask aligner (MA6, SÜSS MicroTec AG, Germany) with an i-line filter (365 nm) was used for the photolithography steps. Structures used to integrate beads (i.e. bead lanes) were etched using a DRIE tool (AMS 200, Alcatel, France) while masking the wafer surface with AZ®6612 positive-tone photoresist (MicroChemicals GmbH, Germany). Following the removal of the photoresist in an O2 plasma asher (600 W, 600 sccm, 5 min), microfluidic structures were fabricated using photolithographic patterning of a 15-μm-thick SU-8 (SU-83010, MicroChem Corporation) using the protocol provided by the supplier. The fabricated wafer was partially diced using a dicing tool after protecting the wafer surface with AZ®6612 photoresist. The wafer was cleaned using acetone and isopropyl alcohol, and dried. Prior to the bead integration, streptavidin-coated, fluorescent beads were incubated with cAbs and rinsed with 1% BSA in PBST (0.01%). Then, about 100 nL of bead suspension (0.5% w/v) in 1% BSA in PBST (0.01%) was introduced at the bead loading pad when the chip surface was still open (not laminated). Beads functionalized with receptors were carried by capillary flow orthogonally to the main channel and got trapped at the bead lane after drying of the solution. More bead suspension can be loaded if needed until the bead lane is completely filled with beads. Following the integration of beads, diced microfluidic chips were sealed by lamination of 50-μm-thick dry film resist (DFR) (DF-1050, Engineered Materials Systems, Inc., USA) using a hotplate at 45 °C and a plastic spatula. The key steps for the fabrication of microfluidic chips and details on bead lane microstructures are provided in Supplementary Fig. 1.
2.3 Protocols for assays
For immunoassays performed on microtiter plates, transparent Costar 96-well microtiter plates were used. Between each step, the plates were washed three times using a plate washer (Sunrise, Tecan). First, 50 μL of a cAb solution (1.0 μg mL−1) was added into each well and incubated for 60 min at room temperature. Then solutions of 5% BSA in PBS was added to block nonspecific binding (50 μL/well) for 30 min. Next, serial dilution of GST-Pf-HRP2-W2 in 1% BSA in PBS was added into each well (starting concentration of analyte was 50 μg mL−1, 50 μL/well). Analyte incubation was allowed to last 60 min at room temperature. Additions of dAb (1.0 μg mL−1) and 2ndAb (1.0 μg mL−1) were followed and each step was incubated for 30 min. For silver staining, a solution of gold conjugate was diluted 1:2000 (v/v) in 1% BSA in PBST and added into each well (50 μL/well). Incubation of the analyte with dAb-2ndAb-conjugated gold was 30 min at room temperature. Silver mixture consisted of silver solution A and silver solution B, which were mixed (1:1, v/v) and added immediately into each well (50 μL/well). The silver development step was 20 min, at room temperature and in the dark (plates covered with an aluminium foil). For standard enzymatic assays, a solution of HRP conjugate was diluted 1:10000 (v/v) in 1% BSA in PBST and added into each well (50 μL/well). This incubation was 30 min at room temperature. Solutions containing blue POD substrate were added (50 μL/well) and the enzymatic reaction was allowed to proceed for 20 min. To measure the end-point absorbance, a plate reader was used with a 570-nm wavelength. Standard deviations of three repeats in a single experiment were shown as error bars.
For immunoassays performed on microfluidic chips, the cAb was enriched 10-fold in concentration from 100 μg mL−1 to 1.0 mg mL−1, then biotinylated. Protocols for the enrichment and biotinylation of cAbs are standard procedures provided by Abcam. A sample containing the analyte in human serum, dAb, 2ndAb, gold conjugated Ab and an aliquot of 0.01% Tween 20 in deionised water were sequentially added to the first loading pad of the microfluidic chip. The silver staining solution and then 0.01% Tween 20 in deionised water were then added to the second loading pad of the chip. Images of fluorescent beads integrated in bead lanes were taken using a fluorescence microscope (Nikon Eclipse 90i, Japan) equipped with a 20× objective and a Texas Red fluorescence filter cube. A LED Lumencor lamp (software SOLA S2 Controller) was used to excite fluorophores. Images were taken using a monochrome CCD camera (DS-1 QM, Nikon) and an acquisition time of 400 ms with ND16 filter. Fluorescence images were analysed using software Fiji (ImageJ). Regions of interest (ROI) comprising the beads before and after silver staining were selected from acquired fluorescence images and the mean fluorescence intensity for each experiment was obtained by subtracting the background signal around an ROI from the mean signal value in the ROI. Standard deviations of six repeats in a single experiment were shown as error bars. For all assays, titration data were fitted using a 4- parameter logistic regression (4-PL) model. The LOD was defined as the lowest concentration of PfHRP2 for which the signal is 3 × standard deviation (99% confidence interval) above the mean signal of samples not containing PfHRP2.
3 Results and discussion
3.1 Validation of IGSS for malaria PfHRP2 detection using a commercial RDT and a microtiter plate
As a first step, we used a commercial RDT to provide a reference for the sensitivity achieved using low cost tests. We chose the commercial malaria RDT Paramax-3 (Zephyr Biomedicals) to detect the biomarker rPfHRP2, because this RDT had passed round 2 testing of the WHO/FIND/CDC product evaluation with 93% panel detection score, which is a criterion taken into account for analytical performance, but also to test inter- and intra-lot variations (World Health Organization; Foundation for Innovative Diagnostics; Centers for Disease Control and Prevention 2009). A serial dilution of rPfHRP2 in 1% BSA in PBS was prepared in accordance with the test protocol used in the round 2 testing with three rPfHRP2 concentrations of 1880 ng mL−1, 188 ng mL−1 and 18.8 ng mL−1, which are the equivalent of parasite densities of 2000 parasites/μL, 200 parasites/μL, and 20 parasites/μL. The test procedure was followed as defined by the manufacturer and we found that the Paramax-3 RDTs detected a concentration of 188 ng mL−1 PfHRP2 and higher, but could not detect a PfHRP2 concentration of 18.8 ng mL−1 (Fig. 1a).
Performance of malaria rPfHRP2 assays using various formats. a An example of a commercial malaria RDT Paramax-3 (Zephyr Biomedicals) used to detect the malarial rPfHRP2 antigen at three concentrations. Absorbance measurements of b an IGSS assay and c an ELISA on microtiter plates for various concentrations of rPfHRP2 in PBS. Insets in (b) and (c) represent the assay architecture for each method. Standard deviations correspond to triplicate of experiments and shown as error bars. Blue lines represent the signal generated from samples containing PfHRP2 and red lines represent the signal generated from samples without PfHRP2. Dotted lines represent the cut-off values of each method (based on the average values of samples without PfHRP2 plus three times standard deviations)
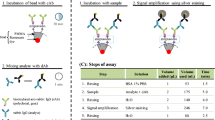
Next, we developed an assay protocol for the IGSS on microtiter plates. The use of microtiter plates is chosen to characterise the IGSS assay for malaria because it is a well-known diagnostic technique in laboratories. This method allows independent tests carried out in the same conditions simultaneously over a wide range of concentration made by serial dilution. IGSS necessitates a dAb conjugated to NPs that can catalyse the electroless deposition of silver, and ideally such an antibody should be directed against PfHRP2, monoclonal (monoclonal antibodies lead to more specific assays), and commercially available. We were not able to identify such a dAb and therefore decided to use additional species/anti-species antibodies to circumvent this issue. We use a monoclonal dAb α-PfHRP2 from mice and a rabbit α-mouse 2ndAb. The gold NPs are then functionalized with a donkey α-rabbit Ab (Fig. 1). The last antibody (α-rabbit Ab) could also be conjugated to HRP to perform an enzymatic assay using a colorimetric readout. Interestingly, the use of 2ndAbs has been suggested in the literature as a mean of improving the sensitivity of assays (Corthell 2014).
We evaluated the sensitivity of the IGSS assay using the above-mentioned antibodies and antigens (Fig. 1b) and compared this sensitivity to that of a well-established reference method, namely an ELISA involving an enzymatic-based signal (Fig. 1c). Both assays were performed on microtiter plates using a bench-top plate reader for colorimetric absorbance measurements. For the immunoassay detecting PfHRP2 on microtiter plates, the LOD of the IGSS assay was 105 ng mL−1, while the LOD of the ELISA was 49 ng mL−1. These results indicate that the IGSS assay is a little less sensitive than ELISA and approximately as sensitive as the commercial malaria RDT.
3.2 IGSS assay protocol developed for the microfluidic chip
We transposed the IGSS assay to an assay on a capillary-driven microfluidic chip (Juncker et al. 2002) for the detection of malaria antigen rPfHRP2 in human serum specimen. For this, we start with two off-chip preparation steps (Fig. 2a). The first step is to coat the fluorescent beads with cAbs prior to their integration to the microfluidic chip. The second step is to incubate rPfHRP2 in human serum with dAbs, 2ndAbs and gold conjugated antibodies for 5 min.
Design of the IGSS assay implemented using capillary-driven microfluidic chips and integrated, receptor-functionalized beads. a There are two steps involved in the off-chip procedure: first, streptavidin-coated fluorescent beads are incubated with biotinylated cAbs for 30 min and second, malaria PfHRP2 in human serum is mixed with a solution containing dAbs, 2ndAbs and gold conjugate Abs for 5 min to form the complex rPfHRP2-dAb before loading the resulting solution to the microfluidic chip. b The two key steps of the on-chip part of the assay are the incubation of the analyte-dAb complex with cAb on bead located in the microfluidic chip, and the signal amplification step during which silver staining occurs in presence of gold nanoparticles conjugated to the 2ndAbs. c Summary of the steps involved in the on-chip procedure, in which each liquid is allowed to flow for a specific time as indicated
The on-chip assay starts with the addition of 2 μL of the pre-incubated solution from above onto a loading pad of the capillary-driven microfluidic chip to let the solution pass over the integrated beads for the capture of the rPfHRP2-complex for 5 min (Fig. 2b). Next, a rinsing solution is added to the chip to remove unbound antibody complexes to avoid non-specific silver staining. Three microliters of silver staining solution are then added to the chip to catalyse the reduction of silver ions using the gold NPs conjugated to the 2ndAbs. The formation of the silver film is then stopped after 7 min by adding 2 μL of deionised water containing 0.01% of Tween 20. The rinsing solution is allowed to flow in the chip for another 2 min before acquiring the fluorescence signal using a fluorescent microscope. These steps are summarized in Fig. 2c and in practical terms, once being added into a loading pad, each solution flows along the microfluidic path due to capillary action. When the solution has flown for the desired time, the excess solution left in the loading pad is removed using a cleanroom tissue and the next solution is added. A capillary pump at the end of the flow path ensures that capillary action is maintained each time a solution is added to a loading pad and the characteristics of the capillary pump were set to support a flow rate of ~40 nL min−1 (Zimmermann et al. 2007). Specifically, with wettable microstructures in the capillary pump, and aqueous samples, a negative pressure of the order of 10 kPa is created at the liquid-air interface that draws the liquid steadily through the flow path with air in the capillary pump escaping through a microvent. We worked out the volume of each solution that needed to pass in the microfluidic chip (Fig. 2c) based on the desired timing of each individual step while keeping the total time below 20 min, which is similar to that of commercial lateral flow tests (Pham et al. 2018a). As the incubation of analyte and dAbs is critical for optimal assay performance, we allocated 5 min for this incubation step. Usually, on commercial malaria RDTs, there is no specific time allocated to allow the binding of analytes in a sample to dAbs, and the addition of a diluent is recommended immediately after the introduction of the sample with an interpretation of the result 15 min after the addition of diluent (World Health Organization 2018b). The sample and silver staining solution should not come in contact with each other in the microfluidic chip: we noticed that Cl− ions contained in the sample react with Ag+ from the staining solution and quickly formed precipitates of AgCl whenever both solutions were consecutively added to the chip. We therefore intercalated a rinse step by adding 2 μL of water to the loading pad and let the rinse solution flow through the chip for 4 min. This rinsing step efficiently prevented non-specific silver staining. The time allocated to the silver staining step on chip is 7 min, which is comparable to an earlier assay we developed for species/anti-species immunoassays (Pham et al. 2018b). This time is also in agreement with the typical duration of silver enhancement steps (Lackie 1996), (Liu et al. 2014). The formation of the silver film attenuates the emitted fluorescence signal of the beads, which allows for optical signal readout by measuring the fluorescence emitted from beads before and after the silver staining step. To avoid non-specific deposition of silver due to light, the microfluidic chip is covered with an aluminium foil during the silver staining step. The remaining 2 min are allocated for stopping the formation of the silver film using a final rinsing step and for keeping the fluorescence signal constant until it is measured using a fluorescence microscope.
The microfluidic chips are microfabricated using a Si wafer using standard photolithography and engraving techniques (Supplementary Fig. 1). Flow paths for the assay are defined in SU-8 (lateral walls), SiO2 (native oxide of silicon substrate, bottom wall), and a dry film resist (DFR) (top wall). The flow path for integrating the beads functionalized with the receptors is defined in the silicon substrate using photolithography and dry etching (Fig. 3a). 100 nL of a bead suspension is applied to a loading pad servicing a microchannel crossing the main flow path of the chip and ending with one narrow outlet (4 μm in width). Beads accumulate in this recessed microchannel, which we term “bead lane”, in a few seconds by capillary assembly and the solution evaporates in ~40 s (Fig. 3b). A sequence of images showing these events are provided in Supplementary Fig. 2, where the images were processed (brightness and background subtraction of microfluidic structures) to make beads and the bead suspension more visible. Salt crystal residues and aggregates of beads may occasionally form in areas where much solution evaporates but these residues do not play a role during assays as they are decoupled from the main flow path and bead lane. Anti-wetting structures patterned around the bead integration zone prevents undesired spreading of the bead solution: these structures are either etched in Si (15 μm) or patterned on the Si surface in SU-8 (15 μm thick) and when the air-liquid meniscus reaches these structures, their geometries create a Laplace pressure barrier and pin the liquid (Arango et al. 2018). Therefore, the bead suspension essentially flows from the droplet to the bead lane through structures etched in the Si layer. This self-assembly process conveniently localizes receptors at a precise location of the chip without the need for specific surface chemistry or patterning of the surfaces forming the flow path. The bead suspension can be placed by hand with a micropipette or using a pipetting robot. No post-processing or rinsing step is needed but instead the chip can be sealed, stored or directly used for an assay.
Integration of beads carrying receptors in bead lanes using capillary assembly. a Simplified illustration of the capillary-driven microfluidic chip (left) and actual layout of a “bead lane” structure (insets on the right) showing the patterns etched in the silicon layer, which can pull a bead suspension pipetted over a pad area through (i) a network of curved structures and (ii) the bead lane itself. b Microscope image showing a droplet of ~100 nL residing on the pad and confined by various anti-wetting structures etched in Si. A fraction of the droplet is drawn by capillary forces toward the bead lane where microbeads accumulate. The solution starts to dry after ~40 s, leaving in place the microbeads and allowing the microfluidic chip to be sealed with a DFR. Illustrations of (c) a flow of solution (PBS containing a blue dye) across a bead lane after the microfluidic chip had been sealed and (d) the sequential flow of 2 solutions (PBS and then DI water) across a bead lane containing microbeads
Lamination uses a DFR with low autofluorescence and is done at 45 °C (Temiz and Delamarche 2014). This sealing technique is chosen for three main reasons. First, this low temperature and fast (< 10 s) lamination does not affect the biological activity of cAbs immobilised on the integrated beads. Second, there is no need to pattern the DFR for the openings of the loading pad and finally this lamination does not require any solvents. The chip then can be singulated by hands and ready for use. After the lamination and singulation, the liquid pipetted to a sample loading pad flows across the integrated beads (Fig. 3c). Multiple liquids that are required for the assay can also be pipetted and displaced by capillary actions (Fig. 3d).
3.3 Chip design
The microfluidic chip is designed as an ensemble of connected microelements (Fig. 4). The chip is ~1 cm2 and comprises five key microfluidic elements: anti-wetting structures, two loading pads, two bead lanes where cAbs are immobilised and the signal is measured, a capillary pump for drawing the liquid at a programmed flow rate, and an air vent. The flow path is 15 μm deep, 100 μm wide and defined by lateral walls in SU-8. These small dimensions of the microstructures lead to short paths for diffusion of analytes and reagents during an assay and efficient rinsing steps owing to the laminar flow of liquids filling the chip. Furthermore, semi-circular ridges around loading pads act as anti-wetting structures, keep liquids pipetted in loading pads and prevent liquid from overflowing loading pad. Volumes of liquids flowed in the chip can be estimated by optical monitoring of the advancement of the solution in the chip. The assay conditions can be easily adapted by either varying the dimensions of the capillary pump, or the addition of flow resistors, or even electrogates for stop-and-go control of liquids filling the chips (Arango et al. 2018).
Image of a microfabricated Si microfluidic chip for IGSS immunoassays for detecting PfHRP2. The flow path starts with 2 parallel loading pads that are surrounded with anti-wetting structures, a connection channel linking the loading pads to “bead lanes”, a capillary pump, and air vent. All microfluidic structures are formed in a 15-μm-high SU-8 photoresist layer excepted for the bead lanes, which are etched in the Si layer. The chip is 15-mm-long and 6.5-mm-wide (97.5 mm2). The insets are SEM micrographs of a bead lane (15-μm-deep, etched in Si and 20-μm-wide) before and after the bead integration
The flow path directs a sample over bead lanes, which are 15-μm-deep and 20-μm-wide structures in Si that hold ~200 beads with a diameter of 5.9 μm, (Fig. 4, inset). The SU-8 lateral walls forming the flow path are patterned so as to be distributed at least 50 μm away from the bead lanes to avoid interferences with the autofluorescence of SU-8 when acquiring the fluorescence signals from the beads before and after the silver staining step. The discontinuity in the flow path does not compromise flow in the microfluidic chip because the chip is sealed and only a few nanolitres of solution can spread laterally toward the areas where the bead suspension is loaded or drawn (Fig. 3c and d).
3.4 IGSS performance
We investigated the performance of the IGSS specifically for a malaria rPfHRP2 assay in human sera. To this end, we used microfluidic chips having fluorescent microbeads coated with α-PfHRP2 antibodies, rPfHRP2 in human sera, solutions and reagents as described earlier. A serial dilution of rPfHRP2 was prepared in human serum. Each of the dilution was incubated with a mouse monoclonal α-PfHRP2 IgG 2a, monoclonal rabbit α-mouse IgG 2a, and a polyclonal gold conjugated donkey α- rabbit IgG for 5 min before use.
Each chip was only used once and fluorescence micrographs of the bead lanes were acquired before and 7 min after adding a silver staining solution to the first loading pad. These experiments were performed at room temperature (24 °C) and the measured optical signal did correlate with the concentration of the rPfHRP2 antigen in the sample (Fig. 5). The obtained data are fitted using a 4-PL model and presented together with some representative fluorescent micrographs of the bead lanes after silver staining in Fig. 5. The LOD of the IGSS malaria assay performed on microfluidic chips was found to be below 6 ng mL−1 PfHRP2. The LOD was defined as being the smallest measurable signal that is above the zero analyte concentration signal plus 3 SD.
Detection of rPfHRP2 using the IGSS assay and capillary-driven microfluidic chips. a Relative change of the fluorescence measured from beads before and after the silver staining step as a function of the concentration of rPfHRP2 spiked in human serum (blue line and data points) and for non-spiked human serum (red lines). The red line represents mean signal measurement for zero analyte concentration (control). The dotted red line represents the limit of detection (LOD) for this IGSS assay. LOD is calculated as the lowest concentration of analyte for which the signal is above the mean signal +3 standard deviation (99% confidence interval) of zero analyte concentration samples. b Fluorescence micrographs showing the attenuation of fluorescence from beads by the stained silver layer
To put the results achieved from this study into perspective, we summarize the LODs of the IGSS assay on capillary-driven microfluidic chips, ELISA and IGSS assay on microtiter plates, and the LODs of some commercial malaria RDTs, together with the desired LOD for the ideal malaria RDTs for elimination settings (Table 1). These figures suggest an excellent sensitivity of our assay given that the assay only needed 20 min and few microliters of sample and reagents.
4 Conclusions
A microfluidic chip prototype for point-of-care (POC) diagnostics was developed based on a unique combination of a fluorescence assay, self-assembled functionalized beads in a capillary-driven microfluidic chip and electroless silver staining technique for the detection of PfHRP2 with a promising LOD for early diagnosis of malaria. Since only the core of the beads is fluorescent, photobleaching and stability issues, which are often found in many fluorescent immunoassays are mitigated. The use of fluorescent beads does not present a limitation for POC applications as many handheld peripheral fluorescence readers have been developed for use at the point of need. The attenuation of fluorescence intensity emitted from the core of the beads due to formation of a silver film alleviates the need for fully transparent devices. The assay achieved a sensitivity below 6 ng mL−1 of PfHRP2, which is in line with the desired sensitivity specified in the TPP for malaria elimination settings. Although not being investigated in this study, the fluctuation in temperature should not cause any adverse effects on silver staining. The short turnover time of 20 min supports the use of this technology in the field where health workers often have to perform many tests in a day, or when patients have to receive treatment within the same visit. The microfluidic prototype reported here relies on capillary forces to facilitate sequential chemical analysis without any active elements. This makes the microfluidic chips compact and easy to use while retaining all advantages of rapid tests. Potentially, automatic reagent and buffer delivery elements can be incorporated so that this device could become a ‘one-step’ test that has been envisioned for the ideal malaria RDT. This work might be extended to the fabrication of disposable polymeric chips using state-of-the-art roll-to-roll, injection molding or hot-embossing techniques. For an objective and effective quantification of the optical signal, a mobile phone reader with automated image processing pipeline can be also built (Cherbuin et al. 2018). In light of the TPP for malaria elimination settings, the prototype reported here represents a step forward and more generally may incentivize the adoption of fluorescent bead-based immunoassays using capillary-driven microfluidics for POC diagnostics applications.
References
Y. Arango, Y. Temiz, O. Gökçe, E. Delamarche, Appl. Phys. Lett. 112, 153701 (2018)
M. Cherbuin, F. Zelder, W. Karlen, Analyst 130 (2018)
J. T. Corthell, in Basic Mol. Protoc. Neurosci. Tips, Tricks, Pitfalls (2014)
S. Das, R.B. Peck, R. Barney, I.K. Jang, M. Kahn, M. Zhu, G.J. Domingo, Malar. J. 17, 1 (2018)
Foundation for Innovative New Diagnostics, HRP2 recombinant panel for malaria diagnostic tests (2017)
A. Jimenez, R.R. Rees-Channer, R. Perera, D. Gamboa, P.L. Chiodini, I.J. Gonzalez, A. Mayor, X.C. Ding, Malar. J. 16, 128 (2017)
D. Juncker, H. Schmid, U. Drechsler, H. Wolf, M. Wolf, B. Michel, N. De Rooij, E. Delamarche, Anal. Chem. 74, 6139 (2002)
P.M. Lackie, Histochem. Cell Biol. 106, 9 (1996)
L. Lafleur, D. Stevens, K. McKenzie, S. Ramachandran, P. Spicar-Mihalic, M. Singhal, A. Arjyal, J. Osborn, P. Kauffman, P. Yager, B. Lutz, Lab Chip 12, 1119 (2012)
R. Liu, Y. Zhang, S. Zhang, W. Qiu, Y. Gao, Appl. Spectrosc. Rev. 49, 121 (2014)
M.L. Mcmorrow, M. Aidoo, S.P. Kachur, Clin. Microbiol. Infect. (2011)
N.M. Pham, W. Karlen, H.-P. Beck, E. Delamarche, Malar. J. (2018a)
N.M. Pham, S. Rusch, Y. Temiz, R.D. Lovchik, H.-P. Beck, W. Karlen, E. Delamarche, Biomed. Microdevices 20(41), 41 (2018b)
Program for Appropriate Technology in Health, Target Product Profile : Point-of-Care Malaria Infection Detection Test (2014)
D.Y. Stevens, C.R. Petri, J.L. Osborn, P. Spicar-mihalic, K.G. Mckenzie, P. Yager, T. Int, Conf. Miniaturized Syst. Chem. Life Sci. 1768 (2008)
Y. Temiz, E. Delamarche, J. Micromech. Microeng. 24, 097001 (2014)
The malERA Consultative Group on Diagnoses and Diagnostics, PLoS Med. 8, e1000396 (2011)
World Health Organization, Universal Access to Malaria Diagnostic Testing: An Operational Manual (2011)
World Health Organization, World Malaria Report 2017 (2017)
World Health Organization, Terms of Reference for the Strategic Advisory Group of Experts on In Vitro Diagnostics (SAGE IVD) (2018a)
World Health Organization, Public Report: One Step Test for Malaria Pf / Pv Ag MERISCREEN Malaria Pf / Pv Ag (2018b)
World Health Organization, Foundation for Innovative Diagnostics, Centers for Disease Control and Prevention, Malaria Rapid Diagnostic Test Performance Round 2 (2009)
G. Xu, D. Nolder, J. Reboud, M.C. Oguike, D.A. van Schalkwyk, C.J. Sutherland, J.M. Cooper, Angew. Chem. Int. Ed. 55, 15250 (2016)
M. Zimmermann, H. Schmid, P. Hunziker, E. Delamarche, Lab Chip 7, 119 (2007)
Acknowledgements
Ngoc M. Pham received an Engineering for Development Scholarship from ETH Global and the Sawiris Foundation for Social Development. Walter Karlen is supported through the Swiss National Science Foundation professorship award 150640 “Intelligent point-of-care monitoring”. Yuksel Temiz and Emmanuel Delamarche thank Elisa Hemmig, Robert Lovchik and Onur Gökçe for discussions and Walter Riess and the IBM Research Frontiers Institute for their continuous support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 833 kb)
Rights and permissions
About this article
Cite this article
Pham, N.M., Rusch, S., Temiz, Y. et al. Immuno-gold silver staining assays on capillary-driven microfluidics for the detection of malaria antigens. Biomed Microdevices 21, 24 (2019). https://doi.org/10.1007/s10544-019-0376-y
Published:
DOI: https://doi.org/10.1007/s10544-019-0376-y