Abstract
This study aims to explain the genetic kinship relationships among populations of the psyllid species collected from several pistachio orchards in Turkey. Agonoscena pistaciae (Burckhardt and Lauterer) (Hemiptera, Psyllidae) is one of the most important pests of pistachio trees (Pistacia vera L.). This species, which continues its biological periods on pistachio trees, has been rapidly spreading in some parts of the world. Although pistachio psyllid leads to significant yield losses in pistachio trees in Gaziantep and Adıyaman in Turkey, in Siirt province such infestation has historically been very rare. However, the population density has significantly increased in Siirt province and districts in recent years. In this study, an internal transcribed spacer (ITS) DNA barcode identifier was used to determine whether these new outbreaks are a simple spread or the evolution of a new A. pistaciae biotype. In this context, by obtaining a 716-bp long ITS gene region by the polymerase chain reaction process, the genetic kinship relationships among populations of A. pistaciae in Gaziantep, Adıyaman, and Siirt provinces, which have significant potential in pistachio production, were revealed. When the data obtained were examined, no genetic differences could be detected among the population members of A. pistaciae spreading in Siirt, Gaziantep, and Adıyaman provinces. It was determined that A. pistaciae living in these regions are the same species. In addition, in the study the Wolbachia sp. type using the 16S ribosomal RNA gene region was determined.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The psyllids, called jumping or leaping plant lice, have spread to many parts of the world (Maryańska-Nadachowska et al. 2018). More than six psyllid species on Pistacia spp. have been identified to date (Mehrnejad 2014). Psyllids are known as hemipteric small insects feeding on phloem veins, and about 4000 psyllid species have been identified worldwide (Burckhardt and Ouvrard 2012). Psyllids negatively affect the physiology, anatomy, biochemistry, and development of plants. In both adult and nymph periods, they absorb plant sap and directly damage it. However, most psyllid species have been observed to cause the most nymphal period damage (Burckhardt 1994). Furthermore, some psyllid species are known as vectors of Candidatus Phytoplasma spp.; these agents cause significant losses to the host (Seemüller et al. 2011; Burckhardt and Ouvrard 2012).
The pistachio psylla (Agonoscena pistaciae [Burckhardt and Lauterer] [Hemiptera, Psyllidae]) that develops on Pistacia atlantica, P. mutica, P. palaestina, P. terebinthus, and P. vera is a serious pest of commercially grown pistachio. It is distributed in Turkey, Greece, Iran, Armenia, and Tajikistan (Burckhardt and Lauterer 1989, 1993; Lababidi and Zebitz 1995; Lauterer et al. 1998). Mehrnejad (2003) has stated that the damage done by this psyllid species in pistachio orchards affects the yield not only for that year but also for the next 2 years.
Agonoscena pistaciae was used by Tokmakoğlu (1973) for the first time with the name Agonoscena targionii (Licht.) in pistachio orchards of Gaziantep province in Turkey. In addition, in previous studies many researchers published the psyllid species under the name Agonoscena succincta (Heeg.) (Çelik 1975; Günaydın 1978; Klimaszewski and Lodos 1977). However, Bolu (1995) determined that the species observed in the pistachio fields in the southeastern Anatolia region is A. pistaciae, not A. targionii.
It has been clearly seen in previous studies that it is both time-consuming and sometimes difficult to identify many important insect species, such as psyllids, based only on morphologic characters. As a matter of fact, according to Pinto and Stouthamer (1994), the identification of small insects by depending on morphologic characteristics is still difficult, and it is essential to use special skills. Therefore, genomic approaches in the diagnosis of taxa currently use diversity among DNA sequences to identify the organism (Boekhout et al. 1994; Wilson et al. 1995). Molecular markers, particularly DNA barcodes, are used to reveal the relationships and divergences of populations from different geographic locations (Li et al. 2020). The identification of living groups and their genetic structures, gene flows, and relationships among different populations is obtained by sequencing the data of cytochrome oxidase I-II, 12S, and 16S genes with internal transcribed spacer (ITS) regions between ITS1 and ITS2. Polymorphisms in nuclear ribosomal RNA (rRNA) ITS regions have been used in many cases to distinguish closely related organisms (Hillis and Davis 1986; Mindell and Honeycut 1990; Hillis et al. 1991). In recent years, several molecular studies have addressed the identity and genetic relationships of Psyllidae family species (Katoh et al. 2014; Xiong et al. 2017; Meng et al. 2018; Chen et al. 2018; Cho et al. 2020). However, no research has been conducted on the molecular genetics of A. pistaciae of the Psyllidae family in our country or, in fact, in the world. Therefore, this study aimed to discover the genetic kinship relationships among A. pistaciae populations displaying distribution in Siirt, Adıyaman, and Gaziantep provinces using ITS DNA barcode identifiers. Moreover, endosymbiotic Wolbachia sp. types were investigated.
An A. pistaciae invasion has appeared in the Siirt province of Turkey in recent years. Producers have been observed taking intense insecticide precautions against this pest. Therefore, the information to be obtained in this research will provide important information for the control of this pest.
In this context, ITS gene region data, the genetic structure of A. pistaciae species, and genetic kinship relationships among different populations were first examined in this study. The genetic data obtained for A. pistaciae will constitute a basic step in subsequent molecular studies.
Materials and Methods
Sampling of Agonoscena pistaciae
Agonoscena pistaciae (Hemiptera: Psyllidae) samples (Fig. 1) were collected randomly (Gaziantep, Adıyaman, and Siirt districts) from a total of 22 locations in Turkey (Table 1).
The pest nymphs were carefully examined under a binocular microscope to ensure that they were not infected with parasitoids. The sufficient number of samples was then stored in 99.5% alcohol until being used in molecular analysis.
Samples of the pest species from each location were labeled and stored at −80 °C until used in experimental stages for molecular diagnosis. The coordinate information of all the sampled gardens was determined by a global positioning system and shown in Table 1.
DNA Extraction Process
Total DNA of A. pistcaiae genotypes was extracted with the DNeasy Blood and Tissue Kit protocol (Qiagen, Tokyo, Japan). This kit was previously used by Lee et al. (2008) and Yang et al. (2004) to detect Cacopsylla cininis and C. qianli species. Extraction was done according to the manufacturer’s instructions. The presence of DNA obtained after DNA isolation was monitored using agarose gel. After this process, the DNA was quantified using the Thermo Scientific NanoDrop 2000 spectrophotometer. A 260/280 ratio of DNA samples was found to be around 1.8–1.9.
PCR Amplification
The DNA obtained was amplified in the ITS2 gene region; DNA amplification was performed in a programmable thermal cycler. Universal primers, NG02955 (ATGAACATCGACATTTCGAACGCACAT) and AB052895 (TTCTTTTCCTCCGCTTAGTAATATGCTTAA), designed by Ji et al. (2003) for the amplification of the ITS fragment, were successfully used.
The polymerase chain reaction (PCR) of ITS was completed under the following conditions: first, an initial denaturation step at 94 °C for 4 min, then a 35-cycle amplification (94 °C for 40 s, 52 °C for 20 s, and 72 °C for 40 s), and then the final extension step for 2 min at 72 °C.
Furthermore, 16S rRNA gene region 27F (AGAGTTTGATCCTGGCTCAG) and 1492R (ACGGCTACCTTGTTACGACTT) sequence primers were used to molecularly identify Wolbachia species. The PCR temperature cycles for the 16S rRNA gene region were adjusted as shown below. First, an initial denaturation step was carried out at 95 °C for 30 sec, then a 35-cycle amplification was done (95 °C for 30 s, 56 °C for 1.30 s, and 68 °C for 1.30 s), followed by the final extension step for 5 min at 68 °C.
At the end of the procedure, the PCR products were stored until electrophoresis at +4 °C. Successful growth on the gel after electrophoresis was stored at −20 °C for sequencing reactions. The amplified products were electrophoresed on 1% agarose gels at 84 V for 45 min with Tris-acetateethylenediaminetetraacetic acid buffer (pH 8.0).
Sequencing and Bioinformatics Analysis
The results obtained from the gel imaging of the products of PCR were sequenced as procurement of services via the commercial company of Sentegen (Ankara, Turkey). Processes for purification of the PCR products of the ITS and 16S rRNA regions were done by the same company. Gene sequencing was conducted both with the forward and reverse primers to check for accuracy of the sequencer and to obtain reliable data. After the sequencing results were obtained, chromatograms of the DNA sequencing results were examined, and 30 bp at the start and end of the fragments were cut by using Sequencer software (version 5.4.6). All sequences were performed using BLASTn (http://www.ncbi.nlm.nih.gov/blast/), the National Centre for Biotechnology Information (NCBI) database, to determine whether the remaining gene sequence belonged to the organism. Then, to create a phylogenetic tree, MEGA X software was used (Kumar et al. 2018), and the quality fragments were aligned by the ClustalW algorithm. To create a tree from this aligned data, the phylogenetic tree was obtained by the neighbor-joining method (Saitou and Nei 1987) embedded in the MEGA X program.
Results and Discussion
In this study, the genetic structure of Agonoscena pistaciae and genetic relationships among different populations were revealed as a result of molecular sequencing and phylogenetic analysis with ITS gene region data. Moreover, the presence of an intracellular bacterial group (Wolbachia sp.) (Werren et al. 2008) that infects a large number of arthropods was investigated with 16S rRNA gene region data. For this purpose, the following analyses were carried out respectively. Overall, our present study did not detect any significant genetic differences among A. pistaciae populations in Turkey. The results can lead to significant new perspectives for A. pistaciae control. Although A. pistaciae is the most invasive species on pistachio, there is no molecular research available to study its genetic relationships and population structure in our country.
Total DNA Extraction
The isolation process was done using a DNA isolation kit of Agonoscena pistaciae samples collected from pistachio orchards in Siirt, Gaziantep, and Adıyaman and their districts. The DNA of the isolated samples was verified by electrophoresis on a 1% agarose gel imaged with the SmartView Pro Imager (Fig. 2). The DNA quality of all samples was found to be appropriate, and as a quantification parameter, a 260/280 ratio of all samples was found to be between 1.8 and 1.9.
PCR Amplification of ITS Gene Region
To amplify the ITS gene region by using the quality genomic DNA, PCR was performed. In this analysis, 716-bp ITS gene regions of 11 insects from 11 locations (location numbers 3, 6, 7, 8, 9, 10, 11, 13, 14, 16, 19) were obtained from the PCR process, and a 100-bp DNA leader was used to determine and confirm the size of the PCR products (Fig. 3).
Internal transcribed spacer 2 gene region of Agonoscena pistaciae polymerase chain reaction results. M marker (DNA leader; 100 bp), 1 Kurtalan, 2 Gaziantep, 3 Nizip, 4 Eruh, 5 Şirvan, 6 Besni, 7 Tut, 8 Gölbaşı, 9 Oğuzeli, 10 Adıyaman Tepecik, 11 Nurdağı, W water control, the red arrow shows the length of ITS region
PCR Amplification of 16S rRNA Gene Region
To amplify the 16S rRNA gene region, PCR was performed. Endosymbiotic Wolbachia bacteria 1429 bp long were obtained from nine insects from nine locations representing Siirt, Gaziantep, and Adıyaman provinces (location numbers 2, 3, 6, 8, 10, 11, 12, 13, 17) as a result of PCR processing (Fig. 4).
After the electrophoresis viewing process, the PCR products belonging to the samples giving positive results were sent to the company, where the sequencing was performed with the gene-specific forward and reverse primer pairs. The samples were sequenced with both reverse and forward primers by using the Sanger sequencing device for the ITS and 16S rRNA gene regions. The results of the sequence were displayed using the ChoramasPro software, and necessary editing was done by superimposing the forward and reverse sequences. A section of sequenced fragment is given in Fig. 5.
Bioinformatics Analysis
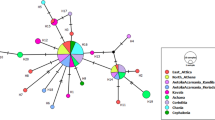
The sequences of the ITS gene region were entered in the NCBI database as each sample separately, and the results were then analyzed in detail with BLASTn. The sequences of the ITS2 gene region of the samples were found to be 95% similar to Pseudophacopteron alstonium from the Psyllidae family as a result of BLAST analysis. The phylogenetic tree was created by the neighbor-joining method using the MEGA X software with the data for the ITS gene region. Detailed molecular kinship relationships among individuals were revealed using this phylogenetic tree (Fig. 6). When the phylogenetic tree created in the study was examined, Pseudophacopteron alstonium from the Psyllidae family was evaluated as an external group. It was seen that the genetic diversity among the sequencing data of A. pistaciae collected from Gaziantep, Siirt, and Adıyaman provinces was found to be very low. The locations related to these data are clustered in two distinct groups. On the one hand, when bootstrap values were analyzed, A. pistaciae collected from Gaziantep, Siirt, and Adıyaman provinces was separated from the samples collected from the Şirvan, Nizip, Besni, and Tut locations, with a low value of bootstrap (61%) of the samples collected at the Kurtalan location. On the other hand, Gölbaşı was separated from the samples collected from the Eruh, Gaziantep, Adıyaman Tepecik, Nurdağı, and Oğuzeli locations, with a low value of 54% (Fig. 6). Considering the morphologic data, it was concluded that these results are quite consistent. While the morphologic data were evaluated, a serious characteristic that would reveal differences among these samples could not be determined.
The high kinship or similarity detected among individuals can be explained by the relatively short geographic distances between the respective populations and the possibility that these populations migrate easily through the wind. The absence of any high mountains or barriers between the geographic regions is also thought to have led to a genetic similarity among the group populations. In addition, when geographic distances among populations were considered, it was found that groups were separated from each other with a low bootstrap value of 54% in terms of genetic distance, although the distance between Gölbaşı and Eruh was the longest (519 km). When the phylogenetic tree was examined, it was observed that the highest discrimination among the populations was found between the Şirvan population, with 76% bootstrap value, and the populations located in Nizip, Besni, and Tut (Fig. 6). The lowest distinction occurred between the Nurdağı and Oğuzeli populations, with a bootstrap value of 27%. Molecular markers are frequently used in determining the genetic relationships of plant and animal species (Brown et al. 2020; İnak et al. 2020; Li et al. 2020) because it is thought that parameters such as morphologic and biochemical properties are weak in revealing the relationships. Until now, studies on genetic relationships among A. pistaciae populations, which are pistachio leaf psyllids, have not been found in the literature.
The ITS gene region has been used with the reliable high-resolution feature in intraspecies differentiation in many previous studies (Faghihi et al. 2020; Ghafar et al. 2020; Naz et al. 2020; Sumner-Kalkun et al. 2020). The same gene region has been commonly used by many researchers to reveal the phylogenetic relationships of many species belonging to the Psyllidae family, which includes plants such as pear, Indian devil’s tree, citrus, and potatoes, as well as pistachio (Ajene et al. 2020; Cho et al. 2020; Liu et al. 2006; Kang et al. 2012; Peccoud et al. 2013). When viewed from this aspect, the current study can be seen as the first report to infer the molecular genetic relationships by using the ITS gene region among spreading A. pistaciae populations in the three main pistachio cultivation regions of Gaziantep, Adıyaman, and Siirt. When the neighbor-joining phylogenetic tree used for this purpose was examined, no significant molecular relationship differences were detected in the population levels spreading in the three different regions. In this case, A. pistaciae species, which spread in these locations, was determined to be a similar species. Considering the morphologic data, a distinction among the species in completed microscopic studies was not observed. As a reason for this, it is estimated that there is no geographic speciation parameter that will create different speciation among regions. For instance, it is thought that the absence of a geographic barrier between Gaziantep and Siirt that would prevent migration of the species may have caused this situation to occur. As a matter of fact, Jackson et al. (2009) stated that the northward migration in the spring is supported by the monsoon winds from the Gulf of Mexico as a reason for the genetic difference in Bactericera cockerelli populations, but there may be geographic barriers between Guatemala and Mexico to prevent this migration. In particular, in B. cockerelli the northward migrations were generally facilitated by the winds (Lopez et al. 2004). Furthermore, Liu et al. (2006) and Jackson et al. (2009) reported this effect of the winds.
In addition, with the 16S rRNA barcode identifier used in the study, endosymbiont Wolbachia species were obtained. These intracellular Wolbachia species have recently been noted to infect a high number of insects, mites, isopods, and filarial nematodes (Stouthamer et al. 1999). As a matter of fact, Wolbachia species have been previously identified in the psyllid collections in the western United States and Mexico by using many molecular markers (mt COI, ITS2, ISSR) (Liu et al. 2006). These Wolbachia species infect the reproductive tissues of arthropods in the long term and also change reproduction in their arthropod host (Werren 1997). Wolbachia species have long been considered as a potential biocontrol agent for insects and the pathogens they transmit (Popovici et al. 2010).
Conclusion
In conclusion, no significant genetic differences in the population of individuals in Siirt, Gaziantep, and Adıyaman provinces could be detected. It was also determined that the A. pistaciae pests in these regions were found to be the same species. That result also supports the reliability of the data obtained with parameters such as the ITS gene region, which was often used in many previous scientific studies, and the neighbor-joining tree, which is the phylogenetic analysis method. In addition to these results, endosymbiotic Wolbachia bacterial species were identified in this study.
It is thought that this study will constitute a basic step for the future of molecular relationship studies of A. pistaciae, which have not been done until now in the molecular sense. Moreover, the genetic information obtained is valuable and important, as it will contribute to the determination of insecticide resistance developed by A. pistaciae populations in different regions.
References
Ajene IJ, Khamis F, Ballo S, Pietersen G, Van Asch B, Seid N, Mohamed S (2020) Detection of asian citrus Psyllid (Hemiptera: Psyllidae) in Ethiopia: a new Haplotype and its implication to the proliferation of Huanglongbing. J Econ Entomol. https://doi.org/10.1093/jee/toaa113
Boekhout T, Kurtzman CP, O’donnell K, Smith MT (1994) Phylogeny of the yeast genera Hanseniaspora (anamorph Kloeckera), Dekkera (anamorph Brettanomyces), and Eeniella as inferred from partial 26S ribosomal DNA nucleotide sequences. Int J Syst Evol Microbiol 44(4):781–786
Bolu H (1995) Güneydoğu Anadolu Bölgesinde Antep fıstığı (Pistacia vera L.)’nda Yaprak Pisillidi (Agonoscena pistaciae Burck. and Laut.) (Homoptera: Psyllidae)’nin Farklı Fıstık Çeşitlerinde Populasyon Değişimi ve Biyolojisi Üzerinde Araştırmalar. Çukurova Üniversitesi Fen Bilimleri Enstitüsü, Adana, p 54 (Yüksek Lisans Tezi)
Brown JW, Aarvik L, Heikkilä M, Brown R, Mutanen M (2020) Euliina (Lepidoptera: Tortricidae) ile olan ilişkisinin teyidi ile Cochylina’nın moleküler filogenisi. Sistematik Entomol 45(1):160–174
Burckhardt D (1994) Psylloid pests of temperate and subtropical crops and ornamental plants (Hemiptera, Psylloidea): a review. Trends Agric Sci Entomol 2:173–186
Burckhardt D, Lauterer L (1989) Systematics and biology of the rhinocolinae (Homoptera: Psylloidea). J Nat Hist 23(3):643–712
Burckhardt D, Lauterer P (1993) The jumping plant-lice of Iran (Homoptera: Psylloidea). Rev Suisse Zool 100(4):829–898
Burckhardt D, Ouvrard D (2012) A revised classification of the jumping plant-lice (Hemiptera: Psylloidea). Zootaxa 3509(1):34
Çelik MY (1975) Gaziantep İlinde Antep fıstığının zararlıları ve bunların faydalı böcekleri üzerinde çalışmalar vol 9. Gıda-Tarım ve Hayvancılık Bakanlığı, Zirai Mücadele ve Zirai Karantina Genel Müdürlüğü Bitki Koruma Araştırma Yıllığı (Özet), pp 43–44
Chen P, Liu Q, Qiao X, Wang J, Zhang T (2018) Identification and phylogenetic analysis of pear psyllids (Hemiptera: Psyllidae) in Chinese pear orchards. J Econ Entomol 20:1–6
Cho G, Malenovský I, Burckhardt D, Inoue H, Lee S (2020) DNA barcoding of pear psyllids (Hemiptera: Psylloidea: Psyllidae), a tale of continued misidentifications. Bull Entomol Res. https://doi.org/10.1017/S0007485320000012
Faghihi F, Hosseini-Chegeni A, Edalat H, Banafshi O, Telmadarraiy Z, Sedaghat MM (2020) Molecular identification of some Haemaphysalis species (Acari: Ixodidae) using mitochondrial and nuclear evidences in parts of Iran. Syst Appl Acarol 25(5):809–820
Ghafar A, Gasser RB, Rashid I, Ghafoor A, Jabbar A (2020) Exploring the prevalence and diversity of bovine ticks in five agro-ecological zones of Pakistan using phenetic and genetic tools. Ticks Tick Borne Dis. https://doi.org/10.1016/j.ttbdis.2020.101472
Günaydın T (1978) Güneydoğu Anadolu Bölgesinde antepfıstıklarmda zarar yapan böcek türleri, tanınmalan, yayıhşları ve ekonomik önemleri üzerinde araştırmalar. Ege Üniversitesi Ziraat Fakültesi Bitki Koruma Bölümü (Basılmamış )uzmanlık tezi, İzmir
Hillis DM, Davis SK (1986) Evolution of ribosomal DNA: fifty million years of recorded history in the frog Genus Rana. Evolution 40(6):1275–1288
Hillis DM, Moritz C, Porter CA, Baker RJ (1991) Evidence for biased gene conversion in concerted evolution of ribosomal DNA. Science 251(4991):308–310
İnak E, Çobanoğlu S, Sade E, Tixier MS (2020) Molecular characterization of phytoseiid mites in Turkey based on the internal transcribed spacer (ITS) region, with a new record for the country. Exp Appl Acarol 81:201–213
Jackson BC, Goolsby J, Wyzykowski A, Vitovksy N, Bextine B (2009) Analysis of genetic relationships between potato psyllid (Bactericera cockerelli) populations in the United States, Mexico and Guatemala Using ITS2 and Inter Simple Sequence Repeat (ISSR) Data. Subtrop. Plant Sci 61:1–5
Ji Y, Zhang D, He L (2003) Evolutionary conservation and versatility of a new set of primers for amplifying the ribosomal internal transcribed spacer regions in insects and other invertebrates. Mol Ecol Notes 3(4):581–585.
Kang AR, Baek YJ, Lee SH, Cho YS, Kim WS, Han YS, Kim I (2011) Geographic homogeneity and high gene flow of the pear psylla, Cacopsylla pyricola (Hemiptera: Psyllidae), detected by mitochondrial COI gene and nuclear ribosomal internal transcribed spacer 2 Animal Cells and Systems. J Animal Cell Syst 16(2):145–153
Katoh H, Inoue H, Uechi N, Fujikawa T, Miyata S, Iwanami T (2014) Analysis of the genetic population structure of Cacopsylla chinensis (Hemiptera: Psyllidae) with mitochondrial DNA markers in Saga and Yamaguchi prefectures, Japan. Jpn Agric Res Q 48:413–417
Klimaszewski SM, Lodos N (1977) New informations about jumping plant Iice of Turkey (Hornoptera Psylloidea). Ege Üniv Ziraat Fak Derg 14(2):259–267
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Lababidi MS, Zebitz CP (1995) Preliminary study on the pistachio psyllid (Agonoscena targionii Licht.)(Psyllidae: Homoptera) and its associated natural enemies in some regions of Syria. Arab J Plant Prot 13(2):62–68
Lauterer P, Broumas T, Drosopoulos S, Souliotis C, Tsourgianni A (1998) Species of the genus agonoscena, pests on Pistacia and first record of A. pistaciae in Greece. Anal Inst Phytopathol 18(2):123–128
Lee HC, Yang MM, Yeh WB (2008) Identication of two invasive Cacopsylla chinensis (Hemiptera: Psyllidae) lineages based on two mitochondrial sequences and restriction fragment length polymorphism of cytochrome oxidase I amplicon. ec 101(4):1152–1157
Li Y, Dong Y, Xu Q, Fan S, Lin H, Wang M, Zhang X (2020) Genetic differentiation and evolutionary history of the circumpolar species Ophiura sarsii and subspecies Ophiura sarsii vadicola (Ophiurida: Ophiuridae). Cont Shelf Res. https://doi.org/10.1016/j.csr.2020.104085
Liu D, Trumble JT, Stouthamer R (2006) Genetic differentiation between eastern populations and recent introductions of potato psyllid (Bactericera cockerelli) into western North America. Entomol Exp Appl 118(3):177–183
Lopez B, Favela S, Ponce G, Foroughbakhch R, Flores AE (2013) Genetic variation in Bactericera cockerelli (Hemiptera: Triozidae) from Mexico. J Econ Entomol 106(2):1004–1010
Maryańska-Nadachowska A, Kuznetsova VG, Golub NV, Anokhin BA (2018) Detection of telomeric sequences and ribosomal RNA genes in holokinetic chromosomes of five jumping plant-lice species: First data on the superfamily Psylloidea (Hemiptera: Sternorrhyncha). Eur J Entomol. https://doi.org/10.14411/eje.2018.061
Mehrnejad MR (2003) The influence of Host Species on Some Biological and Behavioural Aspects of Dibrachys boarmiae (Hymenoptera: Pteromalidae), Parasitoid of Kermania pistaciella (Lepidoptera: Tineidae). Biocontrol Sci Technol 13(2):219–229
Mehrnejad MR (2014) Pest problems ın pıstachıo producıng areas of the world and their current means of control. Acta Hortic 1028:163–169
Meng L, Wang Y, Wei WH, Zhang H (2018) Population genetic structure of Diaphorina citri Kuwayama (Hemiptera: Liviidae): host-driven genetic differentiation in China. Sci Rep 8(1):1–15
Mindell DI, Honeycut RL (1990) Ribosomal RNA in vertebrates: evolution and phylogenetic applications. Annu Rev Ecol Syst 21(1):541–566
Naz S, Chaudhry FR, Rizvi DA, Ismail M (2020) Phylogenetic analysis of astigmatid mites sarcoptes scabiei and dermatophagoides farinae using ITS‑2 as a genetic marker. Life Sci 1(2):5–5
Peccoud J, Labonne G, Sauvion N (2013) Molecular test to assign individuals within the cacopsylla pruni complex. Plos One 8(8):e72454
Pinto JD, Stouthamer R (1994) Systematics of the trichogrammatidae with emphasis on trichogarmma. In: Wajnberg E, Hassan SA (eds) Biological control with egg parasitoids. CABI, Wallingford, pp 1–28
Popovici J, Moreira LA, Poinsignon A, Iturbe-Ormaetxe I, McNaughton D, O’Neill SL (2010) Assessing key safety concerns of a Wolbachia-based strategy to control dengue transmission by Aedes mosquitoes. Mem Inst Oswaldo Cruz 105(8):957–964
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Seemüller E, Schneider B, Jarausch B (2011) Pear decline phytoplasma. In: Hadidi A, Barba M, Candresse T, Jelkmann W (ed) Virus and virus-like diseases of pome and stone fruits. APS press, pp 77–84
Stouthamer R, Breeuwer JAJ, Hurst GDD (1999) Wolbachia pipientis, Microbial manipulator of arthropod reproduction. Annu Rev Microbiol 53:71–102
Sumner-Kalkun JC, Sjölund MJ, Arnsdorf YM, Carnegie M, Highet F, Ouvrard D, Kenyon DM (2020) A diagnostic real-time PCR assay for the rapid identification of the tomato-potato psyllid, Bactericera cockerelli (Šulc, 1909) and development of a psyllid barcoding database. PLoS ONE 15(3):e230741
Tokmakoğlu C (1973) Antep fıstığı (Pistacia vera) zararlısı Agonoscena targionii ucht. Böceğin biyolojisi ve mücadelesi ile ilgili bazı tespitler. Bitki Koruma Bülteni 13(2):67–72
Werren JH (1997) Wolbachia run amok. Proc Natl Acad Sci 94(21):11154–11155
Werren JH, Baldo L, Clark ME (2008) Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol 6:741–751
Wilson MR, Dizinno JA, Polanskey D, Replogle J, Budowle B (1995) Validation of mitochondrial DNA sequencing for forensic casework analysis. Int J Leg Med 108(2):68–74
Xiong X, Wu L, Xin T, Wang J, Zou Z, Xia B (2017) The complete mitochondrial genome of Diaphorina citri (Hemiptera: Psyllidae) and phylogenetic analysis. Biochem Syst Ecol 70:230–238
Yang MM, Huang JH, Li F (2004) A new record of Cacopsylla species (Hemiptera: Psyllidae) from pear orchards in Taiwan. Formos Entomol 24(1):213–220
Acknowledgements
We are thankful to Associate Professor Dr. Behcet İNAL (SIU) for his technical support and to the Yüzüncü Yıl University Scientific Research Project Unit (project number FDK-2017-5951) for funding the current project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
H. Dilmen, M.S. Özgökçe, and B. İnal declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Dilmen, H., Özgökçe, M.S. & İnal, B. Molecular Characterization of Agonoscena pistaciae Burckhardt and Lauterer (Hemiptera Psyllidae) Populations Spreading in Southeast Anatolia. Erwerbs-Obstbau 64, 717–724 (2022). https://doi.org/10.1007/s10341-022-00697-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10341-022-00697-4