Abstract
The occurrence and localization of endophytic actinomycetes within diverse organs of host plants provide ecological information that can be used to evaluate the significance of their spatial habitats. The aim of this study was to isolate and characterize endophytic actinomycetes in different organs of Quercus serrata. For this purpose, actinomycete isolates were obtained from surface-sterilized tissues of both symptomless seedlings and aged trees of Q. serrata and rhizosphere soil of the sampled seedlings. Thirty-five isolates with the ability to sporulate, including 4 from leaves of the aged trees, 10, 6, and 15 from leaves, stems, and roots of the seedlings, respectively, and 8 soil-derived isolates, were selected and characterized. The 16S rDNA nucleotide sequence analyses revealed that all of them belonged to the genus Streptomyces. According to a neighbor-joining phylogenetic tree constructed based on the results, the isolates of plant origin were divided into three major clades with high bootstrap values of 98 or 99 %, whereas eight soil-derived isolates were located at different positions from those of the endophyte isolates. Moreover, two larger clades were formed, one of which contained isolates derived only from aboveground parts, while the other contained isolates from all of the organs. These results suggest that the endophytic streptomycetes in Q. serrata may differ in their habitat positions (i.e., either above- or belowground parts).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Actinomycetes are prokaryotic microbes that belong to the phylum Actinobacteria, and typically possess mycelium and spore forms (Williams et al. 1989). Most actinomycetes were once believed to survive in environments such as the soil, rhizosphere, and pond and lake sediments as saprophytes, by degrading organic materials for nutrition (Ishiyama et al. 2004). Since Brunckhorst (1886) isolated the genus Frankia from nonlegume root nodules in 1886 (Okazaki 2003; Hasegawa et al. 2006), some actinomycetes have been considered to be parasitic, symbiotic, or saprophytic endophytes. A few endophytic actinomycetes, such as Streptomyces scabies, a causal pathogen of potato scab, are pathogenic to host plants (Loria et al. 1997) and locally or systemically colonize plant tissues, causing serious disease in the host plant, while others latently reside in the internal tissues of nonsymptomatic plants, establishing intimate associations with host plants. Endophytic actinomycetes are known to influence the physiology of host plants in diverse ways: some of them have the potential to improve and promote the growth of host plants (Meguro et al. 2006), as well as to reduce disease symptoms caused by plant pathogens (Kunoh 2002; Nishimura et al. 2002; Shimizu et al. 2000, 2001, 2006) and/or various environmental stresses (Hasegawa et al. 2004, 2005). Many researchers have reported that secondary metabolites produced by endophytic microbes and rhizobacteria in or on their host plants often alter the physiology of the host plants, rendering them resistant to biotic or abiotic environmental stresses (van Loon et al. 1998; Shimizu et al. 2001). Endophytes that colonize plant interiors usually derive nutrition and protection from the host plants. In return, they confer profoundly enhanced fitness to the host plants by producing a variety of bioactive metabolites. Growth stimulation of plants by endophytes can be a consequence of nitrogen fixation, the production of phytohormones, or biocontrol of phytopathogens—achieved through the production of antibiotics or siderophores, nutrient competition, or the induction of systemic disease resistance (Hasegawa et al. 2006; Shimizu 2011). Aside from interest in this interference from the endophytic actinomycetes in plants, their production of antibiotics and other secondary products has attracted pharmacologists and agrochemists, as proven by the recent discovery of several novel bioactive compounds produced by actinomycetes of plant origin (Bieber et al. 1998; Igarashi et al. 2000, 2002; Sasaki et al. 2001a, b). All of these reports clearly show that endophytic actinomycetes possess a variety of ways to affect plants in nature and to produce diverse bioactive compounds that are beneficial to human, animal, and plants.
Although research into endophytic actinomycetes has a relatively short history, over the last few decades many researchers have attempted to isolate them from a wide range of cultivated and wild plants (de Araújo et al. 2000; Nishimura et al. 2002; Coombs and Franco 2003; Taechowisan et al. 2003; Cao et al. 2004; Conn and Franco 2004; Gu et al. 2007; Tian et al. 2007; Zin et al. 2007; Shimizu et al. 2000, 2009). Nevertheless, the ecology of endophytic actinomycetes in natural forests is poorly understood, because most earlier works concentrated on herbaceous crops, wild plants, and/or ornamental woody plants, as reviewed by Shimizu (2011). Because naturally regenerated forests, which are affected by the ever-changing environment, include complicated microbial communities with great ecological diversity, there must be a good chance that they contain as-yet undiscovered species of endophytic actinomycetes that produce useful bioactive compounds and/or have unexpected positive effects on individual trees. Information on the diversity of endophytic actinomycetes in forests and their organ specificity is desirable, not only to aid in the screening of strains in order to find new biocontrol agents but also to elucidate their ecological roles, which could be useful in forest management, such as in attempts to biocontrol the pathogen of Japanese oak wilt disease (Seo et al. 2011). These earlier papers led us to isolate endophytic actinomycetes from above- and belowground organs of Quercus serrata, a common tree species in secondary forests in the warm- and cool-temperate regions of Japan (Ito and Kawasato 1978), for a series of ecological studies on forest tree-associated microbial communities. Thus, the aim of this study was to obtain and characterize the actinomycetes present in different organs of Q. serrata. The preferential distributions of actinomycetes within the host organs were determined based on the locations of the isolates in planta.
Materials and methods
Sample collection
In this study, we collected Q. serrata seedlings in order to obtain three parts: leaves, stems, and roots. In addition, only the leaves of aged trees were investigated to ensure that they were from high above ground level. Three one- to two-year-old symptomless seedlings of Q. serrata and 3–4 twigs, each with healthy leaves, from three aged trees were collected from the Experimental Forest of Mie Prefectural Research Institute (34°36′N, 136°21′E, 250 m asl) in Mie Prefecture, Japan, in April 2008. In this forest, Q. serrata and Q. glauca are the dominant species, and individual trees are spaced 10 m apart. For reference, soil samples from the seedling rhizosphere were also collected and mixed. The actinomycetes in all of these samples were isolated within 48 h of collection.
Isolation of endophytic and rhizosphere-inhabiting actinomycetes
The surfaces of leaves, stems, and roots that had been separated from the seedlings and intact leaves from the aged tree twigs were washed in running tap water. Roots were additionally washed in distilled water with an ultrasonic cleaner machine (VC-1, AS ONE Corp., Japan) to dislodge any soil particles and organic debris on their surfaces. Subsequently, the surfaces of the samples were sterilized by submerging them into 70 % ethanol (v/v) for 1 min and then sodium hypochlorite (1 % w/v available chlorine) for 3 min (Okazaki 2003). After being rinsed twice with sterilized distilled water, they were dried on an autoclaved filter paper. The samples were then cut into small pieces of size ca. 3 × 3 mm2 and a total of 120 segments from each organ were placed on water-agar plates that contained 10 mL of Heritage (a fungicide from Zeneca, UK) diluted 30-fold and 10 mL of amphotericin B (an antibiotic, 50 mg in 10 mL of DMSO) in 1 L of medium. They were incubated at 30 °C for three months or more. Colonies emerging from the pieces during the three months of incubation were counted and then transferred with sterilized toothpicks onto membrane filters (mixed cellulose ester, pore size 0.2 μM) (Polsinelli and Mazza 1984) placed on the surface of IMA-2 medium (Shimizu et al. 2000). The plates were then incubated at 30 °C for at least one month. In this method, only actinomycetes grew underneath the membrane filters—bacteria and/or fungi did not. The actinomycetous colonies were independently suspended in 15 mL of 10 % glycerol–10 % DMSO, and then their spores were collected by filtering this through a cotton sheet on a funnel. To disperse the spores evenly, 2 mL of the filtrate were gently sonicated in a sonic water bath for 10 min. The resultant spore suspension was serially diluted with sterilized distilled water. Small aliquots of the diluted suspensions were spread on IMA-2 agar plates and incubated at 30 °C for about 14 days. Visually selected independent colonies were streaked onto IMA-2 agar plates, incubated as described above, and obtained as purified isolates. These steps were repeated until the isolates were purified. The purified isolates were grown on IMA-2 agar plates and incubated at 30 °C for an additional month. To specify plausible actinomycete isolates, only the sporulating isolates were used for subsequent experiments.
The effect of the surface-sterilization protocol described above was validated in a separate experiment as follows. Ten segments of each surface-sterilized organ tissue were soaked in 5 mL of sterilized distilled water and mixed well with a stirrer for 1 min. Aliquots (0.3 mL) of the wash fluid were dropped onto HV (humic acid–vitamin) agar medium (Hayakawa and Nonomura 1987) and incubated at 30 °C, before being examined under a microscope for any microbial growth (Cao et al. 2004).
To isolate actinomycetes from the soil sample, 10 g of air-dried soil were suspended in 90 mL of sterilized tap water in a flask, and this was vigorously stirred for 10 min. The resultant soil suspension was diluted serially and then incubated on HV agar plates at 30 °C for one month. The actinomycetous colonies were purified in a similar manner to the endophytic actinomycetes (see above), and only those with different visual colony appearances and sporulation were subjected to the subsequent visual and microscopic characterization and 16S rDNA sequence analysis.
Visual and microscopic observations
The isolates were streaked onto yeast–starch agar (Emerson 1941) and incubated at 30 °C for 3–4 weeks. Visual morphological characters were examined according to the traditional criteria: aerial spore-mass color, substrate mycelium color, soluble pigment color, and spore chain morphology (Shirling and Gottlieb 1966). Based on the spore chain morphology observed via light microscopy, aerial hyphae and associated spore chains of the respective isolates were categorized into the following types: rectiflexibiles (RF; straight or flexuous spore chains, partly in fascicles), retinaculiaperti (RA; spore chains with hooks, open loops, or short, irregular spirals with 1–3 turns extending into spirals with a wider diameter), or spirales (S; spore chains of either closed compact spirals or open, loose, long, stretched spirals).
16S rDNA nucleotide sequence analysis
The isolates were grown on IMA-2 medium for on month at 30 °C. The independent colonies of area ~4 mm2 were picked up with sterilized toothpicks and transferred individually into 1.5 mL centrifuge tubes. Total DNA was extracted from the specimens using the ISOPLANTII kit (Nippon Gene, Tokyo, Japan) according to the manufacturer’s protocol. The 16S rDNA region was amplified by polymerase chain reaction (PCR) using a universal primer set of 27f (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1525r (5′-AAGGAGGTGWTCCARCC-3′), which can amplify almost the full length of the region (ca. 1.5 kbp; Lane 1991). Fifty microliters of PCR reaction mixture consisted of 2 μL of DNA template, 4 μL of 2.5 mM deoxyribonucleotide triphosphate (dNTP), 2 μL of each primer described above (1.0 μM final conc.), 0.25 μL of EX Taq™ DNA polymerase (Takara, Otsu, Japan), 5 μL of 10× Ex Taq buffer, and 34.75 mL of sterilized distilled water. The mixture was amplified using a thermal cycler (TaKaRa PCR Thermal Cycler Dice, model TP600, Otsu, Japan) with a preheating stage at 95 °C for 5 min, and then 30 thermal cycles of 1 min at 95 °C for denaturing, 1 min at 50 °C for annealing, and 1 min at 72 °C for extension. The reaction was kept at 72 °C for 10 min for a final extension.
Successfully amplified PCR products were purified with the illustra™ GFX™ PCR DNA and Gel Band purification kit (GE Healthcare, Chalfont St. Giles, UK). They were then subjected to sequencing reactions using the thermal cycler with dye-labeled dideoxy terminators of the DYEnamic ET Terminator cycle sequencing kit (GE Healthcare). The sequence reaction was conducted with one of 27f, 1525r, or 533f (5′-GTGCCAGCAGCCGCGGTAA-3′; Weisburg et al. 1991) primers. For each isolate, the sequences obtained were manually contiged with the GENETYX v.4.0 software package (Software Development Co., Ltd., Tokyo, Japan). Accession numbers (AB641840–AB641882) for the sequenced data were obtained from the DNA Data Bank of Japan (DDBJ). The sequence for each isolate was compared with those deposited in the DDBJ/EMBL/Genbank database by BLAST homology (Altschul et al. 1997) to evaluate the similarity of actinomycete taxa.
Phylogenetic analyses
Sequences from the isolates were first aligned with ClustalW, and a phylogenetic tree was constructed using the neighbor-joining (NJ) method and the molecular evolutionary genetics analysis (MEGA) program, version 4 (Tamura et al. 2007). Values of higher than 70 % for the replicate trees in which the associated sequences clustered together in the bootstrap (BS) test (1,000 replicates) were shown next to the branches (Felsenstein 1985). All positions containing gaps and missing data were eliminated from the dataset (complete deletion option). Kitasatospora setae (AB184576) and K. azatica (U93312) were used as outgroups.
Results and discussion
No microbes grew from the wash fluids obtained from surface-sterilized specimens during three months of incubation on HV medium, proving that the current surface sterilization method does indeed eradicate epiphytes, which in turn meant that the microbial colonies that emerged from the specimen surfaces were endophytes.
Shimizu et al. (2000) observed that mycelia of endophytic actinomycetes gradually broke through the epidermises of leaves, stems, and roots of Rhododendron sp. and became visible 3–4 weeks after the onset of incubation. In our plant samples, some mycelial colonies of endophytic actinomycetes became visible on the specimen surfaces within one month, but the majority required a longer period of up to three months, especially in leaf tissues. During this period, a total of 138 actinomycetous colonies emerged from the specimen surfaces: 71 isolates from roots, 48 from leaves, and 19 from stems. In concurrence with earlier reports (Verma et al. 2009; Shimizu 2011), the largest number of endophytic actinomycetous isolates was found in roots of Q. serrata. The purification steps allowed only 35 isolates to grow and sporulate on IMA-2 medium for an additional month. Among these sporulating isolates, 4 isolates originated from leaves of the aged tree (hereafter referred to as A-leaf isolates), and 10, 6, and 15 isolates originated from leaves, stems, and roots of the seedlings (hereafter referred to as S-leaf, -stem, and -root isolates, respectively). These 35 isolates were used for the subsequent experiments.
To identify the actinomycetous isolates, Miyadoh (2001) recommended careful examination of their growth and the morphology of mycelia and aerial spores on the medium as the first step towards distinguishing Streptomyces from non-Streptomyces isolates. Because the current isolates were initially selected based on visual colony appearance and sporulation (as described above), there was no reason to believe that each isolate corresponded to an independent strain. To solve this issue, the morphological and cultural characters of each isolate from Q. serrata organs and the soils were compared, as listed in Table 1. Aerial spore-mass colors of the isolates varied from white to gray, green, and red. Substrate mycelium colors also varied, from colorless to black, brown, yellow, and orange. Some isolates produced soluble pigments, others did not. Yellow pigments were most frequently produced by 21 isolates and orange by 10 isolates. These characters were seen in all isolates from different organs. By contrast, spore chain morphology showed a rather isolation source specific pattern: all (4/4) of the A-leaf isolates were of RF type; in S-leaf isolates, 8/10 were of RF type and 2/10 were of S type; in S-stem isolates, 2/6 were of RF type and 4/6 were of S type; in S-root isolates, 1/15 were of RF and RA type, respectively, and 13/15 were of S type. The pattern for the S-root isolates was similar to that for the soil-derived isolates (1/8 were of RF and RA type, respectively, and 6/8 were of S type), as was expected. The RF type was dominant in leaves of aged trees and seedlings, while the S type was dominant in stems and roots of seedlings, as well as in soil. When all of the above characters were compared among the isolates, it was apparent that they shared only a few morphological and cultural characters, suggesting that the respective isolates might be different strains, although more detailed examination of other characters is certainly required before the respective strains can be defined.
Data on the 16S rDNA nucleotide sequences of the respective isolates are listed in Table 2, which also shows the registered strains (along with their accession numbers) that are most similar to the analyzed isolates. The full-length nucleotide sequences of the 16S rDNA regions from the 35 endophytic and 8 soil actinomycetes were all successfully obtained, and ranged from 1,353 to 1,494 bp. All of the sequences obtained very closely matched those of the genus Streptomyces, with match E (“Expect”) values of zero, suggesting that all of the isolates could belong to the genus Streptomyces. According to Thakur et al. (2007), Streptomyces is a large genus that nearly half of all soil-inhabiting saprophytic actinomycetes belong to. In addition, Streptomyces species are most frequently isolated from plant tissues, just like Microbispora species (Sardi et al. 1992; Okazaki 2003; Takahashi and Omura 2003; Inderiati and Muliani 2008; Verma et al. 2009). Therefore, these earlier descriptions are not inconsistent with the current results which indicate that all of the isolates from the plant organs and soil belonged to the genus Streptomyces.
The 16S rDNA nucleotide sequence analyses showed that some isolates had the same or similar sequences to each other. For comparisons of 16S rDNA regions, it is generally accepted that identities of at least 97 % indicate that the nucleotide derives from the same member (Petit et al. 1999). The nucleotide sequences of most isolates highly matched those of streptomycete species (similarity ≥ 97 %; E values = 0; Table 2). However, 11/35 of those of plant origin showed lower matches of 94–96 %. By contrast, all eight isolates from the soil showed higher similarities of 98–99 %. The wider range of similarities seen for the plant-derived sequences may indicate that some of the endophytic streptomycetes in Q. serrata are more unique than the soil-inhabiting ones. As is evident from Table 2, the isolates with sequences that highly matched those of S. collinus subsp. albescens were most frequently obtained from all three organs of the seedlings, suggesting that they might be well adapted to the in planta environment. Nucleotide sequences from the leaf isolates KSL138F50, KSL168F2, and KSL378F56 (from S-leaves) and KAL258F47 and KAL398F43 (from A-leaves) as well as the S-stem isolate KSS348F38 closely matched those of S. scabiei (98–99 % similarity). Although this species is known to be the causal agent of potato scab (Loria et al. 1997), it is also known to be endophytic species in healthy rice and potato stems (Sessitsch et al. 2002; Tian et al. 2007). Thus, the above five isolates from leaves may have acquired the potential to live inside leaves of Q. serrata.
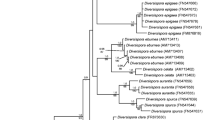
An NJ phylogenetic tree was constructed that incorporated the 43 obtained isolates as well as the registered strains of similar nucleotide sequences for the 16S rDNA region (Fig. 1). The current endophyte isolates were located in different clades but were never nested with the soil-derived isolates. Twenty-one isolates from the three different organs of the seedlings were positioned with other known Streptomyces sequences in a clade with a high BS value of 98 % (group I). Together with two S-root isolates (KSR108A8 and KSR118S17), S. mirabilis (FJ481081) formed a clade with a BS value of 99 % (group III). Another clade comprising S-stem and S-/A-leaf isolates along with other registered Streptomycetes showed a BS value of 99 % (group II). Although 7/8 of the soil isolates appeared to be closely related to group I, the BS value was 70 %, suggesting that the soil isolates should be members of different groups. The current isolates of group I were obtained from both the above- and belowground parts of seedling tissues, whereas those of group II were only from aboveground parts, regardless of age. These results may imply that endophyte isolates preferentially distribute to particular organs, rather than to trees of particular ages (Mano and Morisaki 2008).
Neighbor-joining phylogenetic tree constructed based on an analysis of the 16S rDNA sequences of Streptomyces isolates obtained from Quercus serrata and soil samples. Numbers next to branches indicate levels of bootstrap support based on 1,000 resampled data sets, and are shown if they are ≥70 %. The scale bar represents 0.01 substitutions per nucleotide position. The isolates analyzed in this study are shown in bold. Kitasatospora setae and K. azatica were used as outgroups
Although some researchers (Matsukuma et al. 1994; Takahashi and Omura 2003) have reported the isolation of actinomycetes from woody plant leaf litter, they failed to show whether their isolates were endophytes or epiphytes. By contrast, Sardi et al. (1992) successfully isolated endophytic streptomycetes from the root tissues of Quercus sp. Inderiati and Muliani (2008) reported that the majority of the endophytic actinomycetes recovered from tobacco plants belonged to the genus Streptomyces. Likewise, Sardi et al. (1992) reported that 482 of the 499 isolates obtained from a variety of plants were Streptomyces spp. Interestingly, the majority of the most frequently isolated endophytic Streptomyces species from various countries and regions belong to only a few species, including S. aureus, S. galilaeus, S. caviscabies, S. setonii, S. cyaneus, and S. thermocarboxydus (Shimizu 2011), suggesting that these species could have a higher compatibility with a wide range of plants than other species. Considering that S. collinus subsp. albescens was most frequently isolated in this study but is not present in the above list, it is plausible that this species may be specifically compatible with Q. serrata. However, the isolates studied in the present study were obtained as a result of the specific purification and cultivation process used, before they were morphologically and genetically characterized. Thus, this process may have only dealt with a particular fraction of all of the endophytic actinomycetes present.
Zinniel et al. (2002) noted that the relatively large population of endophytic actinomycetes in roots may indicate that soil-inhabiting actinomycetes can readily move to soil-contacting roots, like endophytic bacteria. In fact, Conn and Franco (2004) demonstrated that the density of endophytic actinomycetes in wheat roots detected by terminal restriction fragment length polymorphism was closely related to that of the soil microflora. Tian et al. (2007) analyzed the 16S rDNA genes of 45 and 33 clones of endophytic actinomycetous isolates obtained from roots and stems, respectively. They found that the clones of the isolates from roots were affiliated with nine known actinomycete genera as well as other unidentified actinomycetes, whereas the clones of the isolates from stems comprised only four known actinomycete genera as well as other unidentified actinomycetes. This result suggests that a wider range of known and unidentified actinomycetes reside inside roots than in the stem region. As Berg et al. (2005) and Ulrich et al. (2008) have suggested, the endophytic actinomycetous community could be a subset of the soil microbial community. However, the current soil-derived isolates never nested in the clades of endophytic isolates. This result may indicate that the latter isolates have independently evolved such that they have adapted to the in planta environment, assuming that they did indeed originally move from the rhizosphere. To our knowledge, few studies have been done on the patterns of occurrence of endophytic actinomycetes in above- and belowground parts. The phylogenetic tree constructed in the present study showed a clade that only contained isolates from the aboveground parts of both Q. serrata seedlings and aged trees. In this respect, this is the first study to infer that endophytic streptomycetes may be organ-specific.
In conclusion, woody plant tissues can harbor various groups of endophytic streptomycetes that are drastically different from those found in the surrounding soil. Moreover, the complex structure of plant tissues may account for the distribution patterns of the endophytes. It is unclear, however, whether the lopsided endophytic colonization seen for this tree (only endophytic colonization by streptomycetes species was observed) is indeed specific to this tree, or whether the environmental conditions of the forest and/or the physiological condition of the tree influence this lopsided colonization and diversity. Further careful ecological studies are required to solve these intriguing questions.
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Berg G, Krechel A, Ditz M, Sikora R, Ulrich A, Hallmann J (2005) Endophytic and ectophytic potato-associated bacterial communities differ in structure and antagonistic function against plant pathogenic fungi. FEMS Microbiol Ecol 51:215–229
Bieber B, Nuske J, Ritzau M, Grafe U (1998) Alnumycin, a new naphthoqionone antibiotic, produced by Streptomyces sp. J Antibiot 51:381–382
Brunckhorst J (1886) Über einige Wuerzelanschwellungen, besonders diejenigen von Alnus und den Elaeagnaceen. Unters Botan Inst Tübingen 2:151–177
Cao L, Qui Z, Dai X, Tan H, Lin Y, Zhou S (2004) Isolation of endophytic actinomycetes from roots and leaves of banana (Musa acuminata) plants and their activities against Fusarium oxysporum f. sp. cubense. World J Microbiol Biotechnol 20:501–504
Conn VM, Franco CMM (2004) Analysis of the endophytic actinobacterial population in the roots of wheat (Triticum aestivum L.) by terminal restriction fragment length polymorphism and sequencing of 16S rRNA clones. Appl Environ Microbiol 70:1787–1794
Coombs JT, Franco CMM (2003) Isolation and identification of actinobacterial from surface-sterilized wheat roots. Appl Environ Microbiol 69:5603–5608
de Araújo JM, da Silva AC, Azevedo JL (2000) Isolation of endophytic actinomycetes from roots and leaves of maize (Zea mays L.). Braz Arch Biol Technol 43:447–451
Emerson R (1941) An experimental study of the life cycles and taxonomy of Allomyces. Lioydia 4:77–144
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Gu Q, Zhen W, Huang Y (2007) Glycomyces sambucussp. nov., and endophytic actinomycetes isolated from the stem of Sambucus adnatawall. Int J Syst Evol Microbiol 57:1995–1998
Hasegawa S, Meguro A, Nishimuta T, Kunoh H (2004) Drought tolerance of tissue-cultured seedlings of mountain laurel (Kalmia latifolia L.) induced by an endophytic actinomycete. I. Enhancement of osmotic pressure in leaf cells. Actinomycetelogica 18:43–47
Hasegawa S, Meguro A, Nishimuta T, Kunoh H (2005) Drought tolerance of tissue-cultured seedlings of mountain laurel (Kalmia latifolia L.) induced by an endophytic actinomycete. II. Acceleration of callose accumulation and lignification. Actinomycetelogica 19:13–17
Hasegawa S, Meguro A, Shimizu M, Nishimura T, Kunoh H (2006) Endophytic actinomycetes and their interactions with host plants. Actinomycetologica 20:72–81
Hayakawa M, Nonomura H (1987) Humic acid–vitamin agar, a new medium for the selective isolation of soil actinomycetes. J Ferment Technol 65:501–509
Igarashi Y, Ogawa M, Sato Y, Saito N, Yoshida R, Kunoh H, Onaka H, Furumai T (2000) Fistupyrone, a novel inhibitor of the infection of Chinese cabbage by Alternaria brassicicola, from Streptomyces sp. TP-A0569. J Antibiot 53:1117–1122
Igarashi Y, Iida T, Yoshida R, Furumai T (2002) Pteridic acids A and B, novel plant growth promoters with auxin-like activity from Streptomyces hygroscopicus TP-A0451. J Antibiot 55:764–767
Inderiati S, Muliani S (2008) Isolation and characterization of endophytic actinomycetes of tobacco plants. J Agrisistem 4:82–100
Ishiyama D, Vujaklija D, Davies J (2004) Novel pathway of salicylate degradation by Streptomyces sp. strain WA46. Appl Environ Microbiol 70:1297–1306
Ito S, Kawasato H (1978) Distribution of secondary forests in Japan. In: Paper on plant ecology to the memory of Dr. Kuniji Yoshioka. Group of plant ecology in Tohoku, Sendai, pp 281–284 (in Japanese)
Kunoh H (2002) Endophytic actinomycetes: attractive biocontrol agents. J Gen Plant Pathol 68:249–252
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–175
Loria R, Bukhalid A, Fry AB, King PR (1997) Plant pathogenicity in the genus Streptomyces. Plant Dis 81:836–846
Mano H, Morisaki H (2008) Endophytic bacteria in the rice plant. Microbes Environ 23:109–117
Matsukuma S, Okuda T, Watanabe J (1994) Isolation of actinomycetes from pine litter layers. Actinomycetologica 8:57–65
Meguro A, Ohmura Y, Hasegawa S, Shimizu M, Nishimura T, Kunoh H (2006) An endophytic actinomycete, Streptomyces sp. MBR-52, that accelerates emergence and elongation of plant adventitious roots. Actinomycetologica 20:1–9
Miyadoh S (2001) Identification protocols of the genera of actinomycetes. In: The Society for Actinomycetes Japan (ed) Identification manual of actinomycetes. Business Center for Academic Societies Japan, Tokyo, pp 9–35 (in Japanese)
Nishimura T, Meguro A, Hasegawa S, Nakagawa Y, Shimizu M, Kunoh H (2002) An endophytic actinomycete, Streptomyces sp. AOK-30, isolated from mountain laurel and its antifungal activity. J Gen Plant Pathol 68:390–397
Okazaki T (2003) Studies on actinomycetes isolated from plant leaves. In: Kurtboke I (ed) Selective isolation of rare actinomycetes. National Library of Australia, Queensland, pp 102–121
Petit JR, Jouzel J, Raynaud D, Barkov NI, Barnola JM, Basile I, Benders M, Chappellaz J, Davis M, Delaygue G, Delmotte M, Kotlyakov VM, Legrand M, Lipenkov VY, Lorius C, Pepin L, Ritz C, Saltzman E, Stievenard M (1999) Climate and atmospheric history of the past 420,000 years from the Vostock ice core, Antarctica. Nature 399:429–436
Polsinelli M, Mazza PG (1984) Use of membrane filters for selective isolation of actinomycetes from soil. FEMS Microbiol Lett 22:79–83
Sardi P, Saracchi M, Ouaroni S, Petrolini B, Borgonovoli GE, Merli S (1992) Isolation of endophytic Streptomycetes from surface-sterilized roots. Appl Environ Microbiol 58:2691–2693
Sasaki T, Igarashi Y, Saito N, Furumai T (2001a) TPU-0031-A and B, new antibiotics of the novobiocingroup produced by Streptomyces sp. TP-A0556. J Antibiot 54:441–447
Sasaki T, Igarashi Y, Saito N, Furumai T (2001b) Cedamycins A and B, new antimicrobial antibiotics from Streptomyces sp. TP-A0456. J Antibiot 54:567–572
Seo MY, Matsuda Y, Nakashima C, Ito S (2011) Taxonomic re-evaluation of Raffaelea quercivora isolates collected from mass mortality of oak trees in Japan. Mycoscience. doi:10.1007/s10267-011-0154-z
Sessitsch A, Reiter B, Pfeifer U, Wilhelm E (2002) Cultivation-independent population analysis of bacterial endophytes in three potato varieties based on eubacterial and Actinomycetes-specific PCR of 16S rRNA genes. FEMS Microbiol Ecol 39:23–32
Shimizu M (2011) Endophytic actinomycetes: biocontrol agents and growth promoters. In: Maheshwari DK (ed) Bacteria in agrobiology (plant growth responses). Springer, Dordrecht, pp 201–220
Shimizu M, Nakagawa T, Sato Y, Furumai T, Igarashi Y, Onaka H, Yoshida R, Kunoh H (2000) Studies on endophytic actinomycetes (1). Streptomyces sp. isolated from rhododendron and its antifungal activity. J Gen Plant Pathol 66:360–366
Shimizu M, Furumai T, Igarashi Y, Onaka H, Nishimura T, Yoshida R, Kunoh H (2001) Association of induced disease resistance of rhododendron seedlings with inoculation of Streptomyces sp. R-5 and treatment with actinomycin D and amphotericin B to the tissue-culture medium. J Antibiot 54:501–505
Shimizu M, Meguro A, Hasegawa S, Nishimuta T, Kunoh H (2006) Disease resistance induced by nonantagonistic endophytes Streptomyces spp. on tissue-cultured seedlings of rhododendron. J Gen Plant Pathol 72:351–354
Shimizu M, Yazawa S, Ushijima Y (2009) A promising strain of endophytic Streptomyces sp. for biological control of cucumber anthracnose. J Gen Plant Pathol 75:27–36
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340
Taechowisan T, Peberdy JF, Lumyong S (2003) Isolation of endophytic actinomycetes from selected plants and their antifungal activity. World J Microbiol Biotechnol 19:381–385
Takahashi Y, Omura S (2003) Isolation of new actinomycete strains for the screening of new bioactive compounds. J Gen Appl Microbiol 49:141–154
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thakur D, Yadav A, Gogoi BK, Bora TC (2007) Isolation and screening of Streptomyces in soil of protected forest areas from the state of Assam and Tripura, India, for antimicrobial metabolites. J Mycol Med 17:242–249
Tian XL, Cao LX, Tan HM, Han WQ, Chen M, Liu YH, Zhou SN (2007) Diversity of cultivated and uncultivated actinobacterial endophytes in the stems and roots of rice. Microb Ecol 53:700–707
Ulrich K, Ulrich A, Ewald D (2008) Diversity of endophytic bacterial communities in poplar grown under field conditions. FEMS Microbiol Ecol 63:169–180
van Loon LC, Bakker PAHM, Pieterse CMJ (1998) Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol 36:453–483
Verma VC, Gond SK, Kumar A, Mishra A, Kharwar RN, Gange AC (2009) Endophytic actinomycetes from Azadirachta indica A. Juss: isolation, diversity, and anti-microbial activity. Microb Ecol 57:749–756
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Williams ST, Sharpe ME, Holt JG, Murray GE, Brener DJ, Krieg NR, Mouldar JW, Pfenning NP, Sneath HA, Staley JT (1989) Bergey’s manual of systematic bacteriology, vol 4. Williams and Wilkins, Baltimore
Zin NM, Sarmin NIM, Ghadin N, Basri DF, Sidik NM, Hess WM, Strobel GA (2007) Bioactive endophytic streptomycetes from the Malay Peninsula. FEMS Microbiol Lett 274:83–88
Zinniel DK, Lambrecht P, Harris NB, Feng Z, Kuczmarski D, Higley P, Ishimaru CA, Arunakumari A, Barletta RG, Vidaver AK (2002) Isolation and characterization of endophytic colonizing bacteria from agronomic crops and prairie plants. Appl Environ Microbiol 68:2198–2208
Acknowledgments
We are grateful to the late Dr. K. Nakanishi of Mie University for his valuable suggestions. We are also grateful to Ms. T. Chikada (Life Science Research Center, Center for Molecular Biology and Genetics, Mie University) for sequencing the samples, and to Dr. H. Shimada (Mie Prefectural Research Institute) and the members of the Laboratory of Forest Pathology and Mycology, Mie University, for their field assistance. This research was partly supported by a Grant-in-Aid for Exploratory Research from JSPS to SI (22658048).
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Thongsandee, W., Matsuda, Y., Shimizu, M. et al. Isolation of endophytic streptomycetes from above- and belowground organs of Quercus serrata . J For Res 18, 179–189 (2013). https://doi.org/10.1007/s10310-012-0337-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10310-012-0337-2