Abstract
l-Arginine is an important amino acid with extensive application in the food and pharmaceutical industries. The efficiency of nitrogen uptake and assimilation by organisms is extremely important for l-arginine production. In this study, a strain engineering strategy focusing on upregulate intracellular nitrogen metabolism in Corynebacterium crenatum for l-arginine production was conducted. Firstly, the nitrogen metabolism global transcriptional regulator AmtR was deleted, which has demonstrated the beneficial effect on l-arginine production. Subsequently, this strain was engineered by overexpressing the ammonium transporter AmtB to increase the uptake of NH4+ and l-arginine production. To overcome the drawbacks of using a plasmid to express amtB, Ptac, a strong promoter with amtB gene fragment, was integrated into the amtR region on the chromosome in the Corynebacterium crenatum/ΔamtR. The final strain results in l-arginine production at a titer of 60.9 g/L, which was 35.14% higher than that produced by C. crenatum SYPA5-5.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Corynebacterium glutamicum is a gram-positive soil bacterium that was isolated in 1957 [1,2,3]. The complete genome sequence of the representative wild-type strain, C. glutamicum ATCC 13032, has been determined and analyzed, greatly aiding our understanding of the molecular biology and physiology of this organism [4]. This bacterium is frequently used in industry for the production of amino acids, such as l-glutamate, lysine, and l-arginine [5]. Corynebacterium crenatum, a subspecies of C. glutamicum, is an l-arginine overproducing strain that was isolated from soil. The mutant strain C. crenatum SYPA5-5, which could produce 32.5 g/L of l-arginine, was obtained by UV irradiation and treatment with nitrosoguanidine (NTG) in our previous study [6, 7]. In recent years, owing to the increased understanding of l-arginine biosynthesis and its metabolic regulation, many metabolic engineering strategies aiming to enhance l-arginine production have been carried out in C. glutamicum [8,9,10] and C. crenatum [11]. These strategies include removing regulatory repressors of l-arginine operon, optimizing NADPH level, disrupting L-glutamate exporter [8], strengthening the preexisting glucose transporter and developing new glucose uptake system, channeling excess carbon flux from glycolysis into tricarboxylic acid cycle [11].
Nitrogen is an essential component of most complex bacterial molecules, including proteins, nucleic acids and several cofactors [12]. To utilize different nitrogen sources and to survive nitrogen-limiting conditions, prokaryotes have developed several mechanisms to adapt their metabolism as the concentration of nitrogen changes [13,14,15]. A sufficient supply of nitrogen is essential for l-arginine synthesis, because there are four nitrogen atoms in the l-arginine molecule, corresponding to a nitrogen content of 32.1% [16].
Ammonium (NH4+) is the preferred nitrogen source for most bacterial, fungal and plant cells, although a high concentration of ammonium is toxic to cells. AmtB is a membrane-bound ammonium transporter protein that is present in most bacteria and is responsible for the uptake of NH4+/NH3 by the ammonium channel [17]. The PII signal transduction protein regulates the activities of many different proteins and sensors of cellular nitrogen status, including enzymes and transcription factors, by interacting with proteins [2, 18,19,20]. The amtB gene is always associated with the gene-encoding PII signal transduction protein GlnK in most bacteria and archaea, suggesting a functional relationship between the encoded proteins [21].
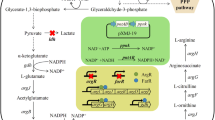
In C. glutamicum, the ATase/GlnK pathway is responsible for the regulation of ammonium assimilation and metabolism [17, 22], and enzymes that are important for these processes are primarily encoded in the operon amtB–glnK–glnD [23]. NH4+ can be assimilated via glutamate dehydrogenase (GDH) or the glutamine synthetase/glutamate synthase (GS/GOGAT) pathway, depending on the availability of ammonium in the medium. At higher NH4+ concentrations(> 5 mM), ammonium is assimilated by GDH; whereas at low concentrations (≤ 1 mM), it is assimilated by GS/GOGAT into l-glutamate [24]. Furthermore, in C. glutamicum, this operon is regulated by the global N-regulatory factor AmtR. The transcriptional regulator AmtR, a member of the TetR family, could regulate at least 35 genes, including those ammonium uptake (amtA and amtB), ammonium assimilation (glnA and gltBD), and urea metabolism (ureABCEFGD), as well as those encoding regulatory proteins (GlnK, GlnD), in response to changes in nitrogen levels [17]. Under conditions of intracellular ammonium starvation, when the ammonium concentration is below 5 mM, GlnK is adenylated by GlnD, and the resulting adenylated-GlnK (GlnK-Amp) subsequently interacts with AmtR, which causes the transcriptional upregulation of nitrogen-regulated genes, thereby activating the intracellular transport of ammonium [25]. Then, NH4+/NH3 can enter the cell through the AmtB ammonium transport channel. Conversely, under conditions of nitrogen surplus, de-adenylated GlnK interacts with the ammonium transporter AmtB, blocking the ammonium flux into the cell and therefore resulting in a GlnK-free state, allowing AmtR to be reactivated and bind to the promoter regions of nitrogen-controlled genes, repressing their transcription.
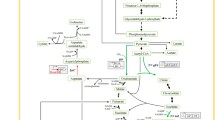
In our previous study on the metabolic engineering of l-arginine production, we focused on carbon flux optimization rather than ammonium uptake and assimilation to improve l-arginine production. Several studies have demonstrated that increasing the expression of genes related to nitrogen metabolism can improve the production of many valuable amino acids [26, 27]. For example, Kim et al. [27] enhanced glutamate production by overexpressing odhI in C. glutamicum. Similarly, Sindelar et al. [28] improved l-lysine production by overexpressing the amtA-ocd-soxA operon in C. glutamicum. Based on these studies, we asked whether an increase in the uptake and assimilation of NH4+ in C. crenatum can improve l-arginine production. Thus, the objective of this study was to increase l-arginine production via increasing NH4+ uptake and assimilation in C. crenatum (Fig. 1). We first observed that NH4+ had a significant effect on l-arginine production in C. crenatum. Furthermore, the effect of knocking out and overexpressing the nitrogen metabolic regulatory factor AmtR on l-arginine production and nitrogen metabolism-related enzyme activities was studied. Based on the qPCR results, we constructed a modified strain that overexpressed the channel protein AmtB, which was beneficial for the uptake of NH4+ and for l-arginine production. In addition, the amtB gene, under the control of a strong PtacM promoter, was integrated at the amtR locus in the Cc–ΔamtR mutant strain, and a subsequent 5-L fermentation assay showed that the l-arginine production was significantly enhanced by increased AmtB expression.
Methods
Strains and plasmids
The amtR (GenBank No: MK034164) and amtB (GenBank no: MK034163) genes, encoding a transcriptional regulator of nitrogen metabolism and an ammonium transporter protein, respectively, were identified in C. crenatum SYPA5-5 (Cc5-5) and were PCR-amplified. Cc5-5, the l-arginine producer obtained by multiple random mutagenesis, was also called as SYPA5-5 [6, 7], and has been deposited in the China General Microbiological Culture Collection Center (CGMCC) under collection number CGMCC no. 0890. E. coli BL21(DE3) was used as the host strain for gene cloning and expression. Cc5-5 was used as the parent strain for the generation of mutants. The shuttle vector pXMJ19 was used for gene expression in Cc5-5 [29]. The suicide vector pK18mobsacB, which harbors the sucrose screening marker sacB, was used for deleting, inserting, and replacing unmarked genes in C. glutamicum, as previously described. All of the strains and plasmids used in this study are listed in Table 1.
Growth medium and culture conditions
Cc5-5 and its recombinant derivatives were routinely cultivated at 30 °C in LBG medium (LB medium supplemented with 7 g/L glucose). For recombinant DNA expression, E. coli BL21(DE3) was cultivated at 37 °C in LB medium. Where appropriate, chloramphenicol was added into the medium (12.5 μg/mL). The modified minimal medium CGXII (containing glucose 50 g, FeSO4·7H2O·10 mg, MnSO4·H2O·10 mg, KH2PO4·1 g, ZnSO4·7H2O·1 mg, K2HPO4·1 g, CuSO4·0.2 mg, MgSO4·7H2O·0.25 g, NiCl2·6H2O·0.02 mg, biotin 0.2 mg, CaCl2·10 mg, 3-(N-morpholino) 42 g, protocatechuic 0.03 mg, and CaCO3 10 g) was used to culture C. crenatum strains for growth, enzyme activity and qPCR experiments. For the l-arginine shake flask fermentation, a single clone of the mutant was cultured on LBG plate for 24 h. Subsequently, a fresh bacterial colony was inoculated into 25 mL of seed medium in a standard 250-mL shake flask at 30 °C for 16 h. Next, 1.5 mL of the seed culture was transferred to 25 mL of the fermentation medium in a standard 250-mL shake flask and was cultured for 72 h at 30 °C with shaking at 200 r/min on a reciprocating shaker. The fermentation medium used for shake fermentation contained the following (per liter): 100 g glucose, 8 g yeast extract, 1.5 g·KH2PO4, 0.5 g·MgSO4·7H2O, 0.02 g FeSO4·7H2O, 0.02 g MnSO4, 20 g CaCO3, and 1–400 mM (NH4)2SO4 (optimized nitrogen concentration). To produce l-arginine via fermentation in 5-L bioreactors, the seed culture (150 mL) was transferred into 5-L stirred fermenters (BIOTECH-5BG, Baoxing Co., China) containing 2.5 L of fermentation medium. The seed medium contained the following (per liter): 50 g glucose, 20 g yeast extract, 20 g (NH4)2SO4, 1 g KH2PO4, and 0.5 g MgSO4·7H2O. The 5-L fermentation medium contained the following (per liter): 100 g glucose, 20 g yeast extract, 1.5 g KH2PO4, 0.5 g MgSO4·7H2O, 0.02 g FeSO4·7H2O, 0.02 g MnSO4, and 40 g (NH4)2SO4, (≈ 300 mM).
DNA manipulation and strain construction
To construct the recombinant strains, genomic DNA from strain Cc5-5 was isolated using a genomic DNA extraction kit (Sangon Biotech, Shanghai, China) and used for PCR amplification. The PCR products and vectors were obtained using a PCR product purification kit and a mini-plasmid isolation kit (Takara, Dalian, China), respectively. The primers used in this study are listed in Table 2. The suicide vector pK18mobsacB was used to introduce modifications into the chromosome of Cc5-5 [29, 30], as described previously.
To generate the amtR knockout strain, upstream and downstream DNA fragments (approximately 1500 bp) flanking amtR were amplified and fused by PCR. Subsequently, the fused fragments were inserted into the EcoRI/XbaI sites of the pK18mobsacB plasmid using T4 DNA ligase, generating the recombinant plasmid pK18–amtR. In addition, the amtR and amtB genes were inserted into the EcoRI/HindIII sites of the expression plasmid pXMJ19, generating the plasmids pXMJ19–amtR and pXMJ19–amtB. All recombinant plasmids were transformed in Cc5-5. The knockout strains obtained after two rounds of homologous recombination were confirmed by colony PCR.
The amtB gene under the control of the strong promoter PtacM was integrated into the Cc5-5 chromosome at the amtR locus. The upstream and downstream regions of amtR using the primers are listed Table 2, and the template was the recombinant plasmid pXMJ19–amtB. Subsequently, the upstream region and the downstream region of amtR were amplified, and the PtacM–amtB fragment was joined with the partial amtR fragment through overlapping PCR to obtain the target segment that generated the integrated gene construct PtacM–amtB. Subsequently, the PtacM–amtB fragment was inserted into the EcoRI/HindIII sites of the pK18mobsacB plasmid using T4 DNA ligase, generating the recombinant plasmid pK18–amtB2. Next, pK18–amtB2 was transferred into the Cc5-5 strain by electroporation. After double crossover recombination, the positive recombinant strain was detected by colony PCR to identify the Cc–amtB2 isolates.
Expression of the AmtR and AmtB proteins in C. crenatum
Recombinant E. coli BL21(DE3) cells were cultured in LB medium at 37 °C to an OD562 of 1.0 (approximately 3 h), after which the cells were induced with 1 mM IPTG and incubated for 12 h at 24 °C. The recombinant C. crenatum strain was inoculated into LBG medium and cultured at 30 °C for 5 h with shaking 180 r/min, after which the culture was induced with 1 mM IPTG, and then the culture was incubated for 12 h. The cells were harvested by centrifugation and then were resuspended in phosphate buffer (pH 7.2) and lysed on ice by sonication to obtain the crude enzyme fraction. The concentrations of the recombinant AmtR and AmtB proteins were determined by SDS–PAGE (12% acrylamide) [31], and the proteins were used for activity assays.
Enzyme activity assays
GDH activity was measured using a Glutamate Dehydrogenase Activity Colorimetric Assay Kit (Solarbio, China) following the manufacturer’s instructions. The Cc–amtR, Cc–ΔamtR, Cc–amtB1 and Cc–amtB2 strains were harvested by centrifugation at 8000 r/min for 10 min after 48 h of cultivation in CGXII medium. One unit of GDH activity was defined as the amount of enzyme that generated one micromole of NADH per minute under the assay conditions. The GS activity was measured using a Glutamine Synthetase Assay Kit (Solarbio, China) following the manufacturer’s directions. One unit of the GS activity was defined as the amount of enzyme that resulted in a change in the absorbance of 0.01 (at 540 nm) per minute under in a 1-mL reaction. The GOGAT activity was measured as described by Tesch et al. [32].
Determination of ammonium concentrations by ion chromatography
A Thermo Scientific ICS-5000+ ion chromatograph(Thermo Scientific,USA) was used to detect the concentration of extracellular NH4+ at 0, 12, 24, 36, 48, 60 and 72 h in the fermentation broth of the Cc–amtB1, Cc–amtB2 and Cc5-5 cultures. Ion chromatography including an IonPac CG12A guard column (4 × 50 mm i.d.) and an IonPac CS12A separation column (4 × 250 mm i.d.). The eluent was obtained using a CH3SO3H eluent generator, and suppression was achieved using a CERS500 4-mm self-regenerating suppressor. The column temperature was set at 30 °C, the eluent flow rate was set at 1.00 mL/min, and the injection volume was 25 µL. To prepare the samples, 50 mL of broth was centrifuged at 1000 r/min for 10 min to remove CaCO3, after which the supernatant was centrifuged at 8000 r/min for 10 min. Next, a high-pressure homogenizer was used to lyse the cells, and cell debris was subsequently removed by centrifugation at 10,000 r/min for 10 min. The supernatants were filtered twice through 0.22-μm filters, and the organic substances in the sample were removed using a Thermo OnGuard II RP column. The instrument control and data collection were performed by a Dionex Chromeleon 6.5 [33].
Real-time PCR
For RNA analysis, 500-μL samples of strains cultured to the exponential phase were collected during batch fermentation in shake flasks for mRNA isolation. RNA was extracted from the collected cells using an RNAiso Plus reagent (Takara, Dalian, China) following the manufacturer’s instructions. cDNA was synthesized using a PrimeScript RT reagent kit (Takara, Dalian, China). Cc5-5 16S rRNA was used as an internal standard [30, 34]. The Ct values of the 16S rDNA gene and those of the amtB, glnK, and glnD genes were obtained by RT-qPCR using a Bio-Rad CFX96 Touch Real-Time PCR Detection System (Bio-Rad Hercules, CA) with SYBR Premix Ex Taq™ II (Takara, Dalian, China). The primer sequences used for RT-qPCR and RT-PCR for assess the integration of PtacM are shown in Table S1. Each assay was replicated three times.
Analytical procedures
Cell growth was monitored by measuring the OD562 (1 OD562 = 0.375 g/L Dry cell weight, DCW) using a spectrophotometer (UNICOTM-UV2000, Shanghai, China) after dissolving CaCO3 with 0.125 M HCl. To quantify substrate consumption and production, 1.5-mL samples of the fermentation culture were harvested and centrifuged. The supernatant was analyzed for glucose using an SBA-40C biosensor analyser (developed by the Biology Institute of the Shangdong Academy of Sciences). The sample supernatants were processed using 0.22-μm filters and centrifuged (10,000 r/min for 10 min at 4 °C). l-Arginine concentrations were determined by HPLC (high-pressure liquid chromatography) on an Agilent 1100 LC system (Agilent Technologies, Waldbronn, Germany) following the procedure described by Xu [6]. All assays were performed in triplicate.
Results
High nitrogen concentrations are essential for l-arginine production
Most microorganisms use ammonium (NH4+) as a preferred nitrogen source, and the nitrogen content in the l-arginine molecule is as high as 32.15%. There are four processes to assimilate the NH4+ in l-arginine biosynthesis (Fig. 1). Therefore, detection of the effect of different concentrations of NH4+ on l-arginine production by Cc5-5 during shake flask fermentation. When 1, 10, 100, 200, 300 and 400 mM (NH4)2SO4 were added into the fermentation medium with an initial glucose concentration of 100 g/L, the cell growth and glucose consumption rate increased as the concentration of NH4+ increased. As shown in Table 3, when an NH4+ concentration of 300 mM was used, the increase in l-arginine production and glucose consumption was the greatest compared with the production in other samples in the 250-mL shake flask cultivation assay. After 72 h of cultivation, the strain cultured with 300 mM NH4+ exhibited an l-arginine production of 31.8 ± 1.8 g/L with a productivity of 0.44 ± 0.05 g/(L·h). For an NH4+ concentration of 400 mM, the observed l-arginine production was 28.2 ± 1.3 g/L, and while the cell growth and glucose consumption rate decreased, the specific l-arginine production increased. As expected, this result indicated that a rich supply of NH4+ is essential for high-efficiency l-arginine biosynthesis. In addition, in fermentation medium, lower amounts of added NH4+ hardly affected strain growth, but excessive NH4+ was unfavourable for strain growth.
Effect of AmtR deletion or overexpression on l-arginine production
AmtR is a global transcriptional regulator of nitrogen metabolism in C. glutamicum that regulates the transcription of at least 35 genes. When nitrogen sources are sufficient, the expression of most of these genes is downregulated [35]. To investigate the effect of AmtR on l-arginine production in C. crenatum SYPA5-5 (Cc5-5), the AmtR deletion and overexpression AmtR mutant strains Cc–ΔamtR and Cc–amtR were constructed, as shown in Table 1; the AmtR deletion mutant strain is verified in Fig. S1. The growth of the constructed strains was normal in LBG medium, similar to the Cc5-5 strain (data not shown). Next, the Cc–ΔamtR, Cc–amtR and Cc5-5 strains were cultivated in the 250-mL shake flask to investigate the effect of AmtR deletion or overexpression on l-arginine production. As shown in Fig. S2, almost no l-arginine was produced during the first 10 h; while after 72 h of cultivation, the production of l-arginine by the Cc–amtR strain (12 ± 1.2 g/L) was only half that observed for the Cc5-5 strain, and the Cc–ΔamtR strain exhibited a 16.67% increase in l-arginine production (30 ± 1.5 g/L) compared to that of the Cc5-5 strain. These results indicated that the deletion of the nitrogen transcriptional regulator AmtR is beneficial to l-arginine production.
To further investigate the function of AmtR in Cc5-5, growth e-related enzymes (GSxperiments and activity assays for nitrogen metabolism, GDH, and GOGAT) regulated by AmtR were carried out in modified CGXII minimal medium containing (NH4)2SO4 at concentrations of 1, 10, and 100 mM. As shown in Fig. 2a, in the presence of different concentrations of (NH4)2SO4, the growth of all three strains increased steadily in medium containing 1 and 10 mM. However, after 36 h of cultivation, cultures containing 1 mM (NH4)2SO4 grew significantly slower than those containing 10 mM (NH4)2SO4. Interestingly, in cultures containing 100 mM (NH4)2SO4, strains grew slowly over the first 24 h; after 24 h, the growth of strains increased steadily. This result suggested that a sufficient concentration of a nitrogen source is essential for the growth of Cc5-5 strain, but excessive NH4+ is detrimental to the growth of the Cc5-5 strain in CGXII medium.
Growth curves for the Cc–ΔamtR, Cc–amtR and Cc5-5 strains (a) and the Cc–amtB1 and Cc–amtB2 strains (b). Strains were cultivated at 30 °C and 200 r/min for 72 h in 250-mL flasks in CGXII minimal medium [supplemented with 1, 10, and 100 mM (NH4)2SO4]. The data represent the mean values and standard deviations obtained from at least three independent measurements
Glutamate and glutamine are the two major donors of NH4+ during the biosynthesis of l-arginine (Fig. 1), and NH4+ assimilation proceeds via glutamate dehydrogenase (GDH) or glutamine synthetase/glutamate synthase (GS/GOGAT), which are regulated by AmtR. The assimilation of NH4+ was observed by comparing the activity of these enzymes in the above three strains. In Fig. 3b, c, when the concentration of (NH4)2SO4 was 1 mM, the GOGAT and GS activities in Cc–ΔamtR strain were two–threefold higher than those observed for the Cc5-5 strain. At 100 mM (NH4)2SO4, the activity of GDH was the highest, 3.5 times higher than that observed for the Cc5-5 strain (Fig. 3a). However, the overexpression of AmtR caused a sharp decrease in the activities of the three assayed enzymes compared with those observed in the Cc5-5 strain. These results show that AmtR plays an important role in the production of l-arginine and in nitrogen metabolism-related enzyme activity in Cc5-5.
Measurements of glutamate dehydrogenase (GDH) (a), glutamate synthase (GOGAT) (b), and glutamine synthetase (GS) (c) for the Cc–ΔamtR, Cc–amtR, Cc–amtB1, Cc–amtB2 and Cc5-5 strains. Strains were cultivated at 30 °C, 200 r/min for 72 h in 250-mL flasks in CGXII minimal medium [supplemented with 1, 10, 100 mM (NH4)2SO4], LBG medium and fermentation medium. The data represent the mean values and standard deviations obtained from at least three independent measurements
Comparative analysis of the transcriptional levels of AmtR-related genes
Deletion of the transcriptional regulator AmtR can increase the activity of NH4+ assimilation-related enzymes and promote l-arginine synthesis when the nitrogen source is sufficient. To investigate the mechanism by which the deletion of the gene-encoding AmtR increased l-arginine production, the transcriptional profiles of genes related to the uptake and assimilation of NH4+ were measured in the Cc–ΔamtR, Cc–amtR and Cc5-5 strains at different concentrations of NH4+. First, the strains were grown in modified CGXII minimal medium containing 1 or 100 mM (NH4)2SO4 as the sole nitrogen source, with cells harvested at an OD562 of between 4 and 6. To accurately evaluate the RT-qPCR results, a series of oligonucleotide primers were tested to determine the PCR efficiency (Table S1), and RiboGreen was used to accurately determine the total amount of RNA used in the real-time PCR experiments. This method allowed for the use of the 16S rRNA gene to standardize the results obtained without the need for additional control genes. As shown in Fig. 4, in cultures containing 100 mM (NH4)2SO4, a comparison of the transcriptional response of the Cc–ΔamtR and Cc5-5 strains revealed that the amtB gene were increased 31.2-fold, while those of the glnK and glnD genes were increased two-fold, respectively. In addition, nitrogen assimilation systems were investigated by measuring the transcriptional level of the genes encoding the nitrogen metabolism-related enzyme GS, GDH, and GOGAT (glnA, gdh, and gltBD). The transcription of the gdh gene in Cc–ΔamtR strain was observed to be increased 20.5-fold compared to that of the Cc5-5 strain; the transcription of the other genes was not significantly changed. In contrast, comparing the Cc–amtR and Cc5-5 strains in the presence of 100 mM (NH4)2SO4, the expression of the amtB gene in the Cc–amtR strain was downregulated 64.2-fold, and that of the glnK and glnD genes was downregulated four-fold, respectively, while that of the gdh gene was downregulated 30.3-fold. In cultures containing 1 mM (NH4)2SO4, the trend was consistent with an insufficient nitrogen supply, but not notably so. The qPCR results indicated that AmtR gives an obvious influence on the ammonium uptake and assimilation.
The transcriptional regulation of AmtR for the amtB–glnK–glnD operon and the genes encoding l-arginine synthesis-related enzymes (GS, GDH, and GOGAT) in different strains. Relative expression levels of amtB, glnK, glnD, glnA, gdh, and gltBD at 1 and 100 mM (NH4)2SO4 concentrations were calculated based on the quantified real-time qPCR results. The error bars denote the standard deviations, and n = 3
Effect of AmtB overexpression on ammonium uptake and l-arginine production
AmtB is an ammonium channel protein involved in ammonium transport, and the amtB gene sequence was compared and analyzed in BLAST searches. The results showed that the Cc5-5 amtB gene sequence was identical to that of C. glutamicum SCgG2 (GenBank: CP004048.1), with the alignment results for C. glutamicum ATCC 13032 showing that the alignment identity of amtB was 81%; while the amino acid sequence alignment showed that the AmtB sequence was 89.95% identical in the two strains. These results showed little difference between the C. glutamicum and Cc5-5 amtB genes. Based on the comparison results and qPCR (Fig. 4), we speculated that the overexpression AmtB would have a great influence on nitrogen assimilation in Cc5-5 strain. First, AmtB was overexpressed in the Cc–ΔamtR strain, resulting in the Cc–amtB1 strain. Next, a strong promoter, PtacM [36], was integrated with the amtB gene at the amtR locus to increase AmtB expression in the mutant strain Cc–ΔamtR. As shown in Fig. S3, compared to that in the Cc5-5 strain, the transcriptional level of the amtB gene in strain Cc–amtB2 increased by 38.3-fold and increased sevenfold compared to that in the Cc–ΔamtR strain. The results suggested that the recombinant strain was successfully constructed and that the expression of the amtB gene was increased by the insertion of the stronger PtacM promoter.
The l-arginine production of the Cc–amtB1, Cc–amtB2 and Cc5-5 strains was compared under shake flask fermentation conditions. As shown in Fig. S2, during the first 48 h, the production of l-arginine was almost the same, but as the fermentation progressed, the Cc–amtB1 and Cc–amtB2 strains exhibited greater l-arginine production than the Cc5-5 strain. After cultivation for 72 h, the Cc–amtB1 strain showed a 23.8% increase in l-arginine production (32.8 ± 1.1 g/L) compared to that of the Cc5-5 strain. The Cc–amtB2 strain produced approximately 36.5 ± 1.2 g/L l-arginine, which was 12.4% more than was produced by the Cc–amtB1 strain. The results suggested that modulation of AmtB overexpression resulted in an improvement in l-arginine production under similar fermentation conditions.
By inserting the PtacM promoter and the amtB gene segment into the amtR chromosomal locus, we overcame deficiencies observed in the plasmid-based AmtB overexpression systems that hampered the generation of derivative strains. This study highlighted the importance of AmtB overexpression in the development of l-arginine-producing strains. Furthermore, in this study, as shown in Fig. 5, the extracellular NH4+ levels in the Cc–amtB1 and Cc–amtB2 strains were notably more quickly reduced than that was observed in the Cc–ΔamtR strain, especially, after 36 h of fermentation. Furthermore, the NH4+ levels were slightly lower in the Cc–amtB2 strain than in the Cc–amtB1 strain. The result indicated that enhancing the ammonium transporter expression can improve the efficiency of NH4+ utilization. Moreover, the effect was stronger with the PtacM promoter integrated into the chromosome than with AmtB overexpression in the Cc–ΔamtR strain.
Comparison of residual NH4+ concentrations in the Cc–amtB1, Cc–amtB2 and Cc5-5 strains under shake flask fermentation. Strains were cultivated at 30 °C and 200 r/min for 72 h in 250-mL flasks in fermentation medium. The data represent the mean values and standard deviations obtained from at least three independent measurements
Further investigations of growth and nitrogen metabolism-related enzyme activity (GS, GDH, and GOGAT) were carried out in the Cc–amtB1, Cc–amtB2 and Cc–ΔamtR strains, which were cultured in different NH4+ concentrations in CGXII minimal medium. The growth rate was almost the same in the presence of 1 and 10 mM NH4+ (Fig. 2b), although the final OD562 values were found to depend directly on the amount of NH4+ added. At 100 mM NH4+, the Cc–amtB2 strain maintained a certain growth advantage over the Cc–ΔamtR strain throughout the growth process. As described above, in the growth experiment, it can be seen that the enhanced expression of the ammonium transporter improved NH4+ consumption. After ammonium is transported into the cells, assimilation proceeds via GDH and GS/GOGAT. As shown in Fig. 3a, as the NH4+ concentration increased, the GDH activity gradually improved. When NH4+ was present at 100 mM, the GDH enzyme activity of the Cc–amtB2 strain was increased 5.2-fold compared to control group, and 1.2-fold compared to the Cc–amtB1 strain. The GS/GOGAT activity was slightly higher at 1 mM NH4+. Furthermore, the three enzyme activities were higher in the fermentation medium than in the CGXII minimal medium, especially for the Cc–amtB2 strain, which had the highest enzyme activity and explained the increase in l-arginine production.
Fed-batch fermentation of Cc5-5 and Cc–amtB2 strains
To maintain the NH4+ concentration at a relatively steady level and relieve the inhibition of growth by a high concentration of nitrogen, a strategy was investigated by continuously adding 25% NH3·H2O in 5-L fed-batch fermentation. Fed-batch cultures were performed to examine the performance of the mutant strains for l-arginine production. Time profiles of l-arginine fed-batch cultures of the Cc5-5 and Cc–amtB2 strains are shown in Fig. 6a, b, respectively. The cell growth of the two strains was similar throughout the whole fermentation period, indicating that the knockout of AmtR and the insertion of the PtacM with the amtB gene segment into the amtR chromosomal locus had no effect on Cc5-5 growth. The Cc–amtB2 strain was also observed to consume glucose more quickly before the first feeding (36 h after incubation) than the Cc5-5 strain (42 h after incubation). At the end of the fed-batch fermentations, the Cc–amtB2 strain produced 60.9 ± 1.31 g/L of l-arginine with a yield of 0.36 ± 0.018 g/g glucose and a productivity of 0.634 g/L/h. In comparison, the fed-batch fermentation of the Cc5-5 strain resulted in 39.5 ± 0.57 g/L of l-arginine with a yield of 0.25 ± 0.02 g/g glucose and a productivity of 0.411 g/(L·h), while the l-arginine production from glucose by the Cc–amtB2 strain was approximately 30.56% higher than that by the Cc5-5 strain in fed-batch fermentation. Amino acids and organic acids analysis of the fermentation broth showed the concentration of lactate, lysine, isoleucine compared with the Cc5-5 strain was decreased slightly and proline was undetectable; specific data are shown in Table S2. This result indicated that the l-arginine yield can be markedly increased by knocking out the nitrogen regulator AmtR and that expressing the ammonium transporter AmtB by integrating a strong promoter on the chromosome in the Cc–ΔamtR strain is a useful strategy to increase the l-arginine yield.
Time course of l-arginine fed-batch fermentations of the Cc5-5 (a) and Cc–amtB2 (b) strains in 5-L fermenters. The filled triangle represents glucose, the filled square represents l-arginine, and the filled circle represents dry cell weight (DCW). The total glucose concentrations in the fed-batch fermentations of the Cc5-5 and Cc–amtB2 strains were 160 and 175 g/L, respectively. The data represent the mean values and standard deviations obtained from three independent cultures
Discussion
With the increased research on nitrogen metabolism in C. glutamicum, it is clear that C. glutamicum has a specific strategy to regulate this process. The most crucial step to increase nitrogen metabolism in this bacterium involved the discovery of the global repressor AmtR, which blocks the transcription of many genes in C. glutamicum during cultivation in medium with sufficient nitrogen [35]. In this study, we explored the effect of the deletion of AmtR and the overexpression of the ammonium transporter AmtB on nitrogen metabolism in Cc5-5, especially with respect to l-arginine production.
To study the function of AmtR in Cc5-5 strain, we constructed amtR gene deletion and overexpression strains. Growth assays revealed that the growth of the Cc–ΔamtR and Cc–amtR strains was slightly slower than that of the Cc5-5 strain before 36 h but resulted in the same biomass in the stationary phase. However, the growth of the strain in the presence of low nitrogen source concentrations (1 and 10 mM (NH4)2SO4) in the early stages of the exponential period was significantly faster than in the presence of a high nitrogen concentration [100 mM (NH4)2SO4]. Obviously, Cc5-5 can avoid the effects of ammonium limitation on the growth rate using intracellular stored nitrogen sources such as glutamate and glutamine [37].
l-Arginine is a basic amino acid, and nitrogen is required for l-arginine biosynthesis [16]. By comparing the enzyme activities of the strains Cc–ΔamtR, Cc–amtR and Cc5-5 at three NH4+ concentrations, we observed that the GDH activity at a high nitrogen source concentration was significantly higher than that at 1 mM (NH4)2SO4, and that GOGAT had almost no activity in the presence of 100 mM (NH4)2SO4. Thus, the high expression of GDH could improve the accumulation of glutamate [38]. Furthermore, glutamate provides nitrogen for the synthesis of the precursor l-arginine [39]. We suspected that this is the reason for the more favorable l-arginine production when the nitrogen source is sufficient. Beckers and Schulz observed that GDH is preferentially used in ammonium-rich medium [40, 41]; while the transcription of gltBD, coding for GOGAT, is completely repressed in this situation, and the transcription of glnA, coding for GS, is maintained at a basal level to provide l-glutamine for growth [42]. When bacterial cells under the limitation of nitrogen, the ammonium assimilation via GDH is not sufficient due to the low affinity of the enzyme. In this situation, the GS/GOGAT pathway is preferentially used for ammonium assimilation [43]. Thus, the AmtR deletion strain exhibited increased ammonium assimilation. In the shake flask fermentation experiment, the final l-arginine production of the Cc–ΔamtR strain was increased by 16.67% compared to that of the Cc5-5 strain.
In a previous study, AmtR repressed the transcription of many genes, including those encoding transporters and genes for ammonium assimilation (amtB, glnK, and glnD) when bacterial cells were grown in nitrogen-rich standard minimal medium. To fully evaluate the performance of the engineered strains Cc–ΔamtR and Cc–amtR strains, the effect of AmtR on the expression of the amtB–glnK–glnD operon and the ammonium assimilation system genes glnA, gltBD, gdh was verified by RT-qPCR. The transcriptional level of the gdh gene increased 20.5-fold in the Cc–ΔamtR strain when cells were grown with 100 mM (NH4)2SO4, a result consistent with the observed enzyme activity. However, it was determined that the overexpression of AmtR had an obvious effect on the ammonium transporter AmtB, resulting in a 64.2-fold downregulation of its expression.
From these results, we concluded that the knockout of the amtR gene could relieve the transcriptional repression of extracellular nitrogen metabolism protein-encoding genes. Furthermore, the overexpression of the ammonium transporter protein AmtB promoted the uptake of NH4+, resulting in a significant increase in l-arginine production. This result indicated that the overexpression of the ammonium transporter AmtB improved the efficiency of nitrogen source utilization. We can also infer from the cell growth results that the strain grows rapidly from 36 to 48 h and that more nitrogen is needed during exponential growth. Considering the instability of the free plasmid, the promoter PtacM was integrated into the amtB gene in the Cc–ΔamtR strain, enhancing the expression of AmtB. Finally, the Cc–amtB2 and Cc5-5 strains were investigated in a fed-batch process. Fed-batch fermentation of the Cc–amtB2 strain resulted in the production of 60.9 ± 1.31 g/L of l-arginine with a yield of 0.36 ± 0.018 g/g glucose and a productivity of 0.634 g/(L h). The fermentation results demonstrated that the high NH4+ concentration of 400 mM is essential for high-efficiency l-arginine production. By deleting the AmtR regulator, which represses the ammonium assimilation corresponding genes under the high NH4+ Cc5-5, the transcriptional level of ammonium transporter AmtB was significant increased. Extracellular NH4+ concentration analysis indicated that the NH4+ consumed more in the engineered strains.
In this study, we have created an environment rich in supply of nitrogen for the l-arginine production, improved the efficiency of nitrogen uptake and assimilation in Cc5-5. Along with the advances made via the continued study of nitrogen metabolism complexities, further enhancement of l-arginine production would be achieved by fine-tuning the adjustment of the expression of ammonium absorption and assimilation genes and through the rebalance of carbon and nitrogen fluxes.
References
Udaka S (1960) Screening method for microorganisms accumulating metabolites and its use in the isolation of Micrococcus glutamicus. J Bacteriol 79:754
Burillo S, Luque I, Fuentes I, Contreras A (2004) Interactions between the nitrogen signal transduction protein PII and N-acetyl glutamate kinase in organisms that perform oxygenic photosynthesis. J Bacteriol 186:3346–3354
Shukuo K, Shigezo U, Masakazu S (2004) Studies on the amino acid fermentation. Part 1. Production of l-glutamic acid by various microorganisms. J Gen Appl Microbiol 50:331–343
Ikeda M, Nakagawa S (2003) The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Appl Microbiol Biotechnol 62:99–109
Jorge JM, Nguyen AQ, Pã©Rez-Garcã AF, Kind S, Wendisch VF (2017) Improved fermentative production of gamma-aminobutyric acid via the putrescine route: systems metabolic engineering for production from glucose, amino sugars, and xylose. Biotechnol Bioeng 114:862–873
Xu M, Rao Z, Yang J, Xia H, Dou W, Jin J, Xu Z (2012) Heterologous and homologous expression of the arginine biosynthetic argC ~ H cluster from Corynebacterium crenatum for improvement of l-arginine production. J Ind Microbiol Biotechnol 39:495–502. https://doi.org/10.1007/s10295-011-1042-4
Xu M, Rao Z, Dou W, Yang J, Jin J, Xu Z (2012) Site-directed mutagenesis and feedback-resistant N-acetyl-l-glutamate kinase (NAGK) increase Corynebacterium crenatum l-arginine production. Amino Acids 43:255–266. https://doi.org/10.1007/s00726-011-1069-x
Park SH, Kim HU, Kim TY, Park JS, Kim S-S, Lee SY (2014) Metabolic engineering of Corynebacterium glutamicum for l-arginine production. Nat Commun 5:4618. https://doi.org/10.1038/ncomms5618
Man Z, Rao Z, Xu M, Jing G, Yang T, Xian Z, Xu Z (2016) Improvement of the intracellular environment for enhancing l-arginine production of Corynebacterium glutamicum by inactivation of H2O2 -forming flavin reductases and optimization of ATP supply. Metab Eng 38:310–321
Shin JH, Sang YL (2014) Metabolic engineering of microorganisms for the production of l-arginine and its derivatives. Microb Cell Fact 13:166
Man Z, Xu M, Rao Z, Guo J, Yang T, Zhang X, Xu Z (2016) Systems pathway engineering of Corynebacterium crenatum for improved l-arginine production. Sci Rep 6:28629. https://doi.org/10.1038/srep28629
Chen QH, Han YL, Yao M, Yan YL, Ping SZ, Wei L (2013) Research progress on structure and evolution of biological nitrogen-fixation gene cluster. J Agric Sci Technol 15:129–138
Merrick MJ, Edwards RA (1995) Nitrogen control in bacteria. Microbiol Rev 59:604–622
Hodgson DA (2000) Primary metabolism and its control in streptomycetes: a most unusual group of bacteria. Adv Microb Physiol 42:47–238
Martín JF (2004) Phosphate control of the biosynthesis of antibiotics and other secondary metabolites is mediated by the PhoR–PhoP system: an unfinished story. J Bacteriol 186:5197–5201
Guo J, Man Z, Rao Z, Xu M, Yang T, Zhang X, Xu Z (2017) Improvement of the ammonia assimilation for enhancing l-arginine production of Corynebacterium crenatum. J Ind Microbiol Biotechnol 44:443
Jakoby M, Krämer R, Burkovski A (1999) Nitrogen regulation in Corynebacterium glutamicum: isolation of genes involved and biochemical characterization of corresponding proteins. FEMS Microbiol Lett 173:303–310
Ninfa AJ, Jiang P (2005) PII signal transduction proteins: sensors of alpha-ketoglutarate that regulate nitrogen metabolism. Curr Opin Microbiol 8:168–173. https://doi.org/10.1016/j.mib.2005.02.011
Arcondeguy T, Jack R, Merrick M (2001) P(II) signal transduction proteins, pivotal players in microbial nitrogen control. Microbiol Mol Biol Rev 65:80–105. https://doi.org/10.1128/mmbr.65.1.80-105.2001
Benelli EM, Buck M, de Souza EM, Yates MG, Pedrosa FO (2001) Uridylylation of the PII protein from Herbaspirillum seropedicae. Can J Microbiol 47:309–314
Thomas G, Mullins J, Merrick M (2000) Membrane topology of the Mep/Amt family of ammonium transport proteins. Mol Microbiol 37:331–344
Buchinger S, Strösser J, Rehm N, Hänßler E, Hans S, Bathe B, Schomburg D, Krämer R, Burkovski A (2009) A combination of metabolome and transcriptome analyses reveals new targets of the Corynebacterium glutamicum nitrogen regulator AmtR. J Biotechnol 140:68–74
Beckers G, Strosser J, Hildebrandt U, Kalinowski J, Farwick M, Kramer R, Burkovski A (2005) Regulation of AmtR-controlled gene expression in Corynebacterium glutamicum: mechanism and characterization of the AmtR regulon. Mol Microbiol 58:580–595. https://doi.org/10.1111/j.1365-2958.2005.04855.x
Endrulat T, Saurer M, Buchmann N, Brunner I (2006) Application of global analysis techniques to Corynebacterium glutamicum: new insights into nitrogen regulation. J Biotechnol 126:101–110
Meier-Wagner J, Nolden L, Jakoby M, Siewe R, Kramer R, Burkovski A (2001) Multiplicity of ammonium uptake systems in Corynebacterium glutamicum: role of Amt and AmtB. Microbiology 147:135–143. https://doi.org/10.1099/00221287-147-1-135
Rehm N, Burkovski A (2011) Engineering of nitrogen metabolism and its regulation in Corynebacterium glutamicum: influence on amino acid pools and production. Appl Microbiol Biotechnol 89:239–248
Kim J, Fukuda H, Hirasawa T, Nagahisa K, Nagai K, Wachi M, Shimizu H (2010) Requirement of de novo synthesis of the OdhI protein in penicillin-induced glutamate production by Corynebacterium glutamicum. Appl Microbiol Biotechnol 86:911–920
Sindelar G, Wendisch VF (2007) Improving lysine production by Corynebacterium glutamicum through DNA microarray-based identification of novel target genes. Appl Microbiol Biotechnol 76:677–689
Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A (1994) Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69
Shu Q, Xu M, Li J, Yang T, Zhang X, Xu Z, Rao Z (2018) Improved l-ornithine production in Corynebacterium crenatum by introducing an artificial linear transacetylation pathway. J Ind Microbiol Biotechnol 45:393–404. https://doi.org/10.1007/s10295-018-2037-1
Schägger H (2006) Tricine–SDS-PAGE. Nat Protoc 1:16. https://doi.org/10.1038/nprot.2006.4
Aa TMDG, Sahm H (1999) In vivo fluxes in the ammonium-assimilatory pathways in Corynebacterium glutaunicum studied by N-15 nuclear magnetic resonance. Appl Env Microbiol 65:1099
Zhu Y, Guo Y, Ye M, James FS (2005) Separation and simultaneous determination of four artificial sweeteners in food and beverages by ion chromatography. J Chromatogr A 1085:143–146
Xu Meijuan, Rao Zhiming, Dou Wenfang, Xu Z (2013) The role of ARGR repressor regulation on l-arginine production in Corynebacterium crenatum. Appl Biochem Biotechnol 170:587–597
Jakoby M, KraMer R, Burkovski A (2010) Nitrogen regulation in Corynebacterium glutamicum: isolation of genes involved and biochemical characterization of corresponding proteins. FEMS Microbiol Lett 173:303–310
Rytter JV, Helmark S, Chen J, Lezyk MJ, Solem C, Jensen PR (2014) Synthetic promoter libraries for Corynebacterium glutamicum. Appl Microbiol Biotechnol 98:2617–2623
Soupene E, He L, Yan D, Kustu S (1998) Ammonia acquisition in enteric bacteria: physiological role of the ammonium/methylammonium transport B (AmtB) protein. Proc Natl Acad Sci USA 95:7030–7034
Huang B, Qin P, Xu Z, Zhu R, Meng Y (2011) Effects of CaCl 2 on viscosity of culture broth, and on activities of enzymes around the 2-oxoglutarate branch, in Bacillus subtilis CGMCC 2108 producing poly-(γ-glutamic acid). Bioresour Technol 102:3595–3598
Takehara M, Hibino A, Saimura M, Hirohara H (2010) High-yield production of short chain length poly(ε-l-lysine) consisting of 5–20 residues by Streptomyces aureofaciens, and its antimicrobial activity. Biotechnol Lett 32:1299–1303
Schulz AA, Collett HJ, Reid SJ (2001) Nitrogen and carbon regulation of glutamine synthetase and glutamate synthase in Corynebacterium glutamicum ATCC 13032. FEMS Microbiol Lett 205:361–367
Beckers G, Nolden L, Burkovski A (2001) Glutamate synthase of Corynebacterium glutamicum is not essential for glutamate synthesis and is regulated by the nitrogen status. Microbiology 147:2961–2970
Nolden L, Ngouoto-Nkili CE, Bendt AK, Krämer R, Burkovski A (2001) Sensing nitrogen limitation in Corynebacterium glutamicum: the role of glnK and glnD. Mol Microbiol 42:1281–1295
Silberbach M, Hüser A, Kalinowski J, Pühler A, Walter B, Krämer R, Burkovski A (2005) DNA microarray analysis of the nitrogen starvation response of Corynebacterium glutamicum. J Biotechnol 119:357–367
Acknowledgements
This work was funded by the National Natural Science Foundation of China (31770058), Natural Science Foundation of Jiangsu Province (BK20181205), the National Key Research and Development Program of China (2018YFA090039), the Research Project of the Chinese Ministry of Education (113033A), the Fundamental Research Funds for the Central Universities (JUSRP51708A), the national first-class discipline program of Light Industry Technology and Engineering (LITE2018-06) and the 111 Project (111-2-06).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xu, M., Li, J., Shu, Q. et al. Enhancement of l-arginine production by increasing ammonium uptake in an AmtR-deficient Corynebacterium crenatum mutant. J Ind Microbiol Biotechnol 46, 1155–1166 (2019). https://doi.org/10.1007/s10295-019-02204-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-019-02204-3