Abstract
Control of biopharmaceutical processes is critical to achieve consistent product quality. The most challenging unit operation to control is cell growth in bioreactors due to the exquisitely sensitive and complex nature of the cells that are converting raw materials into new cells and products. Current monitoring capabilities are increasing, however, the main challenge is now becoming the ability to use the data generated in an effective manner. There are a number of contributors to this challenge including integration of different monitoring systems as well as the functionality to perform data analytics in real-time to generate process knowledge and understanding. In addition, there is a lack of ability to easily generate strategies and close the loop to feedback into the process for advanced process control (APC). The current research aims to demonstrate the use of advanced monitoring tools along with data analytics to generate process understanding in an Escherichia coli fermentation process. NIR spectroscopy was used to measure glucose and critical amino acids in real-time to help in determining the root cause of failures associated with different lots of yeast extract. First, scale-down of the process was required to execute a simple design of experiment, followed by scale-up to build NIR models as well as soft sensors for advanced process control. In addition, the research demonstrates the potential for a novel platform technology that enables manufacturers to consistently achieve “goldenbatch” performance through monitoring, integration, data analytics, understanding, strategy design and control (MIDUS control). MIDUS control was employed to increase batch-to-batch consistency in final product titers, decrease the coefficient of variability from 8.49 to 1.16%, predict possible exhaust filter failures and close the loop to prevent their occurrence and avoid lost batches.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The main goal of any industry is to manufacture a product within prescribed quality specifications. The ease with which this objective is met is directly related to the complexity of the product in conjunction with the ability to adequately control the way in which it is manufactured. Biopharmaceutical production, unlike traditional medicinal products manufactured using consistent chemical and physical techniques, involves biological processes with nonlinear dynamics, inherent batch variability and high sensitivity to minute changes in environmental parameters [22]. In addition, raw materials that can be extremely complex are often variable in composition, which can have an unpredictable and substantial impact on cellular metabolism [17]. Cellular growth and product formation in a bioreactor is recognized as the most complex and significant unit operation in manufacturing a biopharmaceutical and governs the success of the overall process. However, there are still a variety of bioreactor operations that depend on off-line sampling for in-process control. In fact, very few sophisticated analytical measurements are performed in situ and only a handful of critical parameters such as pH, dissolved oxygen (DO) and temperature are monitored in real-time [6]. The clear need to increase process understanding and control led the Food and Drug Administration (FDA) to institute a quality initiative in 2002 that has become known as quality by design (QbD). Soon after, the FDA realized that the advanced control of critical process parameters (CPPs) required to ensure quality would not be possible without adequate and reliable monitoring and, as such, the process analytical technology (PAT) initiative was born in 2004 [9]. It is well understood that the ability to monitor CPP is paramount in developing the required process understanding that enables the advanced process control necessary to achieve enhanced quality in a consistent manner [3]. However, there are a number of challenges that exist when striving to put this concept into practice. First, biopharmaceutical manufacturers are always hesitant to adopt new technologies due to possible regulatory hurdles. The justification required to obtain regulatory approval for process changes can be extensive and is often the reason why process improvements are not made. In addition, while there are a number of guidelines that define QbD and PAT, little published information exists relating to methodology of implementation in a manufacturing setting. Aside from regulatory concerns, there are a number of other very significant challenges related to PAT and implementation of advanced control in bioreactors [6, 12]. These challenges can be divided into the following three broad categories; (1) monitoring, (2) data analysis and integration and (3) advanced control implementation.
Monitoring
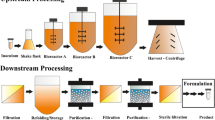
There are various difficulties to overcome related to bioreactor monitoring that range from the physical limitations of the sensors themselves to the complex medium and conditions that exist within the bioreactor. The three phase system of solid cells, liquid media and gas bubbles results in complex hydrodynamics and interactions within the bioreactor that create a challenging environment for monitoring [3]. This is compounded by the transient nature of batch processing, where substrate is being consumed as metabolites are being formed by an increasing number of cells, thereby changing fluid properties such as viscosity and density. The “multiple variable’s” nature not only poses a challenge from a monitoring perspective but can also result in large quantities of data that must be analyzed using multivariate techniques to generate process understanding [12]. There are three types of variables that must be monitored to enable advanced control in bioreactors: physical (such as temperature, pressure, viscosity, agitation, airflow, etc.), chemical (such as pH, dissolved oxygen (DO) and nutritional substrates, etc.) and cell-related or biological (such as total and viable cell density/concentration, host cell proteins, metabolites, CO2 and product, etc.) [3, 19]. Sensors employed to monitor these variables must provide data in sufficient time to describe current conditions within the bioreactor and possibly affect change within the process as needed. Figure 1 shows the difference between in-line, on-line, at-line and off-line sensors/measurements of which in-line are the only sensors capable of real-time data with on-line following close behind. Real-time sensors must be capable of withstanding harsh alkaline and acidic solutions during Clean In Place (CIP) as well as high temperatures during Steam In Place (SIP). Any type of sensor employed must be reliable, accurate and reproducible and should be easy to calibrate, use and maintain. A sensor should also be able to differentiate between background noise and measure process variables with sufficient sensitivity to detect small changes in concentration. When specifically focusing on nutrients and cell-related variables in the liquid phase of the bioreactor, very few sensors exist that meet these challenges other than spectroscopic sensors [of which near infrared (NIR) and Raman spectroscopy dominate] [12, 16, 23, 25].
Examples of in-line, on-line, at-line and off-line monitoring. These are variable data types that are often stored in different locations making integration a common challenge. Standard parameter control and data storage can be accomplished using distributed control systems (DCS), supervisory control and data acquisition (SCADA) systems as well as simple programmable logic controllers (PLC)
Data analysis and integration
With the increase in PAT and advanced monitoring taking place in the industry, there is now a growing challenge of how to make best use of the data that is generated and transform it into process knowledge and understanding [15]. It has been noted that a large part of the future of quality improvement in biomanufacturing will be accomplished by better data analytics of the monitoring that is already in place, making possible more advanced control [11]. The goal of PAT is to identify meaningful data that will lead to process understanding, which ultimately enables process control. This advanced control is based on the link between process knowledge and product quality that is provided through advanced data analytics and ensures a more robust overall process [18]. To perform data analytics, the challenge of data integration from multiple sources must first be overcome. In many cases, data is generated and stored in different locations based on the technology being used, shown in Fig. 1. Standard bioreactor data is often stored in a supervisory control and data acquisition (SCADA) or distributed control system (DCS) while in-line monitoring using NIR spectroscopy or on-line off gas analysis would be stored in another location and can often even be of different data types based on manufacturer software. An integration tool is paramount to enabling the analysis of all types of data simultaneously to build the optimal multivariate analysis (MVA) models for enhanced process understanding and statistical process control (SPC). Data integration between multiple sensors from different manufacturers is still a large challenge today and is a requirement for advanced process control [7]. With these extremely large datasets (big data) MVA must be used to properly make sense of the information [4, 10]. MVA is used in a number of ways with respect to monitoring, ranging from a singular monitoring device, such as with spectra from a NIR probe, to modeling a number of process parameters that would be evolving during the course of a batch. Principal component analysis (PCA) and partial least squares (PLS) models are commonly employed in both situations. Data historian systems are often used to aggregate and store data that is then analyzed by modeling software packages. However, the integration challenge often results in not being able to use generated data effectively, or in some cases, abandoning the advanced monitoring entirely [7]. Once the data has transformed into process understanding the final step in truly integrating PAT is to “close the loop” and feed back into the process to effect changes that will result in a more robust process with more consistent performance and enhanced product quality.
Advanced control implementation
With the understanding that any control methodology needs to be qualified from a regulatory standpoint, there still remains the challenge to make use of data monitoring and analytics in a fully integrated advanced process control strategy. This advanced process control would need to be integrated in such a way as to be able to adjust set points in existing proportional, integral, derivative (PID) controllers that may be under local programmable logic controller (PLC) or DCS control [18]. The use of multivariate models to generate “soft sensors”, where quality is inferred from process measurements, has been in effect for a number of years. However, there is a major challenge in the ability to use those soft sensors to implement process change in a real-time manner, specifically in a manufacturing setting [8, 14]. Advanced control strategies require a platform that can integrate data from standard process parameters, UVA models, external analytical tools (perhaps utilizing PLS models), MVA models (i.e., soft sensors), mechanistic models, external models (for example those developed in Matlab or Python, etc.) as well as predictive models and then utilize the data through control logic that is able to manage alerts and feedback into the process. There are a number of examples in the literature where some parts of this have been achieved. Predictive PLS models using Raman spectra [2] as well as on-line and at-line monitoring of media constituents and cell-based data [24] have been integrated into control logic for feed control in small-scale bioreactors. In addition, a number of soft sensor applications have been reported where MVA models have been used to trigger certain feeds in lab-scale systems [14], however, full integration with closed loop control is currently limited in a manufacturing setting. Various PAT software packages are currently available such as SIPAT from Siemens, syn TQ from Optimal and GE’s Predix that perform a number of these aforementioned required functions. However, there are limitations around integrating third party software tools, incorporating mechanistic models and also implementing rule-based alert management revealing a need for an open platform approach.
The objective of this research is to demonstrate such a platform through the application of PAT to an existing process to increase understanding and subsequently implement an advanced control strategy to obtain more consistent batch-to-batch performance and reduce the potential for failed batches. A pilot scale 30 L Escherichia coli fermentation process producing green florescent protein (GFP) as the target product was selected for this case study due to the high variability in final product concentration and the recent increase in failed batches. Failures were identified based on final GFP titers, which were reduced by 50% compared to the “golden-batch”, however, the cause was not clear due to insufficient understanding of the process. An investigation revealed that the only difference in the failed runs was that a new lot of yeast extract (same manufacturer and supplier) had been used. The process was scaled down to 2 L microbial bioreactors to perform a study that would identify possible contributors and NIR models were generated to track these critical constituents in real-time. After the root cause of the process, failures had been determined, the process was scaled up to 300 L and a control strategy developed to identify and avoid future failures. To decrease titer variability due to induction time, also incorporated in the strategy was automated induction of GFP production based on real-time monitoring of glucose and cell density. Included was a novel advanced control strategy utilizing a multivariate analysis (MVA) model that would make use of process data and univariate statistics to identify deviations from normal operation and predict possible exhaust filter clog failures due to condensate buildup in the filter. The strategy ultimately closed the loop by controlling set-points within predefined limits to save the batch.
Materials and methods
Scale-down 2 L bioreactor study
Once the cause of the final titer failures was determined to be the change in composition of the newer lot of yeast extract (YE), the fermentation process was scaled down from 30 L Sartorius DCU3 stainless bioreactors to 2 L Sartorius BIOSTAT B plus glass bioreactors to perform a media study. Scale-down criteria included aeration rate (vvm or vessel volumes per minute) that was maintained at 0.5 vvm, bioreactor aspect ratio (height/diameter) that was maintained at 1.3, and bioreactor geometry (bioreactor diameter/impeller diameter) that was maintained at 2.5. Temperature was controlled at 30 °C and dissolved oxygen (DO) was controlled to 50% by PID control of agitation, identical to the method used in the larger scale process. This ensured that k La differences between scales was accounted for due to the automated agitation speed increase to match bioreactor oxygen transfer rates to culture oxygen uptake rates. Three media compositions were prepared and the bioreactor runs were performed in triplicate. Each media composition contained the same proprietary basal concentrations of salts, glucose and antifoam (identical to the original process) with three runs conducted using 12 g L−1 of the older lot of YE (identical to the concentration used in the original process), three runs using 12 g L−1 of the newer lot of YE and three runs using 16 g L−1 of the newer lot of YE. The inoculum (E. coli BL21 DE3 genetically modified to produce GFP when induced with lactose or Isopropyl β-d-1-thiogalactopyranoside, IPTG) was prepared by inoculating a 1 L shake flask containing 400 mL of growth media with a l mL vial from the working cell back. The flask was incubated overnight (~ 16 h at 30 °C and 200 rpm) until an OD600nm of ~ 6 was reached. Approximately 60 mL (target starting OD600nm = 0.2) was aseptically transferred into each bioreactor that had been autoclaved and allowed to cool overnight. Prior to inoculation controllers were initiated and the DO probe was calibrated to 100% air saturation.
Batch monitoring
A combination of different monitoring systems were used over the course of the batch: in-line included DO, pH, agitation, airflow and temperature; at-line included glucose using an enzymatic YSI 2700 glucose analyzer (Yellow Springs Instruments) and optical density (OD600nm) using a bench-top Thermo Scientific spectrophotometer (Genesys 20); off-line included amino acid analysis using an Acquity UPLC system from Waters with a PDA detector. Samples were taken aseptically at regular intervals for at-line and off-line measurements. Samples were spun down in a centrifuge for 2 min at 14,000 rpm and the supernatant was used for glucose measurements as well as to blank the spectrophotometer for optical density measurements. One mL of supernatant from each sample was frozen for subsequent analysis by the standard UPLC method as described by Waters [1].
Data analytics
All the data were tabulated and SIMCA from Umetrics was used to analyze the results. A multivariate batch evolution model (BEM) was generated to investigate the relationship between all the variables in a single context. Using time as a maturity variable, a PLS model, instead of a PCA model, was generated from the three-way process measurements array comprised of the number of batches, the process variables in each batch and batch time. The PLS model decomposed the maturity variable vector (y) and the observation data matrix (X) into scores (T), loadings (P and q), weights (W) and residuals (E and f) as follows:
A batch level PLS model (BLM) was generated in a similar fashion to determine the sources of variation in relation to performance attributes or parameters such as yield of cells on glucose (Y x/s), product concentration (g L−1) and maximum specific growth rate (µ max). In this case, the model decomposed the batch performance parameter matrix, of which only one value of each exists per batch, and the batch-wise data matrix into scores, loadings, weights and residuals. The BLM model was used for batch-to-batch comparison as well as to predict performance parameters such as product concentration based on new batch evolution data. This type of prediction capability creates the potential for soft sensor applications, especially in the case that the data analytics tool is utilizing real-time data [22]. The analysis was used to help identify the probable root cause of the failures and the increased understanding resulting from the data analytics helped enable the development of an advanced control strategy.
Scale up to 300 L manufacturing batches
The process was scaled-up to a 300 L bioreactor (equipped with a NIR probe from ABB and in-line AS16-N single channel turbidity probe from OPTEK with a 5 mm path length and operating wavelength between 730 and 970 nm). Three calibration batches were executed at the 300 L pilot scale to build NIR models for in-line monitoring of critical amino acids, glucose and optical density. Samples, coinciding with spectral scans, were taken every 30 min for 7 h for each batch and the appropriate analysis was performed (UPLC for amino acids, YSI for glucose and OD600nm for cell density) so that a correlation could be made with the respective spectra. OPTEK concentration units (CUs) were correlated to the offline OD600nm values by inputting the data into the OPTEK C4000 series photometric converter. A fourth batch was used to validate the spectral calibration models.
Spectroscopy
All sample scans were acquired using an in situ Solvias bubble shedding transflectance probe (12 mm diameter, 230 mm length, 600 µm core and a fixed path length of 1 mm). The probe was connected to an ABB Fourier Transform Process Analyzer Near Infrared FTPA2000-200 series spectrophotometer (Quartz Halogen source) and detection system using a 10.7 m fiber optic cable consisting of high purity fused silica with bidirectional properties that was designed for wavelengths between 200 and 2400 nm. Spectral data was collected at a resolution of 8 cm−1 over a range of wavenumbers between 3800 and 14,000 cm−1. Each spectrum was the average of 1024 scans (background) or 128 scans (samples). Prior to each fermentation batch, the probe was cleaned and allowed to dry before taking a background reading in air.
Chemometrics
GRAMS/AI version 7.0 from Thermo Galactic was used for NIR spectral data collection, spectral processing, and model development. Datasets were created with spectra and their related reference data (generated from the primary methods previously described) and loaded into the GRAMS/AI PLSplus/IQ navigator to create a training data file (tdf). The software was then used to perform spectral preprocessing (i.e., derivatives, baseline corrections, smoothing, normalizations, mean centering) as well as to identify areas of correlation between spectral wavelength regions and constituent concentrations. Pre-processing is required to eliminate unexplainable variation related to sensor noise or scattering effects. After developing a calibration, the software was used to perform statistical analysis on cross-validated data. Cross-validation is the process of removing one sample from the dataset and predicting it using the calibration that is generated from the remaining samples. The predicted values are then compared to the actual value to evaluate the validity of the model. FTSW800 Process software (ABB) was used for the subsequent real-time monitoring for the validation batch and manufacturing runs. Calibration models were loaded into the FTSW800 and the analyte concentrations were automatically calculated from process spectra. These values were sent to the ABB 800xa distributed control system (DCS) and applied materials advanced analytics and control system via a Matrikom OPC tunneler.
Additional 30 L batch runs
The advantage of real-time data was exemplified when using the NIR online analysis of the critical amino acids and glucose. This increased understanding instigated additional analysis of the two different lots of YE to determine the true root cause. In addition to being a major source of amino acids, YE also supplies numerous vitamins, specifically B vitamins. HPLC was used to analyze the YEs for their B vitamin content and based on the variations between lots, additional runs were performed at the original 30 L scale. Since the old lot had been depleted, all runs used the new lot of YE along with the original basal salts, antifoam and glucose concentration. The control consisted of YE at 16 g L−1 and the other two runs were batched at 12 g L−1 with one receiving an additional B vitamin complex mix of 4.1 mg L−1 B2-riboflavin, 48.0 mg L−1 B3-niacin and 2.88 mg L−1 B12-cyanocobalamin when the slowdown in metabolism began to occur (at 4.75 h). An HPLC system (Agilent Ltd.) was used for the analysis and quantitation of vitamins in the two lots of YE as previously described [5].
Advanced control strategy designer
To be able to make use of the advanced monitoring in such a way as to feedback into the process, SmartFactory RX Analytics and Control from Applied Materials Inc. (Santa Clara, CA, USA) was used. The software was able to integrate into multiple data sources such as our DCS (ABB 800xa) system, SCADA (Sartorius MFCS) system and PI (OSIsoft) historian as well as monitoring systems such NIR analyzer (ABB), turbidity probes (OPTEK) and gas mass spectrometer (Thermo Scientific). The unique “drag and drop” strategy engine allows for easy integration of models as well as rule-based logic implementation to manage alerts and alarms. It was also configurable to send notifications or to feedback and change process set-points accordingly. Using the information obtained through the additional monitoring of the scale-down and manufacturing runs, a control strategy was designed based on increased process understanding. The strategy was comprised of simple logic-based case statement blocks that managed model outputs as well as combined multiple sources of data, including the in-line NIR and turbidity data, to help verify YE composition was sufficient for optimal protein production, automate induction at the optimal time, and track univariate (UV) parameters for continuous process verification (CPV). The flow of blocks was determined based on the natural sequence of events required to control the process and will be shown later. The culmination of the strategy was the integration of a predictive multivariate empirical model built with a specific combination of univariate mechanistic statistics. This model demonstrated the complete functionality of the described platform since it not only could detect the potential for condensate buildup in the exhaust filter (which could result in a lost batch) but also effect changes to control parameters to avoid such a failure.
Results and discussion
Deviations in the final protein from the 30 L bioreactor batches began to occur more frequently than acceptable. Figure 2 shows an example of the difference between batches (titers ranging from 0.52 to 0.67 g L−1) that followed our “golden-batch” producing an average of 0.59 g L−1 GFP (left) and batches that exhibited a random decrease in cell growth (slope) near the 5 h mark accompanied by what seemed to be a metabolic shift before continuing the growth phase (right). Glucose was no longer completely consumed by hour 6 and final GFP titer was approximately half of what was expected (0.25–0.33 g L−1). The reason for these failed batches was unknown, which demonstrated a deficiency in process understanding. Failure investigations determined that the only variation between the “good” and “bad” batches was that a new lot of yeast extract (YE) had been used with the poor performing batches. The immediate solution to the problem, which returned batch performance back to the original golden run standard (0.57–0.70 g L−1 with an average titer of 0.63 g L−1), was to increase concentration of the new lot (and all subsequent lots) of YE from 12 to 16 g L−1. The slightly higher average was most probably due to the original YE concentration being too close to the boundary level of limiting performance which also explains why slight variations in YE lots could have such an appreciable effect. Since YE is a major source of nitrogen, in the form of free amino acids, an initial analysis of the two YE lots was performed as seen in Fig. 3. It was clear there were obvious differences, however, the specific source of the failures was not evident and so a scale-down study was designed to investigate the evolution of amino acids throughout each batch.
Scale-down 2 L bioreactor study
In the attempt to identify the source of the variation in fermentation performance, three batch conditions were tested in triplicate at the 2 L scale. Figure 4 shows the averaged trend lines of glucose and optical density for all the bioreactor runs. It is clear that at the original YE concentration, the old lot of YE performs better than the new lot and that performance is increased when using a higher concentration of the new lot. The slightly improved performance of the higher concentration of the new lot indicated that the low concentration of the old lot of YE was on the precipice of being sufficient to ensure glucose was the limiting nutrient. Along with at-line analysis of glucose and optical density, off-line analysis was performed at each time point to determine amino acid concentrations. Combining in-line data from the bioreactor parameters with at-line and off-line data created a very large dataset that required statistical data analytics to elucidate what variables were statistically significant in their contribution to the variance.
Averaged glucose (decreasing trend lines) and optical density (increasing trend lines) data for three batches at each media condition. Yeast Extract new lot at 12 g L−1 (filled diamond), YE new lot at 16 g L−1(filled circle) and YE old lot at 12 g L−1 (filled triangle). Error bars are plus and minus one standard deviation
SIMCA from Umetrics (Umea, Sweden), now owned by Sartorius stedim, was used to analyze the data by generating two models. A principal component analysis (PCA) of all the bioreactor data was performed using time as a maturity variable to determine vectors of scores at each time point for all the variables. This in effect created a partial least squares (PLS) model, which is defined as a batch evolution model (BEM) in the SIMCA software. The plot of all these scores enabled the comparison of each point based on two main principal components describing all the multivariate data at that time. Since scores of the identical principal components were used for each time point, it was possible to make a comparison and identify both expected operation as well as when the process deviated from “normal” operation. Figure 5 is a score plot of each time point from all nine batches. Six of the batches have points that are intermingled together, while points from the other three batches are all grouped together on the bottom right of the plot (as identified by the black circle). Interestingly, these points are all from batches using the original concentration (12 g L−1) of the new lot of YE. Using the software, a statistical analysis was performed that compared these two groupings to identify possible sources of variation. In addition, key performance attributes (KPAs) were calculated for each batch, such as maximum specific growth rate (µ max) and yield of cells on glucose (Y x/s) to analyze how the batch data variations impacted performance. This was accomplished by building PLS batch level models (BLM). Figure 6 shows the complex plot of sources of variation between the three batches of new YE at 12 g L−1 and the other six batches in the top graph and a simplified version on the bottom where variables with minimal sources of variation were removed indicating two main amino acids, alanine and methionine, contribute highly to the source of variation in performance. Contribution of alanine increases dramatically at hour 4.5, while methionine does not vary until hour 5. Further study of these amino acids revealed that methionine was completely consumed by hour 5.5 and that the consumption rate of alanine between 4 and 6 h almost doubled with the new lot of YE (73.5 mM h−1) compared to the old lot (45.7 mM h−1), a greater change than with any other amino acid. This time range coincided with the metabolic shift that occurred in the culture and as such these two amino acids were selected as ideal constituents to monitor in-line using NIR spectroscopy in the 300 L scale production bioreactor. Agitation speed was also identified as a contributor. However, since agitation was automatically increased to control dissolved oxygen (DO) the significance of this variation is a confirmation that respiration was indeed slowing at this time point due to the metabolic shift occurring from a nutrient limitation other than glucose.
The three main contributors to variation between good and bad batches (isolated from all the sources shown in the embedded top left plot) based on a PLS batch level model (BLM) relating in-line, at-line and off-line data to key performance attributes (KPA). Shown is the time range of 4–6 h, where the metabolic shift occurred, alanine (filled diamond), agitation (filled circle) and methionine (filled triangle)
Scale-up to 300 L for NIR model building
To determine the effect of these two amino acids, alanine and methionine, on the utilization of glucose, all three of these parameters were selected to incorporate into an advanced monitoring process analytical technology (PAT) strategy utilizing NIR spectroscopy. Real-time data is essential to elucidate batch performance especially when dealing with microbial fermentations where rapid metabolism shifts occur. Models for each of these constituents were built using Grams AI software. Three batches were executed where samples were taken at regular intervals coinciding with NIR scans. The samples were analyzed to determine the concentration of glucose (YSI analyzer) along with alanine and methionine (UPLC). A matrix was built linking the scans to each constituent concentration and an iterative process was performed to build models that would predict all three. Each PLS model was pre-processed using mean centering with no pathlength correction and a manual baseline. The following spectral regions, in wavenumbers (cm−1), demonstrated the highest correlation to each constituent and were selected for building each respective model: glucose (7117–5987 cm−1), methionine (7425–6195 cm−1) and alanine (6758–6106 cm−1). Figure 7 shows the cross-validated model predictions versus the actual values of all three constituents. The cross-validation was performed by removing one sample from the dataset and using the other samples to build the model and predict the “unknown” sample. These plots indicated a good fit for all three constituents. Further evidence can be seen in the partial residual error sum squared (PRESS) plot in the bottom right image. All constituents had similar shaped plots but only the plot for glucose is shown. This plot shows that as factors are added to the partial least squares (PLS) predictive correlation, there is less error in the prediction. It is important not to select too many factors to avoid modeling noise, thereby increasing the error as seen in the upward drift near the end of the plot. The next step was to validate each model with a dataset that was not used when generating the models. Figure 8 shows a validation batch with predictions every 15 min along with at-line and off-line sample data. Ideally, up to ten batches would be used to generate more robust models, however, in this case, predictions were still acceptable with an average percent error for glucose of 3.29%, alanine of 3.75% and methionine of 19.17% (less acceptable). The results from the in-line NIR scans revealed a different result than what was expected. The initial thought that the amino acids were causing the growth limitation based on the original off-line data was proved to be incorrect when using in-line monitoring. The slowdown in glucose consumption actually occurred first followed by an increased consumption of alanine as seen in Fig. 8. This increased process understanding brought about by the NIR in-line data forced a re-evaluation as to the true growth-limiting nutrient. This illustrates the insight that advanced real-time monitoring can provide during process development, as well as during manufacturing. Previous research into alanine utilization in E. coli indicated that it, along with other key amino acids, is highly linked to B vitamin production, which is very important for cell metabolism and growth rate [13, 20]. While E. coli can produce B vitamins, energy is taken away from growth and protein production, which would also explain the reduction in target protein associated with the failed batches. It should be noted that the original empirical model developed using SIMCA did not predict this possibility. However, these types of models, including PLS chemometric models, are purely data driven and are not suited for extrapolation of any kind. As such, it is critical that the model contains all possible data variation which might be observed in the process whether it be concentration ranges or process variables [21]. The trends in Fig. 8 that show the slowdown of glucose metabolism followed by an increase in alanine consumption, could be explained by a metabolic shift of nutrient utilization required to produce more B vitamins. Further work was, therefore, carried out to investigate this theory at the original 30 L scale.
NIR model calibrations for three constituents, alanine (top left), methionine (top right) and glucose (bottom left). All cross-validated (one sample out at a time) R 2 values were above 0.95 which indicates a relatively good fit for each model. The final plot (bottom right) shows the reduction in error as additional factors are added to the glucose model with a final recommendation of five factors based on the predicted residual error sum squared (PRESS) values
300 L validation batch data with NIR scans (average of 128 scans taken every 15 min) for the following models: glucose (filled diamond), methionine (multiplication sign) and alanine (plus) as well as at-line YSI measurements for glucose (filled square) and offline UPLC measurements for methionine (filled triangle) and alanine (filled circle) determined from manual samples taken at periodic intervals
B vitamin analysis
The first step was to analyze the lots of YE to compare concentrations of B vitamins. Results from HPLC analysis of the two lots can be seen in Fig. 9. The variation suggested that there was a significant difference in content of B vitamins at the same concentration of YE. Three 30 L bioreactors were then batched under identical conditions using the new lot of yeast extract except for the following modifications. One batch contained 16 g L−1of YE, one contained the original 12 g L−1 of YE and the last contained the same 12 g L−1 of YE but a B vitamin complex was prepared to add during the batch between 4.5 and 5 h of run time correlating to the change in metabolism previously observed. The concentration and composition of B vitamins to add was estimated based on the variation between the two lots determined by HPLC as seen in Fig. 9. Since the 30 L bioreactors are not equipped with NIR probes, at-line samples of glucose (YSI analyzer) and optical density (spectrophotometer at 600 nm) were used to determine batch performance. The top graph in Fig. 10 contains the at-line trends from these experiments and confirms that lack of sufficient B vitamins was the actual root cause of the failures. The optimal growth was achieved at 16 g L−1 YE, however, it was evident that at the beginning of the batches all exhibit similar growth until approximately 4.5 h. At this point, the two batches with lower concentration of YE begin to slow and the B vitamin complex was added to one batch at approximately hour 4.75. The additional B vitamins had almost an immediate effect of returning the growth rate back to what was observed at the higher concentration of YE. The bottom graph in Fig. 10 includes the agitation trends for each run and confirms the effect of the additional vitamins. Agitation is controlled based on culture demand and recorded real-time so that it can be used to pinpoint the exact time of shifts in culture oxygen requirements. There was a similar increase in agitation in the two bioreactors with 12 g L−1 YE until the B vitamins were added, and then the respiration increased to closely follow what was observed in the bioreactor with 16 g L−1, as evidenced by the slope of the agitation trend lines. It is clear that the additional B vitamins in the 12 g L−1 run were not sufficient to obtain identical results to the 16 g L−1 bioreactor, based on data in Fig. 10. Cell density was still slightly lower and overall oxygen demand was not as high, however, it is evident that performance was improved in comparison to the standard 12 g L−1 YE batch. With the root cause determined and the corrective action in place using 16 g L−1 of YE, the next step was to utilize the advanced monitoring at the 300 L scale in a control strategy that would make use of this increased knowledge and close the loop to affect greater consistency in product titers from batch to batch.
Comparison of various B vitamin concentrations, determined by HPLC, in two lots of yeast extract at 12 g L−1. The solid bars represent the old lot with acceptable performance and the textured bars represent the new lot with poor performance. (Pantothenic values are shown divided by a factor of 10 for scaling purposes)
Top: glucose (decreasing trend lines) and optical density (increasing trend lines) data for three 30 L batches at identical operating conditions except for the following. 12 g L−1 YE (filled diamond), 16 g L−1 YE (filled circle) and 12 g L−1 YE with additional B vitamins added at 4.75 h (filled triangle). The bottom plot shows the real time agitation speed for each batch, 12 g L−1 YE (bottom trend line) 12 g L−1 with addition of B vitamins at 4.75 h (middle trend line) and 16 g L−1 YE (top trend line). Agitation was in cascade control and automatically changed based on oxygen demands of the culture (increased respiration rate requires higher agitation speeds)
Advanced control strategy
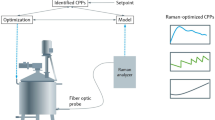
A very common challenge in manufacturing is to make use of advanced monitoring tools in an effective way. To achieve this, the first requirement is a level of integration that links monitoring (including advanced analyzers) to data-analytic modeling tool. This dual functionality of utilizing advanced monitoring tools along with multivariate modeling capabilities to build soft sensors, are both crucial not only to generate process understanding, specifically during the process development stages, but also to allow for advanced control during manufacturing. Once accomplished there is still the requirement for a platform where control strategies can be designed that will have access to all these types of data and be able to close the loop and integrate back into equipment controllers. This is essential to implement the necessary changes that will increase consistency and quality, thereby reducing the number of lost batches. The Analytics & Control (A&C) software package, purchased from Applied Materials, was selected as the platform solution to try and meet this overall challenge. This software achieved the integration of our DCS system from ABB as well as the SCADA system from Sartorious. In addition, PAT tools such as the NIR probe from ABB (for inline measurement of glucose and alanine) and our turbidity probe from OPTEK (for inline cell density) were integrated for use in the final control strategy. The A&C software also includes data analytic capabilities, where MVA models can be generated for soft sensors that can be incorporated into strategies for real-time or even predictive type monitoring and control. The built in strategy designer allowed for easy drag and drop design of a control strategy as shown in Fig. 11. It should be noted that simple control logic can also be configured in the DCS system but this would not have been applicable to bioreactors controlled by the SCADA system or have the capability to integrate soft sensor technology for advanced control purposes. The control strategy shown in Fig. 11 has multiple functionalities. The first was to use A&C to build a univariate analysis (UVA) model around the change of alanine concentration over time (a univariate soft sensor). The model characterized normal consumption rates of alanine when B vitamins were not limiting (based on slope). In the event that there is a rapid decrease of alanine caused by low B vitamin concentrations the model limits will be exceeded and the strategy will send an alarm notification to the operator that the batch may be suspect and additional feed could be added to achieve acceptable protein titers. To date, this predictive control notification has not been required since the current YE concentration is in excess of what is required.
Unique strategy designer for advanced process control that initiates at the start of every batch. Three control workflows are shown including a univariate model monitoring abnormal depletion rate of alanine [1], a set of rules implemented using a case block (taken from the menu on the bottom left and easily configured as shown on the bottom right) that when satisfied trigger induction [2], and a complex control strategy that predicts a filter clog will occur using a multivariate model and closes the loop to effect changes to save the filter and the batch [3]
The second part of the strategy was to utilize the inline glucose and cell density measurements to trigger induction at the optimal concentration and density. Since there are variations from batch to batch in terms of overall fermentation time, it is not optimal to use batch time as the indicator for induction. Glucose concentration and cell density at the time of induction have a high impact on target protein production and normally sampling is done to analyze these parameters at-line, which often results in variation in optimal induction time from batch to batch. Three batches were executed at the 300 L scale to test the induction control where the pump was actuated based on in-line process parameters instead of at-line analysis. Eliminating this variability led to an increase in consistency from batch to batch based on a decrease in coefficient of variation (CV) from 8.49 to 1.16% as well as a higher average GFP titer of 0.69 g L−1. It should be noted that this titer has also been achieved using at-line monitoring for induction, however, not in a consistent manner.
The final part of the strategy was to implement advanced predictive control by not only predicting when an out of limit event might occurs but also to close the loop to change set-points based on a multivariate analysis (MVA) model (a multivariate soft sensor). A MVA model was generated based on gas flow rates and backpressure control output to predict possible internal pressure issues due to condensate buildup in the exhaust filter. The failure is preceded by abnormal output to the back pressure controller as the filter begins to clog. However, backpressure control output varies depending on gas flow changes during processing as well as pressure set-point and total flow. Therefore, an MVA model was necessary to predict when the filter was beginning to clog as can be seen in part three of Fig. 11. The strategy automatically reduces gas flow into the reactor and opens a condensate valve on the exhaust filter to allow the filter to recover before returning control back to the bioreactor controller. Utilizing historical data, the MVA prediction model for abnormal backpressure output was able to predict a filter clog 3.3 h prior to the actual failure allowing for extra time to save the batch. Since no such event has occurred while running the control model, a simulation was performed with water in the bioreactor. Blocking the exhaust filter caused the model to successfully trigger and feedback pre-determined commands to the DCS system via object linking and embedding for process control (OPC) to reduce the inlet airflow and open the exhaust condensate valve to relieve pressure. Real-time multivariate analysis also offers the ability to monitor batch health during the run, relative to the golden standard, from a holistic view rather than relying on a single parameter at a time approach.
Conclusions
The results of this research show that the use of PAT is a powerful tool when seeking to generate process understanding and implement advanced control. It is evident that developing these advanced techniques during process development allows for a much smoother transition during technology transfer to manufacturing. In addition, advanced monitoring and soft sensors have been shown here to increase process understanding by revealing details that would not normally be seen by normal sampling or by end of batch testing. This research elucidates the importance of real-time batch evolution information when performing experimentation to link process parameters to performance and quality attributes. However, this is only part of the equation. This research also elucidates the current gaps that exist in the industry and exemplifies the need for a platform technology that can enable true advanced process control through implementation of the novel and complete system depicted in Fig. 12. The initial requirement is for monitoring (M), however, to make use of all different types of monitoring an integration (I) platform is required. This integration is critical for various types of analyzers and variables but also for various third party modeling tools as well. With the monitoring integrated into one location, data analytics (D) can be performed to generate process understanding (U) and allow soft sensors or predictive models to be built and incorporated, along with other monitoring information, into strategies (S) that enable advanced control (C) or MIDUS Control. The results of this work demonstrate a MIDUS Control platform through the use of analytics and control from applied materials. The platform was able to integrate multiple sources of data and perform analytics in real-time to execute strategies that automated protein induction and decreased batch to batch variability so that “golden-batch” performance could be achieved more consistently. In addition, the platform was configured to detect possible failures with sufficient time to automatically implement process changes and save batches from failure. The potential of this platform is only beginning to be explored and currently more models are being configured around predictive maintenance and probe health. Much work remains to be done in this area, however, the Midas touch, achieved through employing MIDUS control, have been proven to be a reality.
References
Aubin A, Hynes M, Shockcar J (2007) Aquity UPLC for the rapid analysis of amino acids in wine. Waters Appl Note. 720002044:1–8
Berry BN, Dobrowsky TM, Timson RC, Kshirsagar R, Ryll T, Wiltberger K (2016) Quick generation of Raman spectroscopy based in-process glucose control to influence biopharmaceutical protein product quality during mammalian cell culture. Biotechnol Prog 32:224–234
Biechele P, Busse C, Solle D, Scheper T, Reardon K (2015) Sensor systems for bioprocess monitoring. Eng Life Sci 15:469–488
Challa S, Potumarthi R (2013) Chemometrics-based process analytical technology (PAT) tools: applications and adaptation in pharmaceutical and biopharmaceutical industries. Appl Biochem Biotechnol 169:66–76
Chatzimichalakis P, Samanidou V, Verpoorte R, Papadoyannis I (2004) Development of a validated HPLC method for the determination of B-complex vitamins in pharmaceuticals and biological fluids after solid phase extraction. J Sep Sci 27:1181–1188
Chopda VR, Gomes J, Rathore AS (2016) Bridging the gap between PAT concepts and implementation: an integrated software platform for fermentation. Biotechnol J 11:164–171
Graham LJ (2016) Leveraging data analytics innovations to improve process outcomes. Biopharm Int North Olmsted 29:18–22
Hausmann R, Henkel M, Hecker F, Hitzmann B (2017) 25—Present status of automation for industrial bioprocesses. In: Larroche C, Sanromán MÁ, Du G, Pandey A (eds) Current developments in biotechnology and bioengineering. Elsevier, Amsterdam, pp 725–757
Izat N, Yerlikaya F, Capan Y (2014) A glance on the history of pharmaceutical quality by design. OA Drug Des Deliv 2:1–8
Kourti T (2006) The process analytical technology initiative and multivariate process analysis, monitoring and control. Anal Bioanal Chem 384:1043–1048
Langer E (2013) The future of biopharma. Biopharm Int North Olmsted 26:22–24
Lourenço ND, Lopes JA, Almeida CF, Sarraguça MC, Pinheiro HM (2012) Bioreactor monitoring with spectroscopy and chemometrics: a review. Anal Bioanal Chem 404:1211–1237
Magnúsdóttir S, Ravcheev D, de Crécy-Lagard V, Thiele I (2015) Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front Genet 6:148
Mandenius C-F, Gustavsson R (2015) Mini-review: soft sensors as means for PAT in the manufacture of bio-therapeutics. J Chem Technol Biotechnol 90:215–227
Marx V (2013) The big challenges of big data. Nat Lond 498:255–260
Pohlscheidt M, Charaniya S, Bork C, Jenzsch M, Noetzel T, Luebbert A (2013) Bioprocess and fermentation monitoring. Upstream industrial biotechnology: equipment, process design, sensing, control, and cGMP operations. Wiley, Hoboken, pp 1471–1491
Read EK, Park JT, Shah RB, Riley BS, Brorson KA, Rathore AS (2010) Process analytical technology (PAT) for biopharmaceutical products: part I. concepts and applications. Biotechnol Bioeng 105:276–284
Rios M (2014) Analytics for modern bioprocess development. BioProcess Int 12:1–8
Scheper T, Hitzmann B, Stärk E, Ulber R, Faurie R, Sosnitza P, Reardon KF (1999) Bioanalytics: detailed insight into bioprocesses. Anal Chim Acta 400:121–134
Schneider F, Krämer R, Burkovski A (2004) Identification and characterization of the main β-alanine uptake system in Escherichia coli. Appl Microbiol Biotechnol 65:576–582
Swarbrick B, Westad F (2016) An overview of chemometrics for the engineering and measurement sciences. In: Kutz M (ed) Handbook of measurement in science and engineering. Wiley, Hoboken, pp 2307–2407
Ündey C, Ertunç S, Mistretta T, Looze B (2010) Applied advanced process analytics in biopharmaceutical manufacturing: challenges and prospects in real-time monitoring and control. J Process Control 20:1009–1018
Wu H, Read E, White M, Chavez B, Brorson K, Agarabi C, Khan M (2015) Real time monitoring of bioreactor mAb IgG3 cell culture process dynamics via Fourier transform infrared spectroscopy: implications for enabling cell culture process analytical technology. Front Chem Sci Eng 9:386–406
Zhang A, Tsang VL, Moore B, Shen V, Huang Y-M, Kshirsagar R, Ryll T (2015) Advanced process monitoring and feedback control to enhance cell culture process production and robustness. Biotechnol Bioeng 112:2495–2504
Zhao L, Fu H-Y, Zhou W, Hu W-S (2015) Advances in process monitoring tools for cell culture bioprocesses. Eng Life Sci 15:459–468
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vann, L., Sheppard, J. Use of near-infrared spectroscopy (NIRs) in the biopharmaceutical industry for real-time determination of critical process parameters and integration of advanced feedback control strategies using MIDUS control. J Ind Microbiol Biotechnol 44, 1589–1603 (2017). https://doi.org/10.1007/s10295-017-1984-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-017-1984-2