Abstract
Federated learning, an innovative artificial intelligence training method, offers a secure solution for institutions to collaboratively develop models without sharing raw data. This approach offers immense promise and is particularly advantageous for domains dealing with sensitive information, such as patient data. However, when confronted with a distributed data environment, challenges arise due to data paucity or inherent heterogeneity, potentially impacting the performance of federated learning models. Hence, scrutinizing the efficacy of this method in such intricate settings is indispensable. To address this, we harnessed pathological image datasets of endometrial cancer from four hospitals for training and evaluating the performance of a federated learning model and compared it with a centralized learning model. With optimal processing techniques (data augmentation, color normalization, and adaptive optimizer), federated learning exhibited lower precision but higher recall and Dice similarity coefficient (DSC) than centralized learning. Hence, considering the critical importance of recall in the context of medical image processing, federated learning is demonstrated as a viable and applicable approach in this field, offering advantages in terms of both performance and data security.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Artificial intelligence is actively being used in various industrial environments [1,2,3] and various medical fields, including auxiliary diagnosis and health care [4]. Due to privacy policies, the sharing and collection of medical data required for multi-institutional artificial intelligence learning with/from other institutions is restricted [5]. However, training an artificial intelligence model with a single hospital dataset can result in overfitting or biased result [6]. Therefore, it is crucial to find a way to train artificial intelligence models without collecting data from various hospitals [7, 8]. Federated learning is an emerging training method that resolves privacy and security concerns of patient data because it can be trained without collecting raw data from each hospital [9].
Lutnick et al. [10] used federated learning to segment pathological images associated with tissue fibrosis and tubular atrophy and compared its performance with that of centralized learning models; neither client model showed a statistically significant difference. Adnan et al. [11] divided large pathological images into patches, and multiple instance learning (MIL) [12] was applied to classify a bag of patches. In addition, they compared the performance of federated learning models trained with different numbers of clients to see how it affected federated learning. They found that fewer clients resulted better the performance, and the least-client model differed slightly from centralized learning. Terrail et al. [13] applied federated learning to train pathological whole slide images (WSIs) of triple-negative breast cancer (TNBC), a relatively rare data type, from two clusters and showed that federated learning outperformed each cluster model. Cetinkaya et al. [14] used data augmentation techniques to address performance degradation caused by heterogeneity in medical image data, and federated learning with data augmentation improved the performance compared to that without data augmentation.
However, learning the heterogeneous distribution and diversity of data held by clients is difficult for federated learning, and simply using the weight average of each client model can degrade the performance of the artificial intelligence model [15]. Pathological images are also complex, containing not only various histologic patterns and cell types but also heterogeneous staining patterns for each hospital [16, 17]. Therefore, when using federated learning with pathological images, it is necessary to determine whether the model’s performance is degraded and to what extent. In this study, we compared the performances of models for each training method in pathological image segmentation tasks to focus on the effectiveness of federated learning compared to centralized learning. We also aimed to demonstrate the features of the federated learning model using pathological images.
Method
This comparative study utilized hematoxylin and eosin (H&E)-stained pathological whole slide image (WSI) of endometrial cancer from four hospitals. All the pathology slides were acquired from curettage or hysterectomy specimen and were scanned using PANNORAMIC® 250 Flash III scanner (3DHistech) at 40 × resolution. To adhere to the consistent amount of input data for each client model, the training dataset for each hospital was standardized to 66 WSIs per hospital. The number of patients included from each hospital is as follows: Sinchon Severance Hospital (13 patients), Gangnam Severance Hospital (15 patients), and Gachon University Gil Hospital (38 patients). The study was approved by the Institutional Review Boards of each hospital (4–2016-0809, 3–2016-0236, and GBIRB2022-236).
The area suspected to contain cancer lesion were labeled by a pathologist irrespective of data source and the area was regarded as ground-truth of the WSI data (Fig. 1).
We used two labels, one for the pathologist-labeled cancer area and the other for the background which mostly include non-cancerous tissue. The quantity of WSI was increased from 66 to 198 for each hospital by applying horizontal and vertical flip augmentation techniques to the training data.
In anticipation of deploying the trained model in real-world applications across diverse hospital settings, we performed model validation by utilizing data from one of the four hospitals (Kangdong Sacred Heart Hospital) that were not part of the model training process. The validation dataset comprised 14 WSIs from six patients, ensuring its independence from the hospital utilized for model training. The original input data size was 172,032 pixels in width with varying height from 250,000 to 420,000 pixels, encompassing 3 Red, Green, Blue (RGB) channels. Using the bi-cubic algorithm, each image was resized to 512 × 256 pixels before being used as input data. Furthermore, to address apprehensions regarding the potential effects of variances in lesion size among hospital datasets on the federated learning model, we undertook a validation of the distinctions in lesion size (Table 1).
The disparity in the sizes of cancer lesions in training data from three distinct hospitals did not exhibit statistically significant difference (p = 0.90).
The images from each hospital exhibited color distribution heterogeneity from variations in the H&E slide preparation steps, encompassing factors such as fixation, dehydration, sectioning, staining, and the storage conditions of H&E slides for digital scanning. Consequently. To address this issue, we employed vahadane [18] color normalization method (Fig. 2), which leverages structural properties, including sparseness and non-negativity within stained tissue samples, to establish structural color invariance, a distinguishing characteristic not found in previous normalization methods.
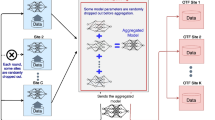
Figure 3 depicts the overall structure of this study. Three of the four hospitals’ data (clients 1, 2, and 3: Sinchon Severance hospital, Gangnam Severance hospital, Gachon University Gil hospital) were utilized for model training. In federated learning, individual hospital datasets (clients 1, 2, 3) were employed for training each respective client model, while integrated datasets from all hospitals were utilized for centralized learning. The fourth hospital’s data (client 4: Kangdong Sacred Heart Hospital) were used for validation and performance comparison of each model training method.
Overview of training and validation to compare each learning method. Federated learning (FL) comprises, a clint model for each individual hospital dataset and an FL model that incorporates the weights from individual hospital client models. Meanwhile, centralized learning (CL) uses all data to train one model. The performances of the differentially trained FL and CL models were compared using validation data
The experimental environment consisted of 40 Intel(R) Xeon(R) Silver 4210R CPUs, 1 NVIDIA A100 GPU, and 128 GB memory capacity. Package versions used were Python 3.8.13, Tensorflow-GPU 2.7.4, CuDNN 7.6.5, cuda-toolkit 11.3.1.
The segmentation model employed in this comparative study is the U-Net [19], a prominent deep learning architecture extensively employed for its effectiveness in image segmentation and medical image processing. U-Net [19] was originally developed with a primary focus on addressing segmentation tasks in the field of medical imaging. Additionally, we made a slight alteration to the original U-Net [19] model by using only two pooling steps in order to prevent the feature size of the extracted cell images from becoming excessively small. Furthermore, considering the heterogeneity observed among client’s datasets and mindful of the potential adverse effects of batch normalization as indicated in Wang [20], we employ group normalization [21] as the normalization method.
The Adam optimizer [22] was used for the centralized learning and client model. Considering potential issues associated with applying FedAvg [23], which simply uses the average of weights among client models in non-convex scenarios due to data heterogeneity among clients, and taking note of the demonstrated performance improvements attributed to adaptive optimizers in federated learning, we introduced the FedYogi optimizer [24] for federated learning model. While sharing similarities with the Adam optimizer in terms of gradient handling, FedYogi [24] has recently exhibited superior performance by mitigating dependency on the magnitudes of its recent gradient.
The batch size, number of epochs, and learning rates of the centralized learning model were set to 9, 40, and 0.001, respectively, and the hyper-parameter of the federated learning model was set the same as for centralized learning.
We employed the two-sample t-test, Mann–Whitney U test to analyze the differences in the performance of the trained models. In addition, the Bland–Altman plot was used to measure the degree of performance difference between the two training methods.
Results
We have compared the performance metric scores (precision, recall, and Dice similarity coefficient) of the centralized learning model and federated learning model using FedYogi optimizer (Table 2).
Federated learning exhibited lower precision by 2.61% (p = 4.26e-04) and higher recall by 5.53% (p = 8.71e-03), higher Dice similarity coefficient (DSC) by 1.64% (p = 0.06) compared with centralized learning, with statistically significant differences observed for precision and recall (p < 0.05) and marginal significance for DSC (p = 0.06).
Figure 4 presents predictions of validation data for the top-level outcomes of each learning method model, alongside the corresponding false-positive rate (FPR: false-positive/actual negative) and false-negative rate (FNR: false-negative/actual positive) based on each area.
Furthermore, we compared the FPR and FNR for the validation data predictions of each learning method model (Table 3).
The FPR (p = 0.79) and FNR (p = 0.77) in the predictions between each learning method model exhibited no statistically significant differences (p > 0.05 for all).
Additionally, Fig. 5, which includes Bland–Altman plot and box plot, was used to visually represent the differences in mean and median values from the DSC perspective.
The subsequent results depict a comparative analysis of the effects of various techniques employed sequentially during the federated learning process, utilizing FedAvg as the baseline. These techniques incorporate augmentation, color normalization, and optimizer.
In order to assess the influence of data augmentation on federated learning, we conducted a performance comparison of federated learning models trained using data both before and after augmentation (Table 4).
Upon comparing the results before and after the application of augmentation, an enhancement in precision, approximately 3.68% (p = 1.80e-07), and a substantial enhancement in recall by approximately 14.94% (p = 4.68e-07) and an increase in the DSC by approximately 11.49% (p = 7.98e-12) were demonstrated. As a result, statistically significant differences were observed for all metrics (p < 0.05 for all). Especially despite being a simple flip augmentation technique, it demonstrated a substantial improvement across all performance metrics.
We also compared the performance of federated learning models using data before and after color normalization to assess the impact of color normalization on federated learning (Table 5).
Upon comparing the performance after adapting color normalization, precision exhibited a reduction of approximately 4.29% with statistical significance (p = 4.51e-07), while recall increased by approximately 2.29% with marginal significance (p = 0.10) and the DSC also increased by 3.47% with statistical significance (3.46e-05).
Lastly, in order to assess the influence of the optimizer on the federated learning process, we conducted a performance comparison of models trained using the previously employed FedAvg and FedYogi optimizers (Table 6).
After adapting FedYogi optimizer exhibited an increase precision by 9.81% (p = 1.08e-26), recall by 12.27% (p = 2.69e-06), and DSC by 10.8% (p = 1.24e-11) compared with using FedAvg optimizer model, with statistically significant differences observed for all three metrics (p < 0.05 for all).
The difference in average performance (DSC) between the two learning methods appears to be approximately 0.02 in Fig. 5a. The median DSC value for federated learning (0.80; range, 0.76–0.83) was higher by 0.04 than that for central learning (0.76; range, 0.66–0.83) and statistically significant differences in the median were observed in Fig. 5b (p = 5.24e-3).
Discussion
In this study, as a response to the increasing demand for federated learning in the field of medical image processing, we conducted a comparative assessment of federated and centralized learning models using the same hyper-parameters, and datasets to verify the effectiveness of federated learning in pathological images.
The federated learning model yielded comparable or better performance on validation set from a DSC perspective and exhibited superior performance in terms of recall. Although the mean DSC of the federated learning (78.51 ± 5.74) was higher than that of centralized learning (75.88 ± 4.83) with marginal significance using two-sample t-test (p = 0.06), the median DSC of federated learning (0.80; range, 0.76–0.83) was significantly higher than that of centralized learning (0.76; range, 0.66–0.83) (p = 5.24e-3). While the t-test results indicate marginal significance in the mean values, the Mann–Whitney U test, which is less sensitive to outliers, confirms that the median DSC is significantly higher in federated learning.
To discover opportunities for further improve the federated learning model from learning what made the federated learning model confused, we compared the histology of the patches according to confusion matrix generated by the federated learning model. The pattern in true positive (TP) could be easily distinguished from the rest of the pattern, whereas the patterns in false positives (FP) and false negatives (FN) were more difficult to distinguish histologically in low-power field view (Fig. 6).
The representative patch images of true positive (TP) contained images from common cancer cell types (endometrioid carcinoma). In contrast, the patches of false positives (FP) were primarily composed of normal tissue that was mostly excluded from the cancer (lesion) labeling and very few patches could have been included for learning. Similarly, the false negative (FN) patches contained a relatively rare cancer cell type (e.g., carcinoma with signet ring cell features), which could have led to a lack of data for learning and led to failure to find distinguishable features.
In conclusion, federated learning method, complemented by a range of techniques, presented higher recall and DSC than centralized learning. Our study highlights that federated learning is an effective approach for deployment in sectors where data security is paramount. It exhibits particular suitability for the domain of medical image processing, where an emphasis on recall and Dice similarity coefficient (DSC) performance holds significant importance. And our research emphasizes the proactive utilization of various techniques to mitigate performance degradation in federated learning models due to heterogeneity in distributed data environments and limited data quantity.
Data Availability
Data are accessible from the corresponding author by request, subject to approval from the institutional review board.
References
Dhalla S, et al.: Semantic segmentation of palpebral conjunctiva using predefined deep neural architectures for anemia detection. Procedia Computer Science 218:328–337, 2023
Kaur A, Kumar M, Jindal M: Cattle identification system: a comparative analysis of SIFT, SURF and ORB feature descriptors. Multimedia Tools and Applications:1–23, 2023
Mohiuddin S, Malakar S, Kumar M, Sarkar R: A comprehensive survey on state-of-the-art video forgery detection techniques. Multimedia Tools and Applications:1–41, 2023
Bohr A, Memarzadeh K: The rise of artificial intelligence in healthcare applications. Artificial Intelligence in Healthcare:25 - 60, 2020
Darzidehkalani E, Ghasemi-Rad M, van Ooijen PMA: Federated Learning in Medical Imaging: Part II: Methods, Challenges, and Considerations. J Am Coll Radiol 19:975–982, 2022
Darzidehkalani E, Ghasemi-Rad M, van Ooijen PMA: Federated Learning in Medical Imaging: Part I: Toward Multicentral Health Care Ecosystems. J Am Coll Radiol 19:969–974, 2022
Rieke N, et al.: The future of digital health with federated learning. NPJ Digit Med 3:119, 2020
Sheller MJ, et al.: Federated learning in medicine: facilitating multi-institutional collaborations without sharing patient data. Sci Rep 10:12598, 2020
Kairouz P, et al.: Advances and Open Problems in Federated Learning. Foundations and Trends in Machine Learning 14:1–210, 2021
Lutnick B, et al.: A tool for federated training of segmentation models on whole slide images. J Pathol Inform 13:100101, 2022
Adnan M, Kalra S, Cresswell JC, Taylor GW, Tizhoosh HR: Federated learning and differential privacy for medical image analysis. Sci Rep 12:1953, 2022
Carbonneau MA, Cheplygina V, Granger E, Gagnon G: Multiple instance learning: A survey of problem characteristics and applications. Pattern Recognition 77:329–353, 2018
Ogier du Terrail J, et al.: Federated learning for predicting histological response to neoadjuvant chemotherapy in triple-negative breast cancer. Nat Med 29:135–146, 2023
Cetinkaya A, Akin M, Sagiroglu, S: Improving Performance of Federated Learning based Medical Image Analysis in Non-IID Settings using Image Augmentation. 2021 International Conference on Information Security and Cryptology (ISCTURKEY):69–74, 2021
Hsu T-MH, Qi, Brown M: Measuring the Effects of Non-Identical Data Distribution for Federated Visual Classification. ArXiv abs/1909.06335, 2019
Farmer ER, Gonin R, Hanna MP: Discordance in the histopathologic diagnosis of melanoma and melanocytic nevi between expert pathologists. Human Pathology 27:528–531, 1996
Lodha S, Saggar S, Celebi JT, Silvers DN: Discordance in the histopathologic diagnosis of difficult melanocytic neoplasms in the clinical setting. J Cutan Pathol 35:349–352, 2008
Vahadane A, et al.: Structure-preserving color normalization and sparse stain separation for histological images. IEEE transactions on medical imaging 35:1962–1971, 2016
Ronneberger O, Fischer P, Brox T: U-Net: Convolutional Networks for Biomedical Image Segmentation. ArXiv abs/1505.04597, 2015
Wang Y, Shi Q, Chang T-H: Why Batch Normalization Damage Federated Learning on Non-IID Data? IEEE transactions on neural networks and learning systems PP, 2023
Wu Y, He K: Group Normalization. International Journal of Computer Vision 128:742 – 755, 2018
Kingma DP, Ba J: Adam: A Method for Stochastic Optimization. CoRR abs/1412.6980, 2014
McMahan HB, Moore E, Ramage D, Hampson S, Arcas BAy: Communication-Efficient Learning of Deep Networks from Decentralized Data. Proc. International Conference on Artificial Intelligence and Statistics: City
Reddi SJ, et al.: Adaptive Federated Optimization. ArXiv abs/2003.00295, 2020
Acknowledgements
We extend our heartfelt gratitude to the members of the biomedical engineering lab for their invaluable guidance and significant contributions to the development of the federated model.
Funding
This work was supported by the Gachon University research fund of 2022 (GCU-2022–202209640001) and National Research Foundation of Korea (NRF), funded by the Korean government (MSIT) (2021R1A2B5B02001915).
Author information
Authors and Affiliations
Contributions
Jisup Kim and Kwang Gi Kim contributed to the study conception and design. Material preparation, data collection and analysis were performed by Jong-Chan Yeom, Jae-Hoon Kim and Jisup Kim. The first draft of the manuscript was written by Jong-Chan Yeom and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethical Approval
This study received approval from the Institutional Review Board (IRB) of Gachon University Gil Medical Center (GBIRB2022-236). Informed consent was waived given the retrospective study design, which presented minimal risk to participants.
Consent to Participate
The study used data devoid of personally identifiable information, and informed consent was waived by the Institutional Review Board (IRB) due to minimal risk posed to participants.
Consent to Publish
The study used data devoid of personally identifiable information, and informed consent was waived by the Institutional Review Board (IRB) due to minimal risk posed to participants.
Competing Interests
The authors declare competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yeom, J., Kim, J., Kim, Y.J. et al. A Comparative Study of Performance Between Federated Learning and Centralized Learning Using Pathological Image of Endometrial Cancer. J Digit Imaging. Inform. med. 37, 1683–1690 (2024). https://doi.org/10.1007/s10278-024-01020-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-024-01020-1