Abstract
The recent availability of genome sequences together with syntenic block information for Brassicaceae offers an opportunity to study microRNA (miRNA) evolution across this family. We employed a synteny-based comparative genomics strategy to unambiguously identify miRNA homologs from the genome sequence of members of Brassicaceae. Such an analysis of miRNA across Brassicaceae allowed us to classify miRNAs as conserved, lineage-, karyotype- and sub-genome-specific. The differential loss of miRNA from sub-genomes in polyploid genomes of Brassica rapa and Brassica oleracea shows that miRNA also follows the rules of gene fractionation as observed in the case of protein-coding genes. The study of mature and miR* region of precursors revealed instances of in-dels and SNPs which reflect the evolutionary history of the genomes. High level of conservation in miR* regions in some cases points to their functional relevance which needs to be further investigated. We further show that sequence and length variability in precursor sequences can affect the free energy and foldback structure of miRNA which may ultimately affect their biogenesis and expression in the biological system.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
MicroRNAs (miRNAs) are a major class of small non-coding RNAs which regulate the expression of a large number of protein-coding genes at transcriptional and post-transcriptional-levels. MiRNAs show sequence complementarity to their respective target sites at seed region and, depending on near-perfect or imperfect complementarity, guide the target transcript for transcriptional cleavage or translational inhibition, respectively (Dugas and Bartel 2004; Axtell et al. 2011; Budak and Akpinar 2015). In plants, miRNA-target module has been repeatedly recruited to perform variety of plant development processes and stress responses (Nazarov et al. 2013). Comparative studies across the plant kingdom has identified several miRNA families which have remained conserved in angiosperms, gymnosperms, ferns, lycopods, and mosses (Zhang et al. 2006). Nevertheless, a large number of species-specific or lineage-specific miRNA families have been identified suggesting that miRNA genes are born and lost at high frequency (Fahlgren et al. 2010; Kantar et al. 2012). These young miRNA genes are weakly expressed, processed imperfectly, and tend to lack targets, and retention of these miRNA in genome is primarily dependent on whether they are able to establish functional relationship with the target gene which are not detrimental to existing regulatory networks but are sources of formation of novel regulatory variations which are advantageous to plant (Cuperus et al. 2011).
Comparative analysis of miRNA across the plant kingdoms can give us an overview of how miRNA genes have evolved in nature for which correct identification of their orthologs and paralogs is a prerequisite. Homology-based approaches which work well for identifying orthologs of protein-coding genes in closely related plant families often fail in the case of miRNA due to their small size and high degree of similarity in the mature region (Li and Mao 2007; Guerra-Assunnção and Enright 2012). Even though a large number of small RNA sequencing projects have been undertaken to characterize novel miRNAs present in an organism (Sunkar and Zhu 2004; Yu et al. 2012; Kurtoglu et al. 2013), such an approach has its own disadvantages. MiRNAs which express in a certain tissue or under specific stress conditions (He et al. 2014) cannot be captured and unambiguously identified via such technique owing to identical or similar mature sequence of members of the miRNA family. It is also difficult to assign them to a particular genomic location using small RNA sequencing and mapping projects.
MiRBase, the largest repository of experimentally determined miRNA, uses different suffixes for miRNA of the same family in a species for designation depending on when they were reported (Kozomara and Griffiths-Jones 2014). Unfortunately, these suffixes often may not reflect true orthology leading to ambiguity in making phylogenetic inferences. Comparative genomics in relation to synteny is widely employed to extrapolate knowledge between related species and to study relationships in an evolutionary context. Synteny analysis was first employed by Nadeu and Taylor (Nadeau and Taylor 1984) and subsequently by Sankoff (Sankoff 2002; Sankoff and Zheng 2012) to study the arrangement and order of genes in closely related organisms and is now widely used between both closely and distantly related organisms. Based on the conserved nature of miRNA, principles of synteny can help resolve ambiguities about orthology and paralogy (Guerra-Assunnção and Enright 2012) and then be used to perform evolutionary analyses. We therefore employed a synteny-based homology search for discovery of miRNA-encoding loci in selected members of Brassicaceae whose genome sequences are available, viz., Arabidopsis thaliana, Arabidopsis lyrata, Capsella rubella, Thellungiella halophila, B. rapa, and B. oleracea. Based on analysis performed by Schranz et al. (2006), 24 genomic blocks (designated as A–X) have been identified which are like building blocks or “lego blocks” of genomes of Brassicaceae. The “blocks” have undergone rearrangements, translocations, inversions, duplications, and deletions and characterize the genomes of present-day Brassicaceae members (Schranz and Mitchell-Olds 2006; Lysak et al. 2007; Schranz et al. 2007; Mandáková and Lysak 2008; Cheng et al. 2013). Using the information of genomic blocks of Brassicaceae, a valuable resource termed “syntenic gene” is available at Brassica database (http://brassicadb.org/brad/searchSyntenytPCK.php) which can be used to find orthologous genes among sequenced species of Brassicaceae. Though this resource is helpful in studying the evolution of protein-coding genes across Brassicaceae, it does not give any information related to miRNA genes. Being an important component of regulatory machinery, study of miRNA genes is rather important and application of syntenic framework for identification of miRNA genes would resolve the orthology and paralogy issues generally encountered in other methods of miRNA identification.
We used two main criteria to choose genomic blocks for study. Firstly, the genomic blocks should contain more number of conserved miRNAs (conservation criteria based on previous studies) so that the comparative study would be comprehensive. Secondly, an attempt was made to analyze at least one complete microRNA family in order to comment on its evolution.
In this respect, two blocks J and R were found to contain a high number of both miRNAs (22 and 24, respectively) and miRNA members from conserved miRNA family (12 and 11, respectively).
These two blocks, J and R, contained two of the three members of the miR164 family, i.e., miR164a and miR164b, respectively; therefore, we also included Q block harboring miR164c to study evolution of this miRNA family.
The miR164 family is one of the most conserved microRNA families, found in almost all land plants. The family comprises three members in A. thaliana, viz., miR164a, miR164b, and miR164c, where they have been shown to have overlapping and redundant roles by mutant and overexpression studies (Baker et al. 2005; Guo et al. 2005; Sieber et al. 2007). miR164, together with its targets (NAC1, CUC1, CUC2, ORE1), forms an important regulatory module that is involved in mediating various plant processes such as lateral root formation, leaf senescence, shoot apical meristem formation, leaf serration, lateral organ development, and ameliorating abiotic and biotic stresses (Laufs et al. 2004; Guo et al. 2005; Nikovics et al. 2006; Jasinski et al. 2010; Koyama et al. 2010; Huang et al. 2012). Even though a large number of studies have analyzed the function of miR164 in A. thaliana, our knowledge of this regulatory molecule in crop plants is limited. Given the important role of miR164 in plant development and adaptation, extension of knowledge regarding function and evolution of miR164 in crop plants of Brassicaceae is an obvious priority. Gaining insights into evolutionary mechanisms through comparative studies would help us understand the history of diversification and evolution of this genetic component which would lay a strong foundation for further functional studies in Brassicaceae. With this perspective in mind, the present study was framed to study and analyze the genomic blocks encompassed by the miRNA members of this family, i.e., J, R, and Q from the sequenced members of Brassicaceae which harbor the members of the miR164 family, viz., miR164a, miR164b, and miR164c, respectively, to understand the dynamics of retention/loss, conservation at p-m and p-m*, and foldback structure.
Methods
Assignment of genomic blocks to miRNA
All the miRNA precursor sequences from Brassicaceae members such as A. thaliana, A. lyrata, C. rubella, T. halophila, B. rapa, and B. oleracea were retrieved from the miRBase registry (www.mirbase.org; version 21; accessed on 28th March 2015) and sorted as per their chromosome number and location. A. thaliana was taken as reference species in this study as it has the highest number of annotated and validated miRNAs. The chromosomal and positional information of genomic blocks in A. thaliana given in Cheng et al. (2013) was used to allocate the miRNA to each genomic blocks.
Identification of miRNA present in J, R, and Q genomic blocks in selected members of Brassicaceae
In the present study, we restricted our identification and analysis of miRNA present in genomic blocks J, R, and Q which contain miR164a, miR164b, and miR164c, respectively. Apart from miRNA information in A. thaliana, we found some additional miRNAs in these blocks which have been reported from A. lyrata but not A. thaliana. According to their chromosomal position, they were appropriately added to the ordered list of miRNA present in these blocks.
Sequences of protein-coding genes present on start and end of J, R, and Q in A. thaliana were used as queries to find the respective homologs in A. lyrata, C. rubella, T. halophila, B. rapa, and B. oleracea genomes using BLASTN with default parameters (e-value 10, max. number of hits 10). The chromosomal location of such orthologous genes were used to define start and end of a genomic block in the respective species. The location of these genomic blocks was also verified from other reports comparing syntenic blocks in Brassicaceae (Schranz et al. 2006; Mandáková and Lysak 2008; Cheng et al. 2013; Parkin et al. 2014).
In order to identify the orthologs of miRNA in J, R, and Q blocks, precursor sequences of miRNA were used as queries to perform BLASTN search on genomes of A. thaliana (Arabidopsis thaliana genome release 9), A. lyrata (Arabidopsis lyrata v1.0), C. rubella (Capsella rubella v1.0), T. halophila (Thellungiella halophila v1.0), B. rapa (B. rapa chromosome v1.5), and B. oleracea (B. oleracea chromosome v1.0) at BRAD database (brassicadb.org) using default parameters [e-value 10, max. number of hits 10]. BLAST results were filtered on the basis of whether they were present on chromosome and coordinates encompassed in a particular genomic block. Further, these BLAST results were analyzed, and, where required, the lengths of BLAST hits were extended in either direction to reach the start and end of query length.
Analysis of mature sequences of miRNA
For analyzing the mature sequence of miRNA, precursor sequences from different species were aligned using Clustal X (version 2.1) (Larkin et al. 2007) program in UGENE (Okonechnikov et al. 2012) and were manually analyzed. The nature and the position of mismatches in mature sequences were recorded vis-à-vis A. thaliana. Any sequence with more than four mismatches as compared to the mature region of miRNA from A. thaliana was removed from analysis. Conservation at both p-m and p-m* was analyzed.
Length polymorphism and secondary structure analysis of precursors
The variation in precursor length across the Brassicaceae was estimated by recording the length of precursor sequences. Minimum free energy of the structures (∆G; Kcal/mol) was calculated using Quikfold program (http://mfold.rna.albany.edu/?q=DINAMelt/Quickfold). The secondary structure of the selected potential miRNA precursor sequences was predicted and generated using mfold (http://mfold.rna.albany.edu/?q=mfold/rna-folding-form) using default parameters (Zuker 2003). The predicted secondary structures were manually compared with those from their orthologs from other species.
Phylogenetic analysis
Phylogenetic and molecular evolutionary analysis was conducted by MEGA version 5 (Tamura et al. 2011). Clustal aligned sequences were subjected to phylogenetic analysis employing maximum likelihood model with Tamura–Nei substitution followed by 1000 bootstrap replicates. Phylogenetic clustering and bootstrapping were performed when at least three homologous sequences were available.
Results
Identification and assignment of miRNA to genomic blocks in A. thaliana
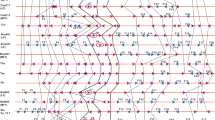
The primary requirement for synteny analysis was the assignment of miRNA to their respective blocks for which we used the positional and chromosomal information of genomic blocks (A–X) of A. thaliana (Cheng et al. 2013; Fig. 1, Supplementary Table 1). Out of 325 miRNA precursors present in A. thaliana, we were able to assign genomic blocks to only 289 miRNA precursors (Fig. 1) and not to the rest due to discontinuity in genomic blocks arising from conflicts in the start and end locations. Even though R block has the highest number of miRNA (24), miRNA gene density was highest in T block (6.72/Mb). The G block has only one miRNA and the lowest miRNA gene density (0.60/Mb) in A. thaliana (Supplementary Table 1). Together, J, Q, and R blocks harbor ca. 20 % of the total miRNA present in A. thaliana. A comparative analysis of organization and distribution of the 24 genomic blocks across the ancestor crucifer type (ACK), translocated proto-calpineae karyotype (tPCK), and modified ACK genome (Supplementary Fig. 1) reveals that the genomic block J is retained as a conserved block with the adjacent I block throughout the three genome types/karyotypes on a gross level. However, Q–R block in ACK (A. lyrata and C. rubella) is related by an inversion event to form R–Q block in the modified ACK (A. thaliana) and is split on separate chromosomes in tPCK (T. halophila) as Q and R blocks. These three blocks thus provide a contrasting evolutionary background.
Diagrammatic representation of position and orientation of 24 genomic blocks and distribution of miRNA mapped on reduced karyotype of Arabidopsis thaliana. Boundaries of the genomic blocks were defined using locus names. Arrows show the orientation of blocks relative to ancestral crucifer karyotype. Black downward pointing arrow indicates similar orientation; black upward pointing arrow indicates that the block is inverted (D, P, V); and red upward pointing arrow indicates that the genomic blocks are in opposite orientation (R, Q, S) but not inverted with respect to ancestral crucifer karyotype (ACK). Thirty-six microRNAs could not be assigned to any particular genomic block due to lack of information (adapted from Schranz et al. 2006, and Cheng et al. 2013)
Identification and analysis of miRNA present in J, R, and Q blocks in genomes of Brassicaceae
We limited our study to members of Brassicaceae with sequenced genomes, viz., A. thaliana, A. lyrata, C. rubella, T. halophila, B. rapa, and B. oleracea, and analyzed the retention status of miRNA present in three genomic blocks J, R, and Q containing miR164a, miR164b, and miR164c, respectively. In A. thaliana, J, R, and Q blocks contain 22, 24, and 8 miRNAs, respectively. In A. lyrata, we found two additional miRNAs (miR3439, miR319d) in J block and one (miR4236) in R block which have not been reported from A. thaliana. These miRNAs were therefore also added to the present study bringing a total number of 24, 25, and 8 miRNAs in J, R, and Q blocks, respectively. Homology-based searches of the miRNA present in J, R, and Q genomic blocks (Supplementary Table 2) led to identification of miRNAs that are conserved in other Brassicaceae genomes (Supplementary Table 3). Out of 57 miRNAs, 26 miRNAs were conserved across all the genomes (Fig. 2). Eighteen miRNAs were only present in A. thaliana, whereas two miRNAs (miR3439, miR319d) were unique to A. lyrata. Instances of lineage-specific gain or loss of miRNAs such as miR417 (J), miR822 (R), miR834 (R), and miR3434 (R) are indicative of several independent events that have occurred in Arabidopsis lineage as these are present only in Arabidopsis species. miR8184 (R block), miR865 (R), and miR4236 (R) were present only in either of the Arabidopsis species and C. rubella, implying that these miRNAs are either “young/recent” in the Arabidopsis-Capsella lineage and then specifically lost from either of Arabidopsis species, or that these could be ancient in nature and lost from one of the Arabidopsis species and other members of Brassicaceae (Fig. 2).
Genome structure of A. thaliana, A. lyrata, and C. rubella represent the ACK, whereas T. halophila, B. rapa, and B. oleracea represent tPCK (Dassanayake et al. 2011; Cheng et al. 2013; Parkin et al. 2014). Our analysis revealed that miRNAs are variably retained in the ACK and tPCK karyotypes. MiRNAs such as miR398c, miR865, miR4236, miR5657, miR8170, and miR8184 were detected in C. rubella and in at least one of the Arabidopsis genome. These miRNAs were not identified in T. halophila and from the two Brassica genomes and can therefore be either considered as ACK-specific miRNAs or lost specifically from the tPCK genomes. In R block, a sub-region between miR398c and miR3434 was found to contain miRNA present only in Arabidopsis and/or C. rubella and hence may be considered as an ACK-specific sub-block (marked in Fig. 2). The retention status of miRNA present in the three genomic blocks is given in Table 1.
Evidence of triplicated nature was clearly visible upon analysis of Brassica genomes. Out of the total 57 miRNAs analyzed, 6 miRNAs were present as three copies, 12 miRNAs were duplicated, and 8 miRNAs were present as single copy in B. rapa. In B. oleracea, 7 miRNAs (miR390a, miR319c, miR160a, miR169b, miR2111b, miR172b, miR156e) are triplicated, 8 miRNAs are duplicated, and 11 miRNAs are present as single copy (Fig. 2). Few miRNAs such as miR169b, miR172b, miR319c, miR390a, and miR2111b were present in all the three sub-genomes of both Brassica species, whereas some miRNAs were found to be preferentially retained in LF sub-genome such as miR403, miR408, miR159c (all in J block), and miR166c (R block) or MF2 sub-genome (miR164b; R block). Seven miRNAs were absent from MF2 sub-genomes of both B. rapa and B. oleracea (miR156j, miR166a, miR164a, miR393a, miR156f, miR398b and miR159c), and the tandemly organized miRNA family miR399def was completely missing from LF sub-genomes. Cumulative preference of retention of miRNA in sub-genome was found to be LF > MF1 > MF2 (Table 1, Fig. 3).
The genomic blocks under study also contain four tandemly arranged miRNA clusters (miR166c-d, miR398b-c, miR399d-e-f, and miR5998a-b), out of which miR399d-e-f cluster was found to be conserved in all the Brassicaceae members. miR166c-d cluster is present in A. thaliana, A. lyrata, C. rubella, T. halophila, and B. oleracea but was partially retained with a single member in B. rapa. miR398b-c cluster was found to be conserved in A. thaliana, A. lyrata, and C. rubella but has only a single member, i.e., miR398b in T. halophila, B. rapa, and B. oleracea. The reorganization of the miR398b-c tandemly arranged family thus can be considered as ACK-specific. miR5998a-b cluster was only found in A. thaliana, suggesting that it is formed by tandem duplication of a young miRNA. We found evidences of two recent events of duplication specific to B. oleracea where miR156d and miR156f have undergone local tandem duplication to form tandem miRNA clusters. In all the analyzed Brassicaceae members, miR156d was detected as a single gene in Q block except in B. oleracea where two copies of miR156d organized in tandem was observed. Similarly, we detected three tandemly arranged copies of miR156f in R block of B. oleracea (Fig. 2).
Members of miR164, namely A, B, and C, are present on J, R, and Q blocks, respectively. Similarly, members of miR166a (J block), miR166c and miR166d (R block), miR156j (J block), miR156d and miR156e (R block), and miR156f (Q block) can be detected across the three blocks. Evidence of local duplication such as miR319c and miR319d (A. lyrata, J block), miR399d, miR399e, and miR399f (J block), miR156d-miR156d (MF1, B. oleracea, R block), and miR156f-miR156f-miR156f (B. oleracea, LF, Q block) are also evident. The synteny (although disrupted) across J, Q, and R blocks indicates that members of the miRNA gene families evolved as a result of either whole genome or segmental duplication in an ancient ancestor. Further, expansion in a genome- or lineage-specific manner occurred as a result of local duplication as exemplified by miR399 d-e-f (across entire Brassicaceae) or miR156d and miR156f (specifically in B. oleracea).
Conservation in mature and miR* region of miRNA across Brassicaceae
The biogenesis of miRNA is a multistep process and involves generation of primary miRNA, precursor miRNA, and finally the mature 20–24 bp miRNA duplex. One strand of the duplex, termed miRNA or guide strand, is incorporated into the RISC complex and brings about post-transcriptional gene silencing (PTGS) by pairing with the target mRNA in a highly sequence-specific manner. Owing to this requirement, both the miRNA and the target binding site in the mRNA are under a higher degree of selection pressure and thus are highly conserved. It was believed until recently that the other strand of the miRNA duplex, termed miR* or passenger strand, is not involved in gene silencing and was degraded. Because of relaxed selection pressure, miR* regions are generally less conserved than mature regions (Guo and Lu 2010a). Studies have however shown that, in certain cases, miR*/passenger strand also has regulatory activity as that of mature miRNA in animal systems (Okamura et al. 2008; Guo and Lu 2010b; Kuchenbauer et al. 2011).
Out of the total 57 miRNAs studied, mature products of 13 miRNA were reported to originate from 5p arm (22.8 %); in 20 miRNAs, it was reported to originate from 3p arm (35.08 %) and in 24 cases the mature product was derived from both 5p and 3p arms (42.1 %; mirBase 21). In cases where miR has been reported to originate from both 5p and 3p arms, the miR species having more number of reads was considered as putative mature miRNA (p-m) and the miR species with lesser number of reads was considered miR*(p-m*). An exception to this is miR2111b, where even though 5p has less number of reads than 3p, it is considered to be mature miRNA (miRBase version 21).
Analysis of mature sequence of miRNA across the Brassicaceae was performed to understand pattern of conservation/divergence in mature sequence. Several miRNA members (miR156e, miR159c, miR160a, miR162b, miR166a, miR166c, miR166d, miR319c, miR393a, miR403, miR408, miR834) did not show any variation in mature sequence. In several instances, mature regions of miRNA showed SNPs and in-dels at specific positions (Supplementary Table 4a–c). For example, both the homologs of miR164a from B. oleracea showed identical nucleotide substitutions at the 6th, 9th, 10th, and 12th positions in the mature region when compared to the rest as an instance of species-specific change (Fig. 4). Similarly, homolog of miR860 in T. halophila and A. thaliana showed substitutions which were unique to these species. Apart from substitutions, SNPs in the form of in-dels were also observed. For example, homolog of miR8170 from C. rubella showed a deletion at the 11th position; a two-nucleotide deletion was observed in a recent tandemly duplicated copy of miR156f (BolmiR156f-2) in B. oleracea; and miR398b from A. lyrata harbors an insertion of two nucleotides in its mature region. Some SNPs in mature sequence were specific to sub-genomes of Brassica; for example, the G/C substitution at the 7th position in miR860 homolog of B. rapa and B. oleracea was specific to LF sub-genome; and SNPs at the 20th position in homologs of miR399e were specific to MF1 sub-genome (Fig. 4). These substitutions/in-dels reflect an evolutionary pattern in being either species-specific (miR860, miR398b, and miR164a, miR156f) or Brassica sub-genome-specific (miR860, miR399e) (Fig. 4, Supplementary Table 4a–c). Some miRNAs such as miR3434 and miR417 were only present in either one or two species and showed high divergence (three to four nucleotides) in their mature region (Supplementary Table 4a–c) suggesting that these are young miRNAs and are rapidly evolving.
Analysis was also done to understand levels of sequence conservation and divergence in the putative miR*(p-m*) region where the mature product has been reported to originate from both 5p and 3p arms. Out of 24 such miRNAs where mature product can arise from both 5p and 3p arms, we could not analyze miR1886 (A. thaliana), miR3439, and miR319d (both A. lyrata) as these miRNA are species-specific. Among the studied miRNA, miR* species in miR408 and miR162b remained invariant; miR390a, miR164b, miR164c, miR172b, and miR162a showed high level of conservation with a single mismatch in their miR* region, whereas in the rest of the 14 miRNAs, the miR* sequence showed low level of conservation (two or more SNPs; Supplementary Table 4a–c). Of the total 21 miR* sequences thus analyzed, 7 miR* representing 33.3 % of the sample can be classified as highly conserved as they had between zero to one mismatches, whereas 14 miR* (equivalent to 66.6 %) showed more than two mismatches and can thus be categorized as divergent. As observed in the case of mature sequences (p-m), SNPs in (p-m*) sequences also reflected an evolutionary pattern (Fig. 5, Supplementary Table 4a–c). Homologs of miR2111b from B. rapa and B. oleracea revealed instances of LF and MF2 sub-genome-specific substitutions at the 11th and 14th positions, respectively. Similarly, miR156d homolog from Brassica harbors SNPs at the 6th and 11th positions which are specific to LF sub-genome, whereas SNP at the 12th position is specific to MF1 sub-genome. Some nucleotide substitutions such as SNP at the 16th position in miR164b and at the 4th position in miR390a were limited to T. halophila, B. rapa, and B. oleracea and thus can be considered as tPCK-specific substitutions. The p-m* region of miR403 and miR822 showed higher conservation as compared to (p-m) implying that the p-m* in these two miRNAs is under high selection pressure (Fig. 5, Supplementary Table 4 a–c).
Length polymorphism and secondary structure analysis
MiRNA is characterized by a low minimum folding energy (MFE), imparted by pairing of bases in its secondary structure which is subject to change by the variability in nucleotide composition or events of in-dels in precursor sequence. In the present study, we found variability in the length of precursor sequences within homologs of Brassicaceae members ranging from 0–37 nucleotides. Some miRNAs such as miR417, miR160a, miR8121, miR156j, miR164a, miR834, miR398b, miR865,miR3434, miR4236, and miR860 showed variation in length that ranged from zero to five nucleotides, whereas in certain other cases (miR159c, mir164b, miR166c, miR166d, miR162b, miR169b) the polymorphism in length was high, ranging from 25 to 36 nucleotides (Supplementary Table 5a–c). With a view to understand a correlation between length polymorphism secondary structure and MFE, we used mfold program (Zuker 2003) to predict the secondary structures and derive the minimum free energy of six homologous miRNAs, namely, miR159c, miR164b, miR166c, miR166d, miR162b, and miR169b, which showed high variation in their length (encircled in Supplementary Table 5a–c). Indeed, in all the cases, length variation led to changes in structure and hence minimum free energy. For example, miR159c homolog from C. rubella (238 nt, dG = −90.1 kcal/mol) is 35 nucleotides longer than its T. halophila (203 nt, dG = −80.1 kcal/mol) homolog primarily due to AU repeats (marked by arrow in Fig. 6) which causes a small hairpin loop-like structure and results in increase in negative free energy. Similarly, miR162b homologs from A. lyrata and C. rubella, miR164b homologs from B. rapa and C. rubella, miR166d homolog from T. halophila and B. oleracea, miR166c homolog from C. rubella, B. rapa, and B. oleracea, and miR169b from A. lyrata and T. halophila showed variation in length leading to formation of extra loops in secondary structure (marked with arrows in Fig. 6) which led to either increase or decrease in free energy of the precursor (Supplementary Table 5a–c).
Apart from the length variation, we also found variability in MFE in homologs of miRNA across Brassicaceae. Precursor sequences of miRNA such as miR156j, miR834, miR5998a, miR5998b, miR156e, miR398c, and miR5657 showed small variation in MFE (0 to 5 kcal/mol), whereas in certain cases such as miR822, miR8184, miR164a, and miR3434, the variability in MFE ranged from −20 to −50 kcal/mol. Our analysis of structures with high variability in MFE (miR822, miR8184, miR164a, miR3434) revealed that a correlation does exist between MFE and foldback structures. miRNA homologs with large difference in MFE also had a variation in their foldback structures (except in case of miR822). The variability in structure was primarily due to unpaired bases which results in the formation of bulges or interior loops and increase in negative free energy of precursor (marked by arrows in Fig. 7, Supplementary Table 5a–c). For example, homologs of miR164a from B. oleracea and A. thaliana show different MFEs of −65.9 and −46.8 kcal/mol, respectively, which is a result of two interior loops and unpaired bases (marked by an arrow in Fig. 7). An exception to the observation was revealed when foldback structures were analyzed for miR822 homologs from A. thaliana and A. lyrata where, despite the large variation in MFE (−31.2 kcal/mol), the secondary structures have not changed considerably (Fig. 7).
Phylogenetic analysis
To gain insights into the evolutionary history and relationship of miRNA precursor sequences, phylogenetic analysis was performed using the maximum likelihood method (Supplementary Figs. 2-3). Shorter length of 80–150 bp and high evolutionary selection pressure does not allow enough substitutions and gaps in sequence for proper resolution of phylogenetic relationships, thus leading to low bootstrap support on branches. Analysis of the phylograms reveals that, in majority of the cases, the precursor sequences cluster according to the species tree with sequences derived from A. thaliana, A. lyrata, and C. rubella grouped together; precursor sequences from the two Brassica species were also grouped according to their genome fraction, i.e., LF, MF1, and MF2 (e.g., blue box; Supplementary Figure 2A B, C, D, E, F, G etc.). Evidence of local duplication leading to expansion of the miRNA gene family can be observed in phylogram of miR156d and miR156f (red box, Supplementary Fig. 2A–B).The sub-genome-specific grouping of miRNAs in Brassica reflects that the triplication event occurred prior to speciation and divergence. We also performed phylogenetic analysis of families of miRNA members present in J, R, and Q blocks, i.e., miR164a-b-c, miR156d-e-f-j, miR162a-b, miR399d-e-f, and miR319c-d. The phylograms clustered the orthologous members, thus confirming their syntenic relationship. An interesting observation is the grouping of miR164a from LF and MF1 sub-genomes from B. rapa and B. oleracea as separate clusters (supported by high bootstrap value), implying this expansion to be a post-speciation event instead of being an outcome of a triplication event that happened at the node of the Brassica lineage before the split of B. rapa and B. oleracea. Similarly, grouping of tandemly duplicated members such as miR156d and miR156f in B. oleracea and miR319c-d clustered with each other indicates that these have arisen as a result of recent genome-specific tandem (green box; Supplementary figure 3A, C, D).
Discussion
Contrasted retention of young versus conserved miRNA
Small RNA sequencing in plant genomes has revealed the presence of a large number of non-conserved miRNAs suggesting that miRNAs are born and lost at high frequency (Fahlgren et al. 2010; Kantar et al. 2012). “Young” miRNAs are known to be lowly expressed and lack in targets and the key factor which decides that their retention in genome is whether such young miRNAs are able to make a functional relationship with a target which is advantageous to plant (Fahlgren et al. 2010). Our homology-based search for homologs of miRNA belonging to J, R, and Q in sequenced genomes of Brassicaceae identified 13 such miRNAs which were restricted to a particular species and 5 miRNA which were lineage-specific, i.e., may represent recently evolved miRNA. In contrast, there exist miRNA families which are deeply conserved across the land plants, and the reason for their extreme conservation is their interaction with their targets and their role in regulating critical developmental processes. Many miRNA members detected in the present study such as miR156, miR159, miR164, and miR169 have been reported to be conserved in the earlier studies (Zhang et al. 2006; Jones-Rhoades 2012). A study published recently to analyze evolution of miRNA in cotton reports that miR156, miR162, miR164, miR172, and miR319 are conserved across Gossypium raimondii, G. arboreum, and G. hirsutum which are present in the J, Q, and R blocks in Brassicaceae, and their conservation status is in accordance with our findings (Xie and Zhang 2015). Apart from computational evidence of conservation of miRNAs, several reports exist that validate functional conservation of miR across plant species (Jasinski et al. 2010), miR165 (Sakaguchi and Watanabe 2012), and miR156 and miR172 (Wang et al. 2011).
Retention of miRNA is dependent on its ancestral karyotype
Several comparative genomic studies have been undertaken to unravel the relatedness of Brassicaceae species with each other (Song et al. 1988; Yogeeswaran et al. 2005). In 2006, Schranz et al. combined decades of knowledge on Brassicaceae comparative genomics and demonstrated that the species in Brassicaceae are made up of 24 genomic blocks (A–X) which have undergone rearrangements to give rise to present-day karyotypes of various Brassicaceae species (Schranz et al. 2006). The analysis of information derived from comparative linkage maps and comparative chromosomal painting techniques showed that A. lyrata and C. rubella represent ACK; A. thaliana represent rearranged/modified ACK; and some of lineage II tribes in Brassicaceae such as Calepineae, Conringieae, and Noccaeeae represent PCK whereas karyotypes of Eutremeae, Isatideae, and Sisymbrieae show an additional translocation in PCK and hence was termed as tPCK (translocated-PCK) (Schranz et al. 2006, 2007; Lysak et al. 2007; Mandáková and Lysak 2008; Cheng et al. 2013). Whole genome sequencing of Thellungiella parvula (a species belonging to Eutremeae) has confirmed its tPCK structure (Dassanayake et al. 2011). Similarly, analysis of genome sequence of B. rapa and B. oleracea also revealed that they represent a triplicated tPCK karyotype with three copies of each genomic block (except G block which is present in two copies) (Cheng et al. 2013; Parkin et al. 2014). In our study, we found that retention of miRNA was dependent on its ancestral karyotype. Some miRNAs were only present in genomes which represent ancestral karyotype ACK (A. thaliana, A. lyrata, and C. rubella) and absent in genomes which represent tPCK genomes (T. halophila, B. rapa, and B. oleracea). Taylor et al. (2014) have combined the miRNA data from miRBase and analyzed in the context of plant kingdom phylogeny for similar analysis (Taylor et al. 2014).
Shen et al. (2015), by combining miRNA and genome sequence data from B. napus (AC), B. rapa (A), and B. oleracea (C), show that the miRNA component of B. napus (AC) can be compartmentalized into the two donor genomes, namely, A and C genomes. Based on such analysis, it was also proposed that miRNA families such as miR156, miR166, and miR171 have undergone B. napus-specific expansion via local segmental duplication; similarly, miRNA families such as miR172, miR395, miR159, and several have undergone gene loss and consequently are smaller (Shen et al. 2015).
A consequence of polyplodization in plants is the opportunity to evolve new genetic networks. Analysis of miRNA and targets from A, D, and AD genomes of cotton (G. arboreum, G. raimondii, and G. hirsutum respectively) shows that miRNA derived from A and D genomes can acquire targets from D and A genomes, respectively (Xie and Zhang 2015). Studies have also provided clear evidence of differential expression pattern of miR and miR* derived from the various polyploid genomes such as in comparative analysis of cotton allopolyploid (G. hirsutum (AD) with G. arboreum (A) and G. raimondii (D); and B. napus amphidiploid in comparison with (AC), B. rapa (A), and B. oleracea (C)) (Shen et al. 2015; Xie and Zhang 2015).
Comparison of precursor sequences across homologs from the J, Q, and R blocks revealed clustering of orthologous sequences albeit with moderate to low bootstrap support in several cases. This may be attributed to low informative polymorphisms across the precursor because of high positive selection pressure. Low bootstrap support in phylograms is indicative of lack of enough character states to resolve the phylogenetic relationships, a problem generally encountered in small sequences and regions which are under high selection pressure as has been observed when analysis of miR165/166 was performed (Barik et al. 2014). Guerra-Assunção and Enright have employed miRNA phylogeny and synteny to understand recent expansion and contraction of miRNA gene families in eight species of animals and also performed comparative phylogeny of targets to investigate functional conservation (Guerra-Assunnção and Enright 2012). In the present study, clustering of precursor sequences based on the sub-genomes is in accordance with the earlier two-step theory to explain genome triplication event that occurred in Brassica (Cheng et al. 2012).
miRNA, like protein-coding genes undergo gene fractionation
In the present study, we analyzed genomes B. rapa and B. oleracea whose genomes are triplicated as compared to A. thaliana and categorized as mesopolyploids (Lysak et al. 2005; Mandáková et al. 2010). Polyploidization is known to be followed by fractionation events where one of the copy of these sub-genomes retains more genes whereas the other sub-genome loses more number of genes as has been seen in the case of maize, wheat, and cotton, to name a few (Chaudhary et al. 2009; Schnable et al. 2011; Zhao et al. 2011; Eckardt 2014). In diploid Brassicas, i.e., B. rapa and B. oleracea), the triplication event led to creation of three such sub-genomes where the gene retention is in the order of LF > MF1 > MF2. One intriguing question is whether miRNA gene-like protein-coding genes also underwent gene fractionation. In our study, it was found that LF sub-genome in both the Brassica species retained at least equal, if not more, percentage of miRNA as compared to MF1 and MF2 counterparts. We observed that, in most of the cases, miRNA members retained by specific sub-genome of B. rapa were also retained by respective sub-genome of B. oleracea which is in accordance with the earlier findings that majority of gene loss observed in two Brassica has occurred prior to their species divergence (Parkin et al. 2014).
Recurring occurrences of tandem duplication of miRNAs in plants
It is now well known that like protein-coding gene families, miRNA families have also expanded via segmental and tandem duplication events (Guerra-Assunnção and Enright 2012; Xiao et al. 2013). In A. thaliana, 18 out of 22 miRNA gene families have been reported to have arisen via tandem duplication (Maher et al. 2006). In the present study, we identified four such tandemly arranged miRNA clusters—miR166cd, miR398bc, miR399def, and miR5998ab which were found to be variably retained in different Brassicaceae species; in addition, two recent events of tandem duplication of miR156d and miR156f in B. oleracea genome were detected. Indeed, such duplications act as a raw material for functional diversification of genes ultimately leading to change in morphology and adaptability of organisms (Cuperus et al. 2011).
Instances of SNPs and in-dels are common in mature region
An important aspect influencing the evolution of miRNA sequences is their interaction with the target RNA. A miRNA precursor is composed of two regions: a stem region harboring mature miRNA and miRNA* and a variable loop region. Mature region of miRNA and seed region of target mRNA are functional units of the PTGS interaction and are therefore under positive selection pressure and highly conserved. The sole reason for this conservation is requirement of high degree of complementary base pairing between the miRNA-mRNA partners which prevents sequence drift during the course of evolution (Ehrenreich and Purugganan 2008). SNPs in the mature region of an miRNA can either destroy or modulate the efficiency of its interaction with the target, lest it can also create a new target. Most miRNAs have more than one target belonging to the same gene family with similar or identical miRNA binding site, and a single nucleotide change in mature miRNA would necessitate a simultaneous compensatory mutation in all of its target genes (Axtell and Bowman 2008). In the present study, several instances of substitutions and in-dels in the mature region were detected. Many of these changes were either species-specific, lineage-specific, karyotype-specific, or sub-genome-specific, implying that such events have followed an evolutionary path and are not random. It is however essential here to understand that substitution at different positional sites of mature miRNA will not have the same effect on miRNA –target interaction. According to the rules, an efficient miRNA target interaction requires effective base pairing at 2–12 base pair of the mature region and only one base pair mismatch is allowed which should not occur at the 10th and 11th base pairs (cleavage site); at the 3′ end (13–21 base pair), the stringency of base pairing is low allowing up to four base pair mismatches, but not two in a row (Ossowski et al. 2008). In the present study as well, we found that most of the mismatches in the mature region lie in the 3′ end of miRNA satisfying the basic rules of miRNA target interaction. A complete understanding would necessitate studying the cognate target sequences from the respective species.
Conservation of miR* region points to their functional importance
One of the essential steps in miRNA biogenesis is processing of precursor miRNA to yield mature miRNA and miRNA*, after which the mature miRNA is recruited into the RISC complex and guides the RISC complex for target degradation whereas miRNA* decays (Cuperus et al. 2011). Even though degradation has been considered as only fate of miRNA* species, this theory has repeatedly faced challenge from small RNA sequencing data where, despite the aggressive and stringent filtering, many miRNA* sequences have been detected above the signal threshold (Cloonan et al. 2011; Llorens et al. 2013; Jagadeeswaran et al. 2010). In support of this proposition, studies have shown that miRNA* species are less abundant than their mature miRNA partners but in physiologically relevant levels can associate with Argonaute proteins and can have inhibitory activity (Okamura et al. 2008). Further, these miRNA* species have been reported to have different targets than their miRNA (mature) counterparts in Drosophila (Marco et al. 2010), A. thaliana, and rice (Manavella et al. 2013; Shao et al. 2013). Expression profiling has shown that both miRNA and miRNA* can co-accumulate in some tissues whereas in others, only one of these two miRNA species accumulate and may perform the necessary physiological function (Ro et al. 2007). Due to growing evidences of the functional role of miRNA* in the biological system, miRBase, the largest repository of miRNA, has started using 5p and 3p for annotation instead of miR and miR*, signifying that the dominant form of mature miRNA can be derived from both 5′ and 3′ arms. In light of this understanding, it is reasonable to believe that in case miRNA* is also performing a regulatory role, it should also follow the rules of miRNA target interaction and therefore should show sequence conservation across the various species. To test this hypothesis, the precursor miRNAs where mature miRNAs from both the arms have been reported in miRBase were also studied. The extreme conservation in miRNA* species across the family in few cases signals their possible regulatory role in the plant system. A similar study in animal miRNAs also showed that, in certain cases, miRNA* sequences are conserved as their mature counterparts which may suggest their functional role in system (Guo and Lu 2010a). A recent study in cotton showed that miR172*, miR390*, miR164*, miR171*, miR2949*, and miR3954* are present in abundance in small RNA fraction and their expressions vary in different tissues (Xie and Zhang 2015). Also, in a study, Kang et al. (2013) found that introduction of an artificial target of miR*(miR-7b*) led to up-regulation of miR-7b* and not miR-7b (mature miRNA), implying that abundance of target transcripts plays a significant role in miRNA arm selection (Kang et al. 2013). In plants, functional activity of miRNA* has not been much investigated, and its study at functional level holds the promise of unraveling another hidden layer of regulation in biological systems.
Extent of secondary structure polymorphism among homologs is dependent on nature and position of SNPs and in-dels in sequences
Unlike the mature region, the loop regions of precursors are variable and are subject to genetic drift. It should be recognized that multiple copies of identical sequences are silenced in genome by RNAi machinery and hence variability in precursors may be required to keep the miRNA machinery active (Tang 2010). The plausible modes of variability in precursor sequences are either by in-dels or by substitutions, both of which ultimately led to change in secondary structure of miRNA. A stable secondary structure is required for an efficient processing of miRNA which indirectly governs the level of expression of miRNA in a system (Mateos et al. 2010). The stability of stemloop is estimated by MFE values which decrease by stacking energy of successive base pairs or increase by destabilizing energy associated with non-complementary bases (Bonnet et al. 2004). To understand the effect of variability or SNPs at a particular position, two types of studies have been performed. The first analysis dealt with exploring the natural variation observed in precursor sequences in plant families and understanding their effects on MFE and secondary structure of precursors (Kusumanjali et al. 2011; Kumari et al. 2012; Shivaraj et al. 2014). The second type of study dealt with generating mutants at various positional locations of precursors and thereafter finding the effect of these mutations by studying their processing in in vivo system (Mateos et al. 2010; Werner and Wollmann 2010). The present study falls in type I category study where we found that increase in precursor length showed a positive correlation with MFE values, and large variation in MFE values resulted due to unpaired bases which formed internal loops, bulges, and unpaired bases in secondary structure of precursors ultimately decreasing MFE of precursor sequences. Collectively, it can be understood that MFE values increase with increasing sequence length if it increases the number of paired bases in precursor (Trotta 2014), and variability by means of substitution can also increase or decrease the free energy depending on whether they create a favorable base pairing or destroy the existing one by formation of internal loops and bulges (Long et al. 2007; Xiong et al. 2013).
Summary/Conclusion
To the best of our knowledge, this is the first report on documentation of lineage-specific such as ACK-specific or tPCK-specific miRNA changes. Although miR164 is one of the most ancient and conserved miRNA families present in the plant kingdom, little information is available on the evolutionary trajectory of the family in the context of genome evolution and functional diversification The present study sampled ca. 20 % of the total miRNAs present in A. thaliana, and extension of such a strategy to other genomic blocks will give us an even larger and more comprehensive view of how miRNAs evolve in nature. Also, our criteria of the absence of miRNA homologs are based on its search in the syntenic/homologous position, and it does not evade the possibility of presence of miRNA at any non-syntenic position in genome as a result of a transposition event.
Abbreviations
- A. thaliana :
-
Arabidopsis thaliana
- A. lyrata :
-
Arabidopsis lyrata
- C. rubella :
-
Capsella rubella
- T. halophila :
-
Thellungiella halophila
- B. rapa :
-
Brassica rapa
- B. oleracea :
-
Brassica oleracea
- LF:
-
Least fractionated
- MF1:
-
Moderately fractionated
- MF2:
-
Most fractionated
- ACK:
-
Ancestor crucifer type
- PCK:
-
Proto-calpineae karyotype
- tPCK:
-
Translocated proto-calpineae karyotype
- MiRNA:
-
MicroRNA
- (p-m):
-
putative mature
- (p-m*):
-
putative miR*
- SNP:
-
Single-nucleotide polymorphism
References
Axtell MJ, Bowman JL (2008) Evolution of plant microRNAs and their targets. Trends Plant Sci 13:343–349. doi:10.1016/j.tplants.2008.03.009
Axtell MJ, Westholm JO, Lai EC (2011) Vive la différence: biogenesis and evolution of microRNAs in plants and animals. Genome Biol 12:221
Baker CC, Sieber P, Wellmer F, Meyerowitz EM (2005) The early extra petals1 mutant uncovers a role for MicroRNA miR164c in regulating petal number in Arabidopsis. Curr Biol 15:303–315
Barik S, SarkarDas S, Singh A et al (2014) Phylogenetic analysis reveals conservation and diversification of micro RNA166 genes among diverse plant species. Genomics 103:114–121. doi:10.1016/j.ygeno.2013.11.004
Bonnet E, Wuyts J, Rouzé P, Van De Peer Y (2004) Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics 20:2911–2917. doi:10.1093/bioinformatics/bth374
Budak H, Akpinar BA (2015) Plant miRNAs: biogenesis, organization and origins. Funct Integr Genomics 15:523–531. doi:10.1007/s10142-015-0451-2
Chaudhary B, Flagel L, Stupar RM et al (2009) Reciprocal silencing, transcriptional bias and functional divergence of homeologs in polyploid cotton (gossypium). Genetics 182:503–517. doi:10.1534/genetics.109.102608
Cheng F, Wu J, Fang L et al (2012) Biased gene fractionation and dominant gene expression among the subgenomes of Brassica rapa. PLoS One 7, e36442. doi:10.1371/journal.pone.0036442
Cheng F, Mandáková T, Wu J et al (2013) Deciphering the diploid ancestral genome of the Mesohexaploid Brassica rapa. Plant Cell 25:1541–1554. doi:10.1105/tpc.113.110486
Cloonan N, Wani S, Xu Q et al (2011) MicroRNAs and their isomiRs function cooperatively to target common biological pathways. Genome Biol 12:R126. doi:10.1186/gb-2011-12-12-r126
Cuperus JT, Fahlgren N, Carrington JC (2011) Evolution and functional diversification of MIRNA genes. Plant Cell 23:431–442. doi:10.1105/tpc.110.082784
Dassanayake M, Oh D-H, Haas JS et al (2011) The genome of the extremophile crucifer Thellungiella parvula. Nat Genet 43:913–918. doi:10.1038/ng.889
Dugas DV, Bartel B (2004) MicroRNA regulation of gene expression in plants. Curr Opin Plant Biol 7:512–520. doi:10.1016/j.pbi.2004.07.011
Eckardt NA (2014) Genome dominance and interaction at the gene expression level in allohexaploid wheat. Plant Cell 26:1834. doi:10.1105/tpc.114.127183
Ehrenreich IM, Purugganan MD (2008) Sequence variation of MicroRNAs and their binding sites in Arabidopsis. Plant Physiol 146:1974–1982. doi:10.1104/pp.108.116582
Fahlgren N, Jogdeo S, Kasschau KD et al (2010) MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana. Plant Cell 22:1074–1089. doi:10.1105/tpc.110.073999
Guerra-Assunnção JA, Enright AJ (2012) Large-scale analysis of microRNA evolution. BMC Genomics 13:218
Guo L, Lu Z (2010a) The fate of miRNA* strand through evolutionary analysis: implication for degradation as merely carrier strand or potential regulatory molecule? PLoS One 5, e11387. doi:10.1371/journal.pone.0011387
Guo L, Lu Z (2010) Expression analysis of MicroRNA cluster based on IsomiRs from high throughput DNA sequencing data 195–197
Guo H, Xie Q, Fei J, Chua N (2005) MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for arabidopsis lateral root development. Plant Cell 17:1376–1386. doi:10.1105/tpc.105.030841
He H, Liang G, Li Y et al (2014) Two young MicroRNAs originating from target duplication mediate nitrogen starvation adaptation via regulation of glucosinolate synthesis in Arabidopsis thaliana. Plant Physiol 164:853–865. doi:10.1104/pp.113.228635
Huang T, López-Giráldez F, Townsend JP, Irish VF (2012) RBE controls microRNA164 expression to effect floral organogenesis. Development 139:2161–2169. doi:10.1242/dev.075069
Jagadeeswaran G, Zheng Y, Sumathipala N, et al. (2010) Deep sequencing of small RNA libraries reveals dynamic regulation of conserved and novel microRNAs and microRNA-stars during silkworm development. 1–18
Jasinski S, Vialette-Guiraud ACM, Scutt CP (2010) The evolutionary-developmental analysis of plant microRNAs. Philos Trans R Soc Lond B Biol Sci 365:469–476. doi:10.1098/rstb.2009.0246
Jones-Rhoades MW (2012) Conservation and divergence in plant microRNAs. Plant Mol Biol 80:3–16. doi:10.1007/s11103-011-9829-2
Kang S-M, Choi J-W, Hong S-H, Lee H-J (2013) Up-regulation of microRNA* strands by their target transcripts. Int J Mol Sci 14:13231–13240. doi:10.3390/ijms140713231
Kantar M, Akpınar BA, Valárik M et al (2012) Subgenomic analysis of microRNAs in polyploid wheat. Funct Integr Genomics 12:465–479. doi:10.1007/s10142-012-0285-0
Koyama T, Mitsuda N, Seki M et al (2010) TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell Online 22:3574–3588. doi:10.1105/tpc.110.075598
Kozomara A, Griffiths-Jones S (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42:D68–D73. doi:10.1093/nar/gkt1181
Kuchenbauer F, Mah SM, Heuser M et al (2011) Comprehensive analysis of mammalian miRNA* species and their role in myeloid cells. Blood 118:3350–3358. doi:10.1182/blood-2010-10-312454
Kumari G, Kusumanjali K, Srivastava PS, Das S (2012) Isolation and expression analysis of miR165a and REVOLUTA from Brassica species. Acta Physiol Plant 35:399–410. doi:10.1007/s11738-012-1082-z
Kurtoglu KY, Kantar M, Lucas SJ, Budak H (2013) Unique and conserved MicroRNAs in wheat chromosome 5D revealed by next-generation sequencing. PLoS ONE 8(7), e69801. doi:10.1371/journal.pone.0069801
Kusumanjali K, Kumari G, Srivastava PS, Das S (2011) Sequence conservation and divergence in miR164C1 and its target, CUC1, in Brassica species. Plant Biotechnol Rep 6:149–163. doi:10.1007/s11816-011-0208-x
Larkin MA, Blackshields G, Brown NP et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi:10.1093/bioinformatics/btm404
Laufs P, Peaucelle A, Morin H, Traas J (2004) MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131:4311–4322. doi:10.1242/dev.01320
Li A, Mao L (2007) Evolution of plant microRNA gene families. Cell Res 17:212–218. doi:10.1038/sj.cr.7310113
Llorens F, Hummel M, Pantano L et al (2013) Microarray and deep sequencing cross-platform analysis of the mirRNome and isomiR variation in response to epidermal growth factor. BMC Genomics 14:371. doi:10.1186/1471-2164-14-371
Long D, Lee R, Williams P et al (2007) Potent effect of target structure on microRNA function. Nat Struct Mol Biol 14:287–294. doi:10.1038/nsmb1226
Lysak MA, Koch MA, Pecinka A, Schubert I (2005) Chromosome triplication found across the tribe Brassiceae. Genome Res 15:516–525. doi:10.1101/gr.3531105
Lysak MA, Cheung K, Kitschke M, Bures P (2007) Ancestral chromosomal blocks are triplicated in Brassiceae species with varying chromosome number and genome size. Plant Physiol 145:402–410. doi:10.1104/pp.107.104380
Maher C, Stein L, Ware D (2006) Evolution of Arabidopsis microRNA families through duplication events. Genome Res 16:510–519. doi:10.1101/gr.4680506
Manavella PA, Koenig D, Rubio-Somoza I et al (2013) Tissue-specific silencing of Arabidopsis SU(VAR)3-9 HOMOLOG8 by miR171a. Plant Physiol 161:805–812. doi:10.1104/pp.112.207068
Mandáková T, Lysak MA (2008) Chromosomal phylogeny and karyotype evolution in x = 7 crucifer species (Brassicaceae). Plant Cell 20:2559–2570. doi:10.1105/tpc.108.062166
Mandáková T, Joly S, Krzywinski M et al (2010) Fast diploidization in close mesopolyploid relatives of Arabidopsis. Plant Cell 22:2277–2290. doi:10.1105/tpc.110.074526
Marco A, Hui JHL, Ronshaugen M, Griffiths-Jones S (2010) Functional shifts in insect microRNA evolution. Genome Biol Evol 2:686–696. doi:10.1093/gbe/evq053
Mateos JL, Bologna NG, Chorostecki U, Palatnik JF (2010) Identification of MicroRNA processing determinants by random mutagenesis of Arabidopsis MIR172a precursor. Curr Biol 20:49–54. doi:10.1016/j.cub.2009.10.072
Nadeau JH, Taylor BA (1984) Lengths of chromosomal segments conserved since divergence of man and mouse. Proc Natl Acad Sci U S A 81:814–818. doi:10.1073/pnas.81.3.814
Nazarov PV, Reinsbach SE, Muller A et al (2013) Interplay of microRNAs, transcription factors and target genes: linking dynamic expression changes to function. Nucleic Acids Res 41:2817–2831. doi:10.1093/nar/gks1471
Nikovics K, Blein T, Peaucelle A et al (2006) The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18:2929–2945. doi:10.1105/tpc.106.045617
Okamura K, Phillips MD, Tyler DM et al (2008) The regulatory activity of microRNA* species has substantial influence on microRNA and 3’ UTR evolution. Nat Struct Mol Biol 15:354–363. doi:10.1038/nsmb.1409
Okonechnikov K, Golosova O, Fursov M (2012) Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28:1166–1167. doi:10.1093/bioinformatics/bts091
Ossowski S, Schwab R, Weigel D (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53:674–690. doi:10.1111/j.1365-313X.2007.03328.x
Parkin IA, Koh C, Tang H et al (2014) Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol 15:R77. doi:10.1186/gb-2014-15-6-r77
Ro S, Park C, Young D et al (2007) Tissue-dependent paired expression of miRNAs. Nucleic Acids Res 35:5944–5953. doi:10.1093/nar/gkm641
Sakaguchi J, Watanabe Y (2012) miR165/166 and the development of land plants. Dev Growth Differ 54:93–99. doi:10.1111/j.1440-169X.2011.01318.x
Sankoff D (2002) Short inversions and conserved gene cluster. Bioinformatics 18:1305–1308. doi:10.1093/bioinformatics/18.10.1305
Sankoff D, Zheng C (2012) Fractionation, rearrangement and subgenome dominance. Bioinformatics 28:i402–i408. doi:10.1093/bioinformatics/bts392
Schnable JC, Springer NM, Freeling M (2011) Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc Natl Acad Sci U S A 108:4069–4074. doi:10.1073/pnas.1101368108
Schranz ME, Mitchell-Olds T (2006) Independent ancient polyploidy events in the sister families Brassicaceae and Cleomaceae. Plant Cell 18:1152–1165. doi:10.1105/tpc.106.041111
Schranz ME, Lysak MA, Mitchell-Olds T (2006) The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci 11:535–542. doi:10.1016/j.tplants.2006.09.002
Schranz ME, Song B-H, Windsor AJ, Mitchell-Olds T (2007) Comparative genomics in the Brassicaceae: a family-wide perspective. Curr Opin Plant Biol 10:168–175. doi:10.1016/j.pbi.2007.01.014
Shao C, Ma X, Xu X, Meng Y (2013) Identification of the highly accumulated microRNA*s in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa). Gene 515:123–127. doi:10.1016/j.gene.2012.11.015
Shen D, Suhrkamp I, Wang Y et al (2014) Identification and characterization of microRNAs in oilseed rape (Brassica napus) responsive to infection with the pathogenic fungus Verticillium longisporum using Brassica AA (Brassica rapa) and CC (Brassica oleracea) as reference genomes. New Phytol 204:577–594. doi:10.1111/nph.12934
Shen E, Zou J, Hubertus Behrens F et al (2015) Identification, evolution, and expression partitioning of miRNAs in allopolyploid Brassica napus. J Exp Bot 66:erv420. doi:10.1093/jxb/erv420
Shivaraj SM, Dhakate P, Mayee P et al (2014) Natural genetic variation in MIR172 isolated from Brassica species. Biol Plant 58:627–640. doi:10.1007/s10535-014-0441-6
Sieber P, Wellmer F, Gheyselinck J et al (2007) Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development 134:1051–1060. doi:10.1242/dev.02817
Song et al. (1988) Brassica taxonomy based on nuclear restriction fragment length polymorphisms (RFLPs). Theor Appl Genet 593–600
Sunkar R, Zhu J (2004) Novel and stress-regulated MicroRNAs and other small RNAs from Arabidopsis. Plant Cell 16:2001–2019. doi:10.1105/tpc.104.022830.The
Tamura K, Peterson D, Peterson N et al (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi:10.1093/molbev/msr121
Tang G (2010) An insight into their gene structures and evolution. Semin Cell Dev Biol 21:782–789. doi:10.1016/j.semcdb.2010.07.009
Taylor RS, Tarver JE, Hiscock SJ, Donoghue PCJ (2014) Evolutionary history of plant microRNAs. Trends Plant Sci 19:175–182. doi:10.1016/j.tplants.2013.11.008
Trotta E (2014) On the normalization of the minimum free energy of RNAs by sequence length. PLoS One 9, e113380. doi:10.1371/journal.pone.0113380
Wang JW, Park MY, Wang LJ et al (2011) MiRNA control of vegetative phase change in trees. PLoS Genet 7:21–25. doi:10.1371/journal.pgen.1002012
Werner S, Wollmann H (2010) Report Structure Determinants for Accurate Processing of miR172a in Arabidopsis thaliana. Curr Biol 1–7. doi: 10.1016/j.cub.2009.10.073
Xiao Y, Xia W, Yang Y et al (2013) Characterization and evolution of conserved MicroRNA through duplication events in date palm (Phoenix dactylifera). PLoS One 8, e71435. doi:10.1371/journal.pone.0071435
Xie F, Zhang B (2015) microRNA evolution and expression analysis in polyploidized cotton genome. Plant Biotechnol J. doi:10.1111/pbi.12295
Xiong X, Kang X, Zheng Y et al (2013) Identification of loop nucleotide polymorphisms affecting microRNA processing and function. Mol Cells 36:518–526. doi:10.1007/s10059-013-0171-1
Yogeeswaran K, Frary A, York TL et al (2005) Comparative genome analyses of Arabidopsis spp.: inferring chromosomal rearrangement events in the evolutionary history of A. Thaliana. Genome Res 15:505–515. doi:10.1101/gr.3436305
Yu X, Wang H, Lu Y et al (2012) Identification of conserved and novel microRNAs that are responsive to heat stress in Brassica rapa. J Exp Bot 63:1025–1038. doi:10.1093/jxb/err337
Zhang B, Pan X, Cannon CH et al (2006) Conservation and divergence of plant microRNA genes. Plant J 46:243–259. doi:10.1111/j.1365-313X.2006.02697.x
Zhao N, Zhu B, Li M et al (2011) Extensive and heritable epigenetic remodeling and genetic stability accompany allohexaploidization of wheat. Genetics 188:499–510. doi:10.1534/genetics.111.127688
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi:10.1093/nar/gkg595
Acknowledgments
This work was supported by grants (BT/PR628/AGR/36/674/2011 and BT/PR14532/AGR/36/673/2010) received from the Department of Biotechnology, Govt. of India, to SD. SD would also like to acknowledge financial grants received under Research and Development Grants from the University of Delhi. Financial assistance as JRF/SRF to Aditi Jain from University Grants Commission, Govt. of India, is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jain, A., Das, S. Synteny and comparative analysis of miRNA retention, conservation, and structure across Brassicaceae reveals lineage- and sub-genome-specific changes. Funct Integr Genomics 16, 253–268 (2016). https://doi.org/10.1007/s10142-016-0484-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-016-0484-1