Abstract
Control of proteolysis is important for plant growth, development, responses to stress, and defence against insects and pathogens. Members of the serpin protein family are likely to play a critical role in this control through irreversible inhibition of endogenous and exogenous target proteinases. Serpins have been found in diverse species of the plant kingdom and represent a distinct clade among serpins in multicellular organisms. Serpins are also found in green algae, but the evolutionary relationship between these serpins and those of plants remains unknown. Plant serpins are potent inhibitors of mammalian serine proteinases of the chymotrypsin family in vitro but, intriguingly, plants and green algae lack endogenous members of this proteinase family, the most common targets for animal serpins. An Arabidopsis serpin with a conserved reactive centre is now known to be capable of inhibiting an endogenous cysteine proteinase. Here, knowledge of plant serpins in terms of sequence diversity, inhibitory specificity, gene expression and function is reviewed. This was advanced through a phylogenetic analysis of amino acid sequences of expressed plant serpins, delineation of plant serpin gene structures and prediction of inhibitory specificities based on identification of reactive centres. The review is intended to encourage elucidation of plant serpin functions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Proteinase inhibitors have arisen on many different occasions throughout the history of life on earth, mostly during the evolution of eukaryotes (Rawlings et al. 2004). Specific proteins that were named as members of the serpin (serine proteinase inhibitor) family (Carrell and Travis 1985) were recognized as homologous proteins more than a quarter of a century ago (Hunt and Dayhoff 1980; Leicht et al. 1982). Among the 68 families of peptidase inhibitors (MEROPS database v7.6Footnote 1), the serpin family (denoted I4) has the largest number of protein sequences in the SWISS-PROT and TrEMBL databases. It is one of only two peptidase inhibitor families found in all domains of life (Archaea, Bacteria and Eukarya), and the only one found additionally in viruses (Rawlings et al. 2004).
Great progress has been made in our understanding of the structure, function and biology of serpins, as explained in several reviews (Gettins 2002; Law et al. 2006; Silverman et al. 2001) and in a recently published monograph (Silverman and Lomas 2007). The structure of serpins is highly conserved, but individual molecules can assume distinct conformations associated with the mechanism by which these proteins function and the regulation of their activity (Pearce et al. 2007; Whisstock et al. 1998). The mechanism of inhibition of target proteinases by serpins involves a remarkable molecular transition unique to the serpin family (Carrell and Evan 1992; Carrell and Owen 1985; Gettins 2007; Law et al. 2006). The majority of serpins are irreversible inhibitors of serine proteinases of the chymotrypsin family; examples of these enzymes from animals include trypsin, thrombin, elastase and blood coagulation factors such as factors Xa and VIIa. A minority of serpins have evolved to inhibit the activity of serine proteinases other than those of the chymotrypsin family (such as subtilisins) and a few serpins inhibit cysteine proteinases (which have a similar catalytic mechanism to the serine proteinases). Some serpins, such as ovalbumin and pigment epithelium-derived factor (PEDF; SerpinF1), have lost their ability to inhibit proteinases during evolution and have taken on other functions (Becerra et al. 1995; Gettins 2002; Law et al. 2006; Stein et al. 1989).
Determination of the presence or absence of serpin genes in major groups of organisms can provide insights into serpin evolution and function. Until the turn of the 21st century, no serpins were known from prokaryotes or unicellular eukaryotes and, thus, the protein family was thought to have arisen after the onset of multicellularity (Irving et al. 2000). When serpins were subsequently discovered in unicellular organisms, the evolutionary history of the protein family had to be reconsidered (Atchley et al. 2001; Buck and Atchley 2005; Irving et al. 2002b, 2004; Krem and Di Cera 2003; Roberts et al. 2004). Considerable interest remains concerning the unresolved origin and evolution of the serpin fold (Whisstock and Bottomley 2006) and the evolution of specific clades of serpins (Xu et al. 2006). With an exponentially increasing number of serpin gene sequences available from an expanding diversity of life forms, it now seems likely that with the exception of most groups of fungi, all multicellular eukaryotic organisms contain serpin genes.
In the early development of plant serpin research, studies on the nature of dominant antigens in beer led to the discovery of an ~43-kDa polypeptide of barley origin (Hejgaard 1977, 1982; Hejgaard and Bog-Hansen 1974; Hejgaard and Kaersgaard 1983; Hejgaard and Sorensen 1975), which was named Protein Z (Hejgaard 1976) and later identified as a barley grain serpin (Hejgaard et al. 1985). Subsequently, more than 20 serpins have been isolated or cloned and produced recombinantly from grains of the Triticeae cereals barley, wheat, rye and oats, and their inhibitory properties characterized (Brandt et al. 1990; Dahl et al. 1996a, b; Hejgaard 2001, 2005; Hejgaard and Hauge 2002; Hejgaard et al. 2005; Ostergaard et al. 2000; Rasmussen et al. 1996; Rosenkrands et al. 1994). Our analysis of the rice genome—the only monocot to have been fully sequenced—indicates the presence of 14 serpin genes (on 3 of the 12 chromosomes), but only 8 of these are associated with evidence of expression. Serpins are also known from soybean, cotton, tomato and numerous other eudicots, but only serpins from the phloem of the cucurbits pumpkin (Yoo et al. 2000) and cucumber (la Cour Petersen et al. 2005), one from Arabidopsis (Vercammen et al. 2006) and one from apple seed (Hejgaard et al. 2005) have been characterized. To date, the only eudicot genomes to be fully sequenced are those of Arabidopsis thaliana (thale cress) (AGI 2000)Footnote 2 and Populus trichocarpa (black cottonwood or western balsam poplar) (Tuskan et al. 2006). Arabidopsis appears to contain six genes (on three of the five chromosomes) that encode full-length serpins. A more detailed discussion of serpins in rice and Arabidopsis appears later in the review.
Expressed serpin genes are also known from a moss (a “bryophyte”) and gymnosperms (conifers); thus, serpins appear to be present throughout the plant kingdom. Despite the ubiquity of plant serpin genes and the well-documented inhibitory properties of plant serpins, very little information concerning their physiological functions is available. Functions in defence have been suggested for those serpins found in high concentrations in cereal and apple seeds (Hejgaard 2001; Hejgaard et al. 2005; Ostergaard et al. 2000), but no systematic studies have been performed to test this hypothesis. Recently, the first demonstration of inhibition of an endogenous proteinase by a plant serpin in vitro was published (Vercammen et al. 2006), but this interaction has not yet been confirmed to be physiologically relevant. A discussion of this interaction will appear later in the review.
This review should serve as a complement to a recently published chapter entitled “Plant Serpins” (Hejgaard and Roberts 2007)Footnote 3 within a monograph on serpins, and thus, explores different themes. Accordingly, the history of plant serpin research and its links to beer proteins are not a focus of this review, but detailed discussions of the phylogeny and expression of serpin genes and the reactive centres of plant serpins are given. The review will explore possible plant serpin functions with the aim of encouraging research in this area. For the benefit of readers relatively unfamiliar with serpins, the following section outlines the unique mechanism by which serpins function as proteinase inhibitors. There is also a discussion on the distribution of serpins across domains and their known functions to place the detailed analysis of plant serpins in context.
The serpin mechanism of proteinase inhibition
The structure and inhibitory mechanism of serpins have been extensively studied. Briefly, inhibitory serpins contain eight to nine α-helices (A-H), three β-sheets (A-C), and a reactive centre loop (RCL; also referred to as a reactive site loop, RSL) containing a specific bait sequence for the serpin’s target proteinase(s) (Gettins 2002; Whisstock et al. 1998; Whisstock and Bottomley 2006) (Fig. 1). It is at the peptide bond immediately after the P1 residue of the bait sequence (the P1–P1′ bond) that the proteinase initiates the cleavage reaction, and it is the identity of the P1 residue that largely determines the inhibitory specificity of the serpin (Gettins 2002). Some serpins have overlapping reactive centres, meaning that adjacent residues can act as the functional P1 residue with distinct proteinases (Dahl et al. 1996a; Potempa et al. 1988). Other residues influence specificity, particularly those immediately N-terminal to P1 (P2, P3, P4) (Ke et al. 1997; Plotnick et al. 1997), but residues on the C-terminal side of the reactive centre, such as P4′ and P5′, have also been shown to have substantial bearing on the kinetics of serpin–proteinase interactions in some cases (Ibarra et al. 2004; Sun et al. 2001). In contrast to the extensive mammalian serpin studies, no experiments on site-directed mutants of plant serpins have been performed to determine the effect of changes to reactive centre residues on proteinase inhibition.
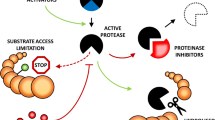
X-ray crystal structures showing steps in the formation of a serpin-–proteinase covalent complex. a A serine proteinase of the chymotrypsin family (above) and a serpin (below) in the native conformation. The enzyme has a reactive site with a high affinity for the reactive centre of the exposed reactive centre loop (RCL; red) of the inhibitor. b An initial (reversible) encounter complex forms between the serpin and proteinase. c The corresponding final irreversible covalent complex forms between the serpin and proteinase. After cleavage at the reactive centre, the RCL (red) has inserted as an additional strand into β-sheet A of the serpin, flinging the covalently bound proteinase from the top to the bottom of the serpin and distorting its structure. The proteinase in (a) and (c) is wild-type bovine trypsin while in (b) it is Ser195→Ala bovine trypsin (used to trap the encounter complex). The serpin in (a) and (b) is human α1-antitrypsin Pittsburg variant, which has P1 Met358→Arg (used to provide a potent inhibitor of trypsin), while in (c) it is normal human α1-antitrypsin. The pdb files used are: (a) 5PTB for the proteinase (Finer-Moore et al. 1992) and 1OO8 for the serpin (Dementiev et al. 2003); (b) 1OPH (Dementiev et al. 2003); (c) 1EZX (Huntington et al. 2000). Images were made using Swiss-PdbViewer (Guex and Peitsch 1997)
After formation of a Michaelis–Menten encounter complex between the proteinase at the “top” of the serpin structure (Peterson et al. 2000; Ye et al. 2001), the reactive centre is cleaved (Wilczynska et al. 1995), and an acyl enzyme intermediate of the proteolytic reaction is formed, linking the two proteins covalently (Lawrence et al. 1995; Matheson et al. 1991; Olson et al. 2001) (Fig. 1). The length of the RCL (from a highly conserved Glu residue, normally at P17 to P1) is then critical in allowing rapid insertion of the RCL into the major β-sheet of the serpin, forming a stability-enhancing antiparallel strand (Zhou et al. 2001). Plant serpins, like their animal counterparts, use a 17-residue RCL length for proteinase inhibition (Dahl et al. 1996a; Hejgaard 2001, 2005; Hejgaard and Hauge 2002; Hejgaard et al. 2005; Ostergaard et al. 2000). The barley serpin HorvuZx (BSZx) was shown to inhibit distinct proteinases (trypsin and chymotrypsin) at overlapping reactive centres, demonstrating that an RCL length of 17 or 16 residues is compatible with inhibitory activity in this case (Dahl et al. 1996a). It is still largely unknown how RCL cleavage triggers conformational changes in the serpin, including RCL insertion into β-sheet A, but substantial insights are being gained from data indicating that the RCL of native serpins flickers between a fully expelled and partially inserted conformation (Tsutsui et al. 2006; Whisstock and Bottomley 2006).
The major conformational change within the serpin structure flings the proteinase, with the Ser of its catalytic triad covalently attached to P1, to the “bottom” of the serpin (Aleshkov et al. 1996; Stratikos and Gettins 1997). This distorts the reactive site of the enzyme (Huntington et al. 2000; Perron et al. 2003), rendering it unable to complete the proteolytic reaction to cleave the bond connecting P1 to the active site Ser (Mellet et al. 1998; O’Malley et al. 1997). The resulting serpin conformation separates the P1 from the P1′ residue by ~70 Å and changes the relative positions of modules within the serpin structure (Fig. 1). The serpin is no longer metastable (or “stressed” = S), but hyperstable (or “relaxed” = R), and cannot function as a proteinase inhibitor again—hence, the serpin is said to have undergone an “S→R transition”. Terms used in the literature for inhibitory serpins include “suicide-substrate inhibitors” and “molecular mouse traps”. The mechanism of inhibitory serpins is reflected in the very high thermal stability of the reactive-centre-cleaved form (T m>100°C) compared to that of the native conformation (~58°C) (Bruch et al. 1988). Thermal melting curves have yet to be determined for any plant serpins but are likely to be similar to those for serpins generally.
The interaction between an inhibitory serpin and a cognate proteinase is normally defined by two values. The first is a constant describing the speed of the association between enzyme and inhibitor, namely the second-order association rate constant, k a, which normally has a value of between 103 and 107 M−1 s−1. The second value is known as the stoichiometry of inhibition, SI, which is the average number of serpin molecules required to irreversibly inhibit one enzyme molecule (Gettins 2002; Schechter and Plotnick 2004). The need to determine an SI derives from the branched nature of the pathway used to describe serpin–proteinase interactions (Patston et al. 1991). For any particular serpin–proteinase combination, an associated SI value of 1.0 would mean that all individual interactions result in irreversible inhibition of the proteinase, whereas an SI value of 4 (for example), would mean that for every one interaction resulting in irreversible inhibition of the enzyme, there are three interactions that result in cleaved inhibitor and free enzyme. Most interactions between plant serpins and mammalian proteinases of the chymotrypsin family have been characterized by k a values between 103 and 106 M−1 s−1 and an SI ~1 (Dahl et al. 1996a, b; Hejgaard 2001, 2005; Hejgaard and Hauge 2002; Ostergaard et al. 2000). In the only case in which a plant serpin has been tested against a large panel of distinct proteinases, recombinant barley serpin HorvuZx (BSZx) exhibited a high degree of inhibitory specificity, demonstrating that plant serpins share this property with their animal counterparts. Examples of HorvuZx–proteinase interactions characterized are those for trypsin (k a = 3.9 × 106 M−1 s−1, SI = 1.0), blood coagulation factor Xa (k a = 6.9 × 103 M−1 s−1, SI = 5.9), and elastase (no inhibitory activity) (Dahl et al. 1996a, b).
Most serpins inhibit proteinases through the kinetic trap described above, which is effectively irreversible. A small number of serpins, however, have been shown to inhibit specific proteinases in a reversible manner. Examples for human serpins are interactions between (1) protein C inhibitor (PAI-3; SerpinA5) and the proteinase scuPA (Schwartz and Espana 1999), and (2) α2-antiplasmin (SerpinF2) and chymotrypsin (Shieh et al. 1989). An example for a plant serpin is the reversible inhibition of trypsin (at P1 Arg) by TriaeZ2a (WSZ2a), a serpin found in wheat grain. Note that TriaeZ2a was found to irreversibly inhibit plasmin at P1, chymotrypsin at P2 Leu and cathepsin G possibly at both sites (Ostergaard et al. 2000). Reversible inhibition by proteinases is discussed in more detail in a review on serpins generally (Gettins 2002).
Several serpins have the ability to form polymers with like molecules through trans RCL insertion (as opposed to insertion of the individual molecule’s own RCL, or cis insertion). More than 40 variants of five human serpins are known to result in misfolding diseases or serpinopathies (Whisstock and Bottomley 2006). As far as we are aware, no misfolding of non-vertebrate serpins, including those from plants, has been reported. The tendency of serpins to form protein polymers will not be further reviewed here except to point out that some plant serpins are known to form complexes through intermolecular disulfide bridges under oxidizing conditions. These complexes have been observed both among serpins and between serpins and β-amylase protein molecules (found together in cereal grain endosperm) and can be broken by addition of reducing agents such as dithiothreitol (DTT) or β-mercaptoethanol (Hejgaard 1977; Hejgaard and Carlsen 1977).
Occurrence and functions of serpins across domains
Serpins have been remarkably adaptive during the evolution of animals. Functions for mammalian serpins—most found through studies of human serpins and their orthologues in the mouse—include the regulation of serine proteinases in haemostasis and blood coagulation by antithrombin (SerpinC1), control of plasminogen activation by plasminogen activitor inhibitor type 1 (PAI-1; SerpinE1), control of inflammation by α1-antitrypsin (SerpinA1) and C1 inhibitor (SerpinG1); inhibition of urokinase plasminogen activator (uPA) by PAI-2 (SerpinB2) and control of fibrinolysis by the inhibition of plasmin by α2-antiplasmin (SerpinF2). Examples from mammals of intracellular serpin function through control of serine proteinase activities include inhibition of cathepsin G by proteinase inhibitor 6 (PI6; SerpinB6) (Sun et al. 1997, 1998), furin by cytoplasmic antiproteinase 8 (PI8; SerpinB8) (Dahlen et al. 1998), and granzyme B by cytoplasmic antiproteinase 9 (PI9; SerpinB9) (Annand et al. 1999; Dahlen et al. 1997; Law et al. 2006; Sprecher et al. 1995). The inhibition of cysteine proteinases is an activity of some viral and animal serpins. Examples studied are the inhibition of caspases 1, 3 and 8 by viral serpin crmA (Komiyama et al. 1994; Ray et al. 1992; Zhou et al. 1997), inhibition of prohormone thiol proteinase by α1-antichymotrypsin (SerpinA3) (Hook et al. 1993), inhibition of cathepsins K, L and S by squamous cell carcinoma antigen 1 (SCCA1; SerpinB3) (Schick et al. 1998), inhibition of caspases 1, 4 and 8 by human serpin PI-9 (cytoplasmic antiproteinase 9; SerpinB9) (Annand et al. 1999), and inhibition of papain-like cysteine proteinases cathepsins L and V by MENT (Irving et al. 2002a). Functions of mammalian serpins that are not based on inhibitory activity include chaperonin activity through binding of procollagen by HSP47 (SerpinH1) and hormone transport and release by corticosteroid binding globulin (CBG; SerpinA6) and thyroxin binding globulin (TBG; SerpinA7) (Law et al. 2006; Zhou et al. 2006).
Some serpins have additional features that facilitate regulation of their inhibitory activity or modulate other biochemical functions of the serpin. A classic example is the vast increase in inhibitory activity of antithrombin (SerpinC1) upon binding to the glycosaminoglycan heparin, the mechanism of which has been elucidated (Dementiev et al. 2003, 2004; Johnson et al. 2006; Li et al. 2004; Rezaie 2006). To date there have been no features of plant serpins reported to regulate inhibitory activity, although several plant serpin genes are predicted to encode proteins with loops and extensions not found in the standard serpin sequence.
N-terminal regions of some animal serpins have been shown to have specific functions; for example, the acidic-residue-rich, hirudin-like N-terminal tail of heparin cofactor 2 (SerpinD1), a mammalian serpin that inhibits thrombin in the presence of glycosaminoglycan, is released for interaction with thrombin as part of the serpin’s inhibitory mechanism (Baglin et al. 2002). Functions of C-terminal extensions (relative to the C-terminus of α1-antitrypsin; SerpinA1) in some animal serpins have been elucidated; for example, the ~50-residue C-terminal extension of α2-antiplasmin contains an RGD sequence that enables binding of integrins, and thus facilitates cell adhesion (Thomas et al. 2007).
A range of animal serpin functions additional to those known from mammals has been defined through studies of model organisms such as Caenorhabditis elegans (Luke et al. 2006; Pak et al. 2004, 2006; Whisstock et al. 1999), Drosophila melanogaster (Han et al. 2000; Initiative 2000; Ligoxygakis et al. 2003; Oley et al. 2004; Reichhart 2005; Robertson et al. 2003) and the agriculturally important pest Manduca sexta (Gan et al. 2001; Jiang and Kanost 1997; Jiang et al. 1995, 1996; Kanost et al. 1989; Li et al. 1999; Zhu et al. 2003; Zou and Jiang 2005). These roles include defence against exogenous proteolytic attack and the regulation of developmental processes—functions that may be shared by some plant serpins (see later detailed discussion).
Among the archaeal and bacterial species whose genomes have been fully sequenced, only a small minority contain serpin genes (Irving et al. 2002b; Roberts et al. 2004; Zhang et al. 2007). Crystal structures and biochemical properties of the serpin from the thermophilic bacterium Thermobifida fusca have been determined (Fulton et al. 2005; Irving et al. 2003), and the inhibitory specificity of a serpin from Bifidobacterium longum, a commensal bacterium of humans, has been characterized (Ivanov et al. 2006). Two serpins from the Firmicute bacterium Clostridium thermocellum ATCC 27405 have been shown to be associated with the cellulosome, a multiprotein complex responsible for the degradation of plant polysaccharides. For both serpins, the serpin sequence is preceded by a dockerin moduleFootnote 4, and the N-terminus of each protein contains a leader sequence and a region homologous to the fibronectin III-like binding domain (Kang et al. 2006). To date, no explanation for the distribution of serpin genes among members of classes of archaea and bacteria has been given. Whether any functions of serpins in prokaryotes are related to those in plants remains unknown, but it is possible that serpins in microbial species known to interact with plants may influence aspects of plant biology.
Serpin genes are present in a substantial number of unicellular eukaryotes, but are notably absent from yeast (Roberts et al. 2004). The only reported cloning, expression and characterization of a serpin from a unicellular eukaryote is that from Entamoeba histolytica, a parasite that causes amoebic dysentery and amoebic liver abscess. The serpin was found to be excreted from the protozoan cytoplasm in the presence of specific mammalian cells (Riahi et al. 2004). Evidence exists for lateral transfer of several genes from anaerobic prokaryotes to Giardia lamblia and Entamoeba histolytica, including genes encoding ferredoxins, nitroreductases, NADH oxidase and alcohol dehydrogenase 3 (Nixon et al. 2002). There have been no studies on the origins of serpins in algae or plants, and lateral gene transfer from bacteria to algal ancestors remains a possibility.
A recent general review of serpins mentioned that serpin genes had not been identified in species belonging to the kingdom fungi (Law et al. 2006), which include numerous filamentous species and yeasts with fully sequenced genomes. A serpin gene (AAR97890)Footnote 5 has been identified, however, in the anaerobic fungus Piromyces sp. E2 isolated from the hindgut of an Indian elephant (P. J. Steenbakkers et al., unpublished). The predicted gene product is similar in primary structure to serpins of prokaryote cellulosomes (Kang et al. 2006), which have an N-terminal extension domain associated with polysaccharide binding or degradation (see above). It is tempting to speculate that horizontal gene transfer from bacteria to the anaerobic fungus may have occurred in this case.
The presence of serpin genes has been established for several green algae. A full serpin gene sequence and corresponding ESTs (e.g., BG856559) are known for Chlamydomonas reinhardtii, a member of the class Chlorophyceae and order Chlamydomonadales (Roberts et al. 2004). A serpin EST (EC751031, encoding 220 amino acid residues) exists for another Chlamydomonadaceae species, Polytomella parva (which has a colourless plastid). At least nine serpin ESTs (e.g., CF258534) have been identified from Acetabularia acetabulum, a member of the class Ulvophyceae and order Dasycladales, resulting in the construction of distinct C-terminal serpin sequences with standard RCL hinge sequences.Footnote 6 The presence of serpin genes in several distinct members of the green algae suggests that serpins might be present in most members of this division (but see comment on Ostreococcus tauri below).
As serpin genes were present in green algae, PSI-BLAST searches were conducted for serpins among all algae to explore serpins from photosynthetic eukaryotes generally. One sequence found was an EST (CF947062) encoding a serpin fragment (187 residues with P2-P1′ RSV) in the photosynthetic dinoflagellate (class Dinophyceae) Alexandrium tamerense (Roberts et al. 2004), a member of the order Gonyaulacales. Other dinoflagellate serpin sequences were found in Karlodinium micrum (e.g. EC159695; P2-P1′ LTA), a member of the order Gymnodiniales. Serpin sequences were also found in Cyanophora paradoxa (EC664221), a member of the class Glaucocystophyceae, and in Isochrysis galbana (EC137223), a member of the class Haptophyceae (the prymnesiophytes, one of the two classes of haptophytes) and order Isochrysidales. With the exceptions of C. reinhardtii and A. tamerense, no algae for which near-full genome information is available (Grossman 2005) appear to contain serpin genes. These organisms include the unicellular red alga Cyanidioschyzon merolae strain 10D (which lives in sulphate-rich, acidic hot waters and has an extremely compact genome), the diatom Thalassiosira pseudonana, the vestigial red algal genome associated with the nucleomorph of the cyptomonad Guillardia theta, and the marine picoeukaryote Ostreococcus tauri, which is thought to represent the type of organism from which all other green algae and ancestors of land plants have descended. The last result is interesting because it suggests that a more advanced green alga than Ostreococcus tauri may have acquired a serpin gene that was then retained through development of the embryophytes (the land plants).
Plant serpin nomenclature
Until associations between function and phylogeny have been established for specific plant serpins, it is not possible to adopt the serpin naming system proposed by Silverman et al. (2001), which is now being used in a substantial number of papers on animal serpins. Under this system, names such as “SerpinP1” would be used for serpins with equivalent functions in different plant species. The number of plant serpins identified is expanding rapidly, mostly due to the availability of EST databases for an increasing number of (mostly economically important) plants. The expansion in numbers of plant serpins has led to a need to examine carefully the system by which plant serpins should be named. Most names of plant serpins have incorporated the letter “Z” to designate sequence similarity with the first plant serpin identified, “barley protein Z” (Hejgaard 1976). Subfamilies within the Z family in specific plant species, such as WSZ1, WSZ2 and WSZx in wheat (Ostergaard et al. 2000), have been assigned based on levels of amino acid identity. More recently, plant serpins have been named according to a two-letter code for the species name and the subfamily, type and variant according to number and letter codes; e.g., MdZ1a1 for a serpin from apple (Malus domestica) (Hejgaard et al. 2005).
For this review, it became necessary to use five-letter codes denoting the species name: three letters for the genus and two for the specific epithet (Table 1). The Z-numbering system used in previous papers was retained; thus, a hypothetical serpin from (say) sorghum (Sorghum bicolor) could be named SorbiZ3b2.Footnote 7 A systematic numbering of serpin subfamilies based on (full-length) sequence homology has been established only for wheat and barley (Dahl et al. 1996a; Ostergaard et al. 2000). The original Zx designation (Rasmussen 1993) was based solely on the conserved reactive centre (P2–P1′ LRS), and the numbering Z1, Z2, etc. in other species is arbitrary. It is possible that some of the serpins named Z1, Z2, etc. are, in fact, “Zx-like” but are not labelled as such. For only a few cereal serpins with higher numbers, such as ZeamaZ6 and OrysaZ9, was a clear affiliation with barley/wheat subfamilies established. For example, OrysaZ9 was so named because it is similar to both HorvuZ9 (BSZ9) and ZeamaZ9. The plant serpin nomenclature developed here is used throughout the review.
Serpins in model plant and green algal genomes
Rice (Oryza sativa), Arabidopsis and Populus trichocarpa Footnote 8 are the only plants with fully sequenced genomes, although several other plant genomes are expected to be completed in the next couple of years, including that of barrel medic (Medicago sativa) and maize (Zea mays). New sequencing technologies such as 454 sequencing are expected to result in an explosion of fully sequenced plant genomes over the next 5–10 years.
A comparison of the (non-transposable-element-related) protein-coding genes in the rice and Arabidopsis genomes has shown that 29% of those in rice have no homologue in Arabidopsis, and 10% of the Arabidopsis genes have no homologue in rice (IRGSP 2005)Footnote 9. These substantial differences in gene complement suggest the possibility of serpin functions and target proteinases specific to either monocots or eudicots. The potent and irreversible inhibition of proteinases by plant serpins in vitro argues strongly that they function in vivo by preventing or regulating proteolytic events. If specific plant serpins did not use their inhibitory capability in vivo, it would be expected that their sequences would have mutated and their inhibitory capability would have been lost, as is the case for ovalbumin (Carrell and Owen 1985).
The following discussions refer to genes selected on the basis that they encode putative functional serpins. In a previous extensive overview of serpins in multicellular eukaryotes, nine serpin (or serpin-like) amino acid sequences in the Arabidopsis genome were included amongst 17 plant serpin sequences used to generate a majority-consensus maximum-parsimony bootstrap tree (Irving et al. 2000). This tree suggested that the Arabidopsis serpins could be subdivided into three groups (plus one orphan sequence). In the new version of the TAIR annotation of the Arabidopsis genomeFootnote 10, 16 sequence hits are obtained by searching for proteins with the query “serpin”, but this result is misleading. Six of these sequences given are too short to be serpins (<340 aa), and three others (of sufficient length) are clearly not serpins. Our analysis using PSI-BLAST searching of the Arabidopsis genome indicates that only 6 of the ~25 serpin genes and pseudogenes are likely to encode functional serpins (Table 2). One of the Arabidopsis serpins, ArathZ5, contains the sequence Val-Thr at P11–P10, and is thus predicted to be a non-inhibitory serpin (Table 2). Another additional Arabidopsis serpin gene, at locus At2g25240, is included in Table 2 because it is expressed at the mRNA level, but is not considered with the six serpins above because it appears to encode a truncated protein lacking helix A, strand 6 of sheet B, and helix B. While some viral and prokaryotic serpins lack specific secondary structural elementsFootnote 11, we are unaware of inhibitory serpins that lack helix B, and therefore conclude that the product of the gene at At2g25240 is not a real serpin. Each of the putatively functional serpins in Arabidopsis lies at a distinct locus; however, several serpin-like genes are found clustered in some cases; for example, At1g64010, At1g64020 and At1g64030 are all loci for serpin-like genes.Footnote 12
As was the case for Arabidopsis, careful interpretation of sequence databases was required to assemble the complement of serpins in rice. O. sativa has 12 chromosomes and a genome size of ~430 Mb (IRGSP 2005). BLAST searching of the rice genome revealed 14 serpin genes, three of which appear to encode non-inhibitory serpins (Table 3). Each of the remaining 11 sequences encodes a serpin with a unique (to rice) reactive centre P2–P1′ sequence (Table 3). OrysaZ5 (Os11g12520) has a C-terminal extension rich in Asp residues, whose function is unknown, but conceivably could be involved in the binding of positively charged species such as Ca2 + ions. The P1 residue diversity evident among both the Arabidopsis and rice serpins indicates a range of inhibitory specificities and suggests corresponding distinct functions. Such diversity of reactive centres is similar to that found for the serpin complement of oat grain (Hejgaard and Hauge 2002), but contrasts with the (mostly) Gln-rich reactive centres of wheat and rye grain (Hejgaard 2001; Ostergaard et al. 2000).
Many of the rice serpin genes are clustered in a region on chromosome 11 (refer to the loci numbers; Table 3), which is the site of a rice blast resistance locus, Pi-CO39(t), that corresponds to the avirulence gene AVR1-CO39 of the fungal pathogen Magnaporthe grisea (Chauhan et al. 2002). Further research is required to determine whether there is a functional link between the serpins encoded near this locus and the product of the avirulence gene. The clustering of serpins at a single locus has been used as evidence of a recent common serpin ancestor (Benarafa and Remold-O’Donnell 2005; Kaiserman and Bird 2005; Rollini and Fournier 1997).
Prediction of serpin localization based on amino acid sequences was performed using the program WoLF PSORT for the rice and Arabidopsis serpinsFootnote 13. The program, which is a major extension to the venerable PSORTII, makes predictions based on both known sorting signal motifs and some correlative sequence features such as amino acid contentFootnote 14. Both ArathZ5 (At1g62170) and ArathZ2 (At2g14540) were predicted to be localized to the nucleus with (particularly for readers familiar with WoLF PSORT) scores of 12 and 8, respectivelyFootnote 15. Recent confocal microscopy-based localization studies of GFP-fusion proteins in Arabidopsis protoplasts have shown that GFP-ArathZ2 is localized to the nucleus, confirming the prediction above. The other fusion proteins examined were GFP-ArathZx (At1g47710), GFP-ArathZ1 (At1g64030) and GFP-ArathZ3 (At2g26390), which were localized to the cytosol, as was GFP alone (the control) (J.-W. Ahn and T.H. Roberts, manuscript in preparation; data not shown). Some animal and viral serpins are known to have nucleocytoplasmic distributions, a fascinating example of which is the chicken erythrocyte serpin MENT, which is involved in chromatin organization (Grigoryev et al. 1999; Irving et al. 2002a; McGowan et al. 2006; Springhetti et al. 2003). Among the rice serpins, OrysaZxa (Os03g41419) and OrysaZ8 (Os11g11760) are predicted by WoLF PSORT to be localized to the cytosol with scores of 12 and 10, respectively. OrysaZ9 (Os11g11500) and OrysaZ6c (Os11g12460) are predicted to be localized to the chloroplast, both with scores of 14. No other Arabidopsis or rice serpins are associated with scores ≥8 with WoLF PSORT. Following up on the prediction of rice serpin localization to the chloroplast, searches were conducted for evidence of serpins in chloroplast-focussed rice proteome studies (Tanaka et al. 2004), but no evidence was found for serpins in this organelle.
C. reinhardtii is a unicellular green alga (a Chlorophyte) and serves as a model system for the study of several processes common to green algae and plants (Harris 2001). Searching using the recently released JGI C. reinhardtii v3.0 databaseFootnote 16 showed that C. reinhardti contains only one serpin geneFootnote 17, confirming results of an earlier search of v2.0 (Roberts et al. 2004). This makes Chlamydomonas particularly attractive to study the effects of RNAi-induced knockdown of serpin gene expressionFootnote 18. The Chlamydomonas serpin has the sequence Arg-Cys-Ala in the canonical P2–P1′ position, but it is possible that the reactive centre is shifted by one residue giving a 16-residue RCL, which may also be the case for serpins from other unicellular organisms (Roberts et al. 2004). The (noncanonical) P2–P1′ sequence of the Chlamydomonas serpin would thus be Leu-Arg-Cys as seen in a large number of LR serpins in plants (Hejgaard and Roberts 2007). Currently, the Chlamydomonas serpin is named ChlreS1, rather than ChlreZx, reflecting the uncertainty in identification of its functional reactive centre.
Phylogenetic analysis of plant serpins
A combination of amino acid sequence alignments and gene structure (intron–exon placement in the gene) has been used to infer serpin phylogeny (Atchley et al. 2001; Buck and Atchley 2005; Irving et al. 2000; Marshall 1993; Scott et al. 1999). Classification of serpins in multicellular eukaryotes resulted in plant serpins being assigned their own clade (clade p) among 16 clades in total (Irving et al. 2000); however, no phylogenetic analysis of a large number (>20) of plant serpins has been reported.
Careful examination of sequence data was required to check the gene structures of the 7 Arabidopsis and 14 rice serpin genes. Unlike all the other rice serpin genes, those at loci Os01g16200 and Os11g11760 are annotated in TIGRFootnote 19 as containing no intron, but there is no evidence that these genes are expressed (Table 3) and, thus, they may be pseudogenes. For Os11g11760, the presence of an intron has been overlooked by the computer-based annotation in TIGR. Our analysis shows that the remaining 12 rice genes contain one intron (in the true serpin-encoding region of the locus in each case).Footnote 20 The intron–exon boundary for all of the genes is conserved (not shown). This boundary corresponds to a region of the serpin that is predicted to lie at the beginning of helix F (hF), after strand 1 of β-sheet A (s1A)—determined using an alignment made to human α1-antitrypsin (not shown). The seven Arabidopsis genes (Table 2) each contains a single intron, and in each case, the boundary corresponds exactly to the conserved position described above for the rice serpin genes. Thus, as far as we are aware, locus Os01g16200 corresponds to the only (pseudo?) gene without a single intron in the standard position in rice or Arabidopsis. The genes for barley serpin HorvuZ4 (BSZ4) (Brandt et al. 1990), maize serpin Zeama9, Populus serpin PoptrZx and others each contains a single intron at the same conserved position as described above, suggesting that this represents the standard gene structure for plant serpins.
We conclude that the common ancestor of all eudicots and monocots is likely to have had one or more serpin genes with a single intron at a position corresponding to a region of the protein lying at the beginning of hF. The split between mono- and eudicots occurred a substantial time after the appearance of the angiosperms: the earliest known fossils of flowers and pollen grains are up to 130 million years old from the Cretaceous (Raven et al. 2005). The question then arises: Are serpin genes with the conserved intron–exon boundary found in plants whose lineage is more archaic than that of the monocots and eudicots? While the monocots and eudicots make up 97% of the extant angiosperm species, 3% of living angiosperms include those that represent lines that arose before the divergence between the monocots and the (combined) magnoliids-eudicots; thus, it would be interesting to determine the serpin gene complement and structure of one or more of these plant species. The Chlamydomonas serpin gene (4,650 bp total) contains eight introns (including two in the 3′ UTR) and seven protein-encoding exons. This gene structure is very different from those observed in plants, suggesting that the Chlamydomonas serpin and plant serpin genes are not closely related. The implications for this with regard to plant serpin evolution are unclear, but may be resolved by examining serpin gene structures in a greater number of algal species more closely related to the embryophytes.
A list was compiled of 60 full-length serpin sequences—all of which correspond to genes known to be expressed—from ~35 species of plants and one species of green alga (C. reinhardtii) by BLAST searching EST databases. Most of the sequences were constructed from overlapping ESTs, leaving open the possibility that a small minority of the full-length sequences might represent unnatural chimeras (the judgement was that this risk was justified in the analysis). Serpins expressed on the basis of proteomic or microarray experiments were included in the case of Arabidopsis and rice. The full-length plant/algal sequences were aligned using ClustalX (Thompson et al. 1997).Footnote 21 An msf file was generated for import into PAUP (v. 4.0b 10) (Swofford 2002). All sites (columns) in the alignment with gaps in any sequence were excluded from the alignment and the remaining sites aligned using parsimony (default settings in PAUP). The Chlamydomonas serpin was used as the outgroup due to the evolutionary distance between unicellular green algae and plants and the distinct gene structure for the Chlamydomonas serpin. Of the 487 total characters in the alignment, 45 characters were identical in all sequences, 83 variable characters were parsimony-uninformative, and 359 characters were parsimony-informative. The resulting heuristic treeFootnote 22 was bootstrapped using 1,000 trials.
Concerning names of the serpins used for the analysis (Fig. 2), many are denoted Zx (see Plant Serpin Nomenclature section above). GlymaZ1 (P2-P1′ GCA) is clearly Zx-like in sequence other than the reactive centre, exhibiting 91% sequence identity to GlymaZx (P2–P1′ LRS). The Arabidopsis serpin encoded by the gene at At2g14540 was not included in the phylogenetic analysis (despite EST evidence for expression) because it lacks sequence corresponding to the surface loop linking helixI1 to strand 5A. Due to exclusion of sites (columns) with gaps in construction of the phylogenetic tree, inclusion of ArathZ2 (At2g14540) in the alignment would have resulted in a substantial reduction in the number of sites available for construction of the tree.
Phylogenetic tree of full-length amino acid sequences of the products of expressed plant serpin genes. The sequences were aligned using ClustalX and a parsimony tree constructed with 1,000 bootstrap trials as detailed in the text. Bootstrap values are given in the tree. Names for serpin sequences are based on rules given in the text and in Tables 1, 2 and 3. Evidence for expression is based on corresponding ESTs
The resulting phylogram consists of two major branches (comprising eudicot and monocot sequences, respectively) two minor branches and a single sequence representing the outgroup Chlamydomonas serpin (Fig. 2). One minor branch comprises gymnosperm (Pinus) and bryophyte (moss) sequences. The other minor branch comprises OrysaZ9 and ZeamaZ9 sequences. The pitchfork-shaped eudicot branch indicates that PAUP was unable to resolve these sequences to create a phylogeny within this branch (in contrast to the monocot branch), despite running 1,000 bootstrap trials. Within the eudicot (major) branch, there is evidence for (expected) recent gene duplication; e.g., among the Solanum tuberosum (potato) sequences. Some sequences from different genera appear as ‘leaves’ on the same ‘twig’ of the tree: BranaZx and ArathZx, as do TarkoZx and LacsaZx. This may have been expected based on our knowledge of the ubiquity of the Zx serpin in the plant kingdom. It may be more interesting that SoltuZ1a and LycesZ1 also appear as leaves on the same twig, suggesting that it may not only be Zx that is conserved among plants.Footnote 23 The location of OrysaZ1 (Os01g56010) in the tree was unexpected. It seems to be behaving as an “ophan” sequence as does ZeamaZ6.
Within the monocot major branch, the sequences at the top are all non-Zx sequences, and the clustered wheat serpin sequences suggest recent gene duplications. Like some cases in the eudicot major branch, there are different genera represented as leaves on a single twig; e.g., HorvuZx and TriaeZxa. The sequences in the bottom of the branch are mostly Zx, and include OrysaZxb and SacofZ1 (not Zx-like). For the gymnosperm/bryophyte minor branch, the white spruce, Sikta spruce, loblolly pine, and maritime pine Zx sequences all cluster very close together on one sub-branch.
In conclusion, most of the plant serpins cluster with species, but the exceptions suggest possible conserved functions in specific groups of plants.
Serpin gene expression in seeds
Research on the nature of the dominant antigens in beer lead to the discovery of ~43-kDa polypeptides of barley origin, which were named Protein Z (Hejgaard 1977, 1982; Hejgaard and Bog-Hansen 1974; Hejgaard and Kaersgaard 1983; Hejgaard and Sorensen 1975). The protein was shown to be highly resistant to pH extremes and to boiling (30 min at pH 7) (Hejgaard 1982), which is a feature of the hyperstable, cleaved form of inhibitory serpins (Carrell and Owen 1985; Gettins 2002; Stein et al. 1989). Immunoelectrophoresis of extracts made from barley grains during the first 4 days of germination showed that Protein Z was present in two different, immunochemically related forms. These two antigens were shown to be products of distinct structural genes on barley chromosomes 4 and 7 (Nielsen et al. 1983) and were named BSZ4 and BSZ7, respectively (here HorvuZ4 and HorvuZ7). This research represented the first investigation of plant serpin gene expression and began to demonstrate the complexity of the serpin complement in plants before genomic information was available. Use of a combination of thiol extraction, ammonium sulfate precipitation, and ion exchange chromatography allowed for the isolation of highly pure Protein Z. The protein was identified as a member of the serpin family through observation that its primary structure was similar to those of α1-antitrypsin, antithrombin and ovalbumin (Hejgaard et al. 1985). This breakthrough allowed the techniques and theoretical understanding of animal serpins to be brought to bear on the study of barley Protein Z and its equivalents in other plants. Barley and wheat serpins were the first plant serpins to be made recombinantly, enabling simpler purification based on Ni-NTA binding of N-terminal 6×His tags (Dahl et al. 1996a; Rasmussen et al. 1996).
The isolation, purification and inhibitory specificity of grain serpins from several monocot crop species—barley, wheat, rye and oats—have been extensively studied. Purification of serpins from mature grains of barley, wheat and rye has been facilitated by “thiol extraction” of flour after exhaustive protein extraction with a buffer lacking thiol reagent (which alone gives a “salt extract”). The thiol extract is dominated by serpin and β-amylase isoforms, and approximately half the total amount of each serpin contained in the mature grain is differentially extracted using this method (Hejgaard 1976, 1978; Kaersgaard and Hejgaard 1979). Unfortunately, thiol extraction of flour from other grains, such as oat, does not result in a partial purification of the serpins and, thus, a distinct protocol for partial purification has to be used (Hejgaard and Hauge 2002). Further purification of serpins from thiol extracts or ammonium sulfate fractions has been successful using a combination of thiophilic adsorption chromatography (using a negatively sloping Na2SO4 gradient), and anion exchange chromatography, the latter often involving rechromatography with slightly altered buffer conditions. Native PAGEFootnote 24 provides an extremely useful tool to determine the expression of serpins in grains of wheat, barley and rye and can be used to compare varieties (including ancestral forms) within these species (unpublished results). Separation of the various serpin forms (either in a thiol extract or during purification) with native PAGE relies on small differences in pI (all pI~5) between the isoforms.
Two-dimensional gel electrophoresis (2-DE) studies have added to our knowledge of the expression of serpins in cereal grains. Finnie et al. (2004a, b) documented variation in serpin spot number and intensity between barley cultivars in 2-DE experiments; for example, seeds of cv. Mentor collected 4 weeks before harvest contained several additional abundant serpin spots compared with cultivars Meltan, Barke and Morex. Another study showed differences in HorvuZ7 (BSZ7) protein spots between cultivars Barke and Golden Promise (Finnie et al. 2004b). An enzyme-linked immunosorbent assay (ELISA)-based study of 93 cultivars of barley showed variation in the abundance of HorvuZ4 and HorvuZ7 in mature grain and in malt, but no correlation of this variation with the industrial use of each cultivar (feed versus malt) was evident (Evans and Hejgaard 1999). Barley serpins continue to be of interest to scientists studying malting, brewing and beer quality (Curioni et al. 1995; Evans and Sheehan 2002; Gorinstein et al. 1999; Lusk et al. 1995; Mills et al. 1998; Perrocheau et al. 2005; Yokoi et al. 1994).
Proteomic studies of the barley grain at various developmental stages have confirmed that specific serpins are major components of the endosperm in mature grain (Finnie et al. 2002, 2004a; Finnie and Svensson 2003; Ostergaard et al. 2002, 2004). The abundance of HorvuZ4 and the number of corresponding spots on 2D gels was found to increase gradually during the entire period of grain development (Finnie et al. 2002), but HorvuZ7 and HorvuZx were not detected in these experiments. In repeated Western blotting experiments using anti-rBSZx antibodies (Rasmussen 1993), HorvuZx could not be detected in flour extracts, but this serpin was finally detected as a minor spot in 2D gel electrophoresis (Ostergaard et al. 2004).
Through analysis of barley grain serpins isolated and studied from a wide range of cultivars and EST sequences encoding barley serpins, the diploid genome of Hordeum vulgare is estimated to contain around seven serpin genes, of which several are known to be expressed in the endosperm. Expression of serpin genes (various Protein Z isoforms) in barley grain was found to be highest during the later stages of grain filling and influenced by nitrogen nutrition (Giese and Hejgaard 1984). Alleles conferring high-lysine content of barley grain protein were shown to be associated with expression of these serpin genes. Expression of the gene encoding HorvuZ4 was shown to be enhanced by the allele lys1 (Hiproly barley) and repressed by lys3a (Bomi mutant 1508) (Hejgaard and Boisen 1980; Sorensen et al. 1989); note that β-amylase is similarly affected by these two alleles (Kreis et al. 1987). Calculations showed that serpins could contribute up to 5% of the total grain lysine in normal barley varieties and more than 7% in some high-lysine varieties (Hejgaard and Boisen 1980; Nielsen et al. 1983).
Immunohistochemistry has been particularly important in establishing expression of serpins in tissues of barley. Readers are referred to figures 2 and 4 in Roberts et al (2003) to illustrate the various tissues of barley grain, roots and embyro, and the relative levels of serpin expression in these tissues. Serpins from both HorvuZ4 and HorvuZ7 subfamilies have been localized to specific tissues in the mature grain by immunomicroscopy. In cv. Alexis, serpins were present in the central and peripheral starchy endosperm, the subaleurone layerFootnote 25 and (in lower concentrations) in the living aleurone (Roberts et al. 2003). No antibody labelling was seen in the testa, pericarp and husk, but serpins were detected in the scutellum and epithelial cells. Using a barley cultivar (Pirkka) lacking HorvuZ4 and a monoclonal antibody raised against a particular form of HorvuZ7 absent in cv. Alexis, this specific serpin was shown to be localized to a subcellular layer immediately adjacent to the aleurone, with only weak labelling in the endosperm and aleurone detected. Serpin labelling in longitudinal sections of barley cv. Alexis embyronic roots was strong in the root cap and coleorhyza (protective structures), while transverse sections revealed labelling in the apical meristem (Roberts et al. 2003).
The expression of 3,500 Arabidopsis genes has been examined during development of seeds (aged between 5 and 13 days) using a microarray (Ruuska et al. 2002) and the details published in an accompanying tableFootnote 26; however, none of the genes encoding the putatively inhibitory serpins in Arabidopsis were included in this study. In proteome maps at a websiteFootnote 27 dedicated to the Arabidopsis seed proteome and its changes during germination, no serpins have been reported. This was unexpected, as some serpin genes are known to be expressed in other eudicot seeds including apple (Hejgaard et al. 2005). Most of the apple serpin MdZ1b found in the mature seed was immunolocalized using a monoclonal antibody to the cotyledons; the remainder to the endosperm (Hejgaard et al. 2005). A few layers of nucellusFootnote 28 appeared to contain no serpin. Cotyledon labelling was clearly intracellular and seemed to be associated with protein bodies (cell walls were free of label). An indirect competition ELISA was used to estimate the content of serpin MdZ1b as 5–26 μg per seed (Hejgaard et al. 2005).
Serpin gene expression in vegetative tissues
Purification of a serpin from barley roots and analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), electroblotting and N-terminal sequencing identified it as HorvuZ4 (Roberts et al. 2003). This was supported by RCL cleavage experiments, which also indicated that the major serpin from leaf was HorvuZ4. The lack of detection of serpins in the leaves and roots of barley by rocket immunoelectrophoresis experiments suggested that the serpins are in low concentrations (generally) in these tissues relative to concentrations in the mature grain.
Microarray analyses provide valuable information on serpin expression. For example, PlexDBFootnote 29, which is a valuable resource for analysis of plant gene expression generally, includes BarleyBaseFootnote 30, which contains a data set describing 21,439 barley (cv. Morex) genes in 15 tissues sampled throughout development (Druka et al. 2006). Searching the probe set using the HorvuZx protein sequence (using tblastn) found a probe matching this serpin, Contig1878_atFootnote 31. A corresponding plot of Affimetrix hybridization value vs barley tissue indicated that the gene encoding HorvuZx is expressed in nearly all tissues. In contrast, a probe set for HorvuZ4 (BSZ4), Contig1877_atFootnote 32, indicated expression only in the caryopsis 16-DAP sample and the endosperm 22-DAP sampleFootnote 33. These data support an earlier study of barley serpin gene expression using northern dot blot and reverse transcriptase-polymerase chain reaction (RT-PCR) analysis (Roberts et al. 2003). Immunohistochemistry experiments on barley vegetative tissues showed the embryonic leaf and coleoptile were labelled with serpin antibody, while in the undifferentiated vascular tissue, serpin was not detected. Serpins were localized in longitudinal sections of 10-day roots to the apical meristem, cortex and vascular cylinder. Transverse sections revealed labelling in some phloem cells and less-intense labelling in surrounding cortex and the pith (Roberts et al. 2003).
For Arabidopsis, Affimetrix microarray data are available for most of the putative serpin genes at TAIRFootnote 34. The expression of eight Arabidopsis genes encoding serpins or proteins with serpin-like sequences (six in Table 2 plus genes at loci At1g61710 and At2g35580) across 2,352 arrays and 199 experiments was investigated using the Meta-Analyzer tool in GenevestigatorFootnote 35 (Table 2). Using Genevestigator, the ratio of signal intensity value (SIV) in stressed plants to SIV in control plants grown under 78 different stress conditions was calculated for each of the eight genes. The Digital Northern analysis revealed that some genes were not expressed above background noise levels (SIV p values > 0.05) for any of the experiments. For these genes, any deviation of the ratios observed was due to random noise. Significant expression was found, however, for a number of serpin genes and experiments (Table 2). One example is that of a 16-fold increase in the expression of the gene at At2g26390 in roots after 6 h of salt stress, which dropped back to a twofold increase after 24 h of salt stress.
Evidence from Genevestigator for upregulation of the gene at At1g47710 under cold treatment (Zimmermann et al. 2004) (Table 1) is supported by three independent microarray experiments: expression (1) increased 13-fold in the leaves and fivefold in the roots of plants exposed to 4°C over a 27-h period (Kreps et al. 2002), ranking it the 45th most highly upregulated cold-response gene out of ~8,100 studied; (2) increased sevenfold in plants grown at 4°C for 24 h (Fowler and Thomashow 2002); (3) increased tenfold in all tissues in plants grown at 4°C for 24–28 h (Hannah et al. 2005).
The Arabidopsis serpin gene at At1g64030 (P2-P1′ GCS) was found to be one of 65 “highly plastic” genes with variable expression patterns among five Arabidopsis accessions (Col-0, C24, Ler, Ws-2, and NO-0) (Chen et al. 2005). Further information was provided by Dr. Tong Zhu, SyngentaFootnote 36 (Table 2). ArathZx (the At1g47710 gene product) was identified in an extract from siliques (seed pods), but was not found in extracts from any other tissues (Giavalisco et al. 2005). Despite using a protein extraction procedure very similar to that used by Giavalisco et al. (2005), ArathZx was not identified in extracts from leaves of plants grown in the cold (see preceding paragraph) using 2-D gel electrophoresis (Tom Joss, Macquarie University Honours thesis 2006). Proteins that were successfully identified from leaf protein 2-D gels (ibid) were matched to an Arabidopsis silique protein gel of Giavalisco et al. (2005)—for which a visible molecular weight marker was not given. This comparison demonstrated that a protein identified by Giavalisco et al. as ArathZx had migrated at a rate corresponding to that of a ~30 kD protein. As the predicted molecular weight of ArathZx is 42.6 kD, it appears that the protein was present in the siliques in a degraded form too small to be an inhibitory serpin or was partially cleaved during the experiment.
For apple (Malus domestica), analysis of EST libraries constructed from a range of vegetative and reproductive tissues at various stages of development showed that five distinct serpin genes (of two subfamilies with ~85% amino acid sequence identity) were expressed in developing and mature fruits (including seeds), vegetative buds, mature and senescing leaves, vascular tissues and cell cultures. Seven of the 31 EST libraries examined were found to contain a proportion of serpin ESTs different from that expectedFootnote 37, but application of the Bonferroni statistical correction left only one library as containing a frequency of serpin ESTs significantly different from the other libraries (which might itself have been due to chance). The conclusion was that the apple tissue EST libraries in which serpins were found covered 80% of the total ESTs sequenced (160,000), but there was no statistically significant differential expression of serpins during development (Hejgaard et al. 2005).
Leaf expression of a gene encoding a serpin from spotted knapweed (Centaurea maculosa)Footnote 38 was shown to be upregulated twofold (a small increase) at the mRNA level after treatment with the fungal toxin Nep1 at 5 μg ml−1 [plus 0.1% (v/v) Silwet-L77] compared to treatment with Silwet-L77 alone (Keates et al. 2003). Nep1 is an extracellular protein produced by Fusarium oxysporum in liquid cultures. Leaves harvested 15, 60, or 240 min after treatment showed approximately equal levels of increased expression by Northern blotting. The target proteinase(s) of the knapweed Zx-like serpin (P2-P1′ LRS) were not identified and, thus, the biochemical pathways in which the serpin participates remain unknown.
The discovery of an inhibitory serpin, named CmPS-1 (P2-P1′ IVS, 389 aa) (CucmaZ1 in this review; Fig. 2), among the large number of proteins present in the phloem sap of pumpkin (Cucurbita maxima) (Yoo et al. 2000) showed for the first time that some plant serpins have the potential to act as long-distance signalling proteins. Low-molecular-weight proteinase inhibitors have also been identified in the phloem sap (Christeller et al. 1998; Murray and Christeller 1995; Walz et al. 2004). A study using pumpkin and the closely related species cucumber (Cucumis sativus) established that (1) the phloem serpins were localized exclusively in the sieve elements (not in the companion cells); (2) the serpins accumulated over time in the phloem, confirming earlier results (Yoo et al. 2000), and (3) through heterografting experiments the serpins were mobile in the phloem (la Cour Petersen et al. 2005).
Diversity and conservation of serpin reactive centres in plants and green algae
A consequence of the hypervariability of serpin RCL sequences (Barbour et al. 2002; Creighton and Darby 1989; Hill and Hastie 1987; Inglis and Hill 1991) is that identical (or very similar) reactive centre sequences found in serpins among different groups of organisms suggest conserved functions for these inhibitors. Complementing this, reactive centres unique to specific phylogenetic branches of the plant kingdom could be due to RCL hypervariability allowing rapid evolution of new functions for specific tissues or for responses to specific environmental conditions.
We used BLAST searches to assemble a list of ~130 P4–P4′ reactive centre sequences from ~65 species of plant serpins. The sequence P4–P4′ is likely to cover the most important residues in determining inhibitory specificity for most serpins. The list of reactive centres was assembled by alignment of highly conserved residues in the proximal hinge of the RCL (P17 Glu; P14 Thr; P8 Thr/Ser) to predict the identity of P4–P4′.Footnote 39 This prediction method is not 100% reliable, as some serpins are capable of inhibiting proteinases using a 16-residue RCL (Zhou et al. 2001). The P4–P4′ alignment was analyzed to determine whether the reactive centre sequences could be resolved into groups and whether any of these groups were found in only particular clades of plants (e.g., the monocots) (Table 4). In assignment of the groups, the P1 residue was given the most importance; thus, the sequences were grouped initially based on physico-chemical properties of the P1 residue, and subsequently using the identity of the P2 and then P3 residues (Table 4).
The analysis showed that the positively charged residue Arg is by far the most common P1 residue among plant serpins and that a large number of serpins have Leu at P2 (Table 4). The “LR serpins”, which are those with P2–P1′ Leu-Arg-X, where X is a small residue (Ser, Cys, Ala, Gly), are very widespread (perhaps universal) in the plant kingdom (Table 4). Nearly 40% of the LR serpins have a positively charged residue at P3, and most of the remainder have hydrophobic or small/polar residues at this position. Those LR serpins with a positively charged residue at P3 are all from eudicots. There are two well-studied examples of LR serpins. Barley serpin HorvuZx (BSZx) employs overlapping reactive centres to efficiently inhibit a range of serine proteinases of mammalian origin with both trypsin- and chymotrypsin-like specificity at P1–P1′ Arg-Ser and P2–P1 Leu-Arg, respectively (Dahl et al. 1996a, b). The other well-studied LR serpin is ArathZx (AtSerpin1) encoded by the gene at At1g47710 (Vercammen et al. 2006), which is also discussed in more detail below. The ubiquitous LR serpins (all named Zx, along with Zx-like serpins with conservative substitutions at P2; e.g., Phe) are expected to have a conserved function in plants, which may involve regulation of one or more endogenous proteinases (see later).
Smaller residues (Gly, Ala, Ser, Cys) and intermediate neutral/hydrophobic (Thr, Val) residues and large aliphatic (Leu) or aromatic (only the strictly nonpolar Phe) residues are also found at P1 (Table 4). Nine of the sequences have P1 Cys (a substantially higher proportion than would be expected by chance), and it is interesting to compare this with the frequency of P1 Cys in serpins from other life forms. No prokaryotic serpins (of 36 known) have P1 Cys (Zhang et al. 2007), but six of ten serpins examined in unicellular eukaryotes, including C. reinhardtii, do have Cys at this position if a 17-residue RCL length is assumed. P3 is variable, whereas almost all of the residues at P4 are aliphatic, as expected based on the general requirement of serpins for hydrophobic residues at even-numbered positions P4–P8. Consistent with serpins generally, almost all of the P1′ residues are small residues, with exceptions including Arg and Gln. Pro is far more common at P3′ and P4′ than would be expected by chance.
The sequence Gly-Cys is common in the region of the reactive centre of plant serpins (Table 4), much more so than in other positions in these proteins. It is found in only a small number of serpins in animals and apparently in no serpins in prokaryotes (Zhang et al. 2007). The sequence Cys-Gly is also common in the region of the reactive centre among serpins in many plant species. A sequence of three or more proline residues is found in the distal RCL sequence of many of the GC/CG serpins, and also in some other plant serpins (but in very few animal serpins) (Table 4).
Analysis of serpin sequences in wheat grain showed that the majority contained the reactive centre P1–P1′ Gln-Gln or X-Gln (Ostergaard et al. 2000), while reactive centres in most rye grain serpins contained P2–P1′ Gln-Gln-Ser or X-Gln-Ser (Hejgaard 2001). From both barley and wheat grain, a serpin with a sequence of three Gln residues at the reactive center has been isolated (Hejgaard 2005). Wheat, rye and barley grain contain prolamin storage proteins whose primary structure includes repeat sequences that are mimicked by the reactive centres of the grain serpins. For example, the repeated octapeptide PQQPFPQQ in the N-terminal domain of γ-gliadin contains two PQQ sequences that are identical to the P2–P1′ sequence of WSZ1b (Ostergaard et al. 2000). Gln-rich reactive centres are not restricted to the related monocot cereals mentioned here, but are found also in eudicots. Overlapping sequences identified from fiber EST libraries of cotton (Gossypium arboreum) could be assembled to encode a serpin with putative P3–P1′ Gln-Gln-Gln-Gln (Hejgaard 2005); this serpin has an extended N-terminal region, but no start Met has been identified. To our knowledge no viral, prokaryotic or animal serpins with confirmed inhibitory activity contain Gln at P1.
The information contained in Table 4 should allow researchers with knowledge of the proteolytic specificity of a plant or plant-pest proteinase to identify serpins with matching (or near matching) reactive centre residues.
Target proteinases for plant and algal serpins
The serpin mechanism of inhibition can function with any endopeptidase that employs a covalent acyl intermediate; i.e., serine and cysteine proteinases of whatever fold. The first demonstration that a plant serpin could inhibit a serine proteinase of the chymotrypsin family by formation of a covalent complex was made with TriaeZ1a (WSZ1a) and chymotrypsin (Rosenkrands et al. 1994). Since then, more than 20 plant serpins—either purified from tissues or recombinant proteins from grains of cereals of the Poaceae family (wheat, barley, rye and oat) and from eudicots pumpkin, apple and Arabidopsis—have been shown to irreversibly inhibit one or more members of this family of proteinases in vitro. Second-order association rate constants (k a) for these serpin–proteinase interactions range from ~103 to ~106 M−1 s−1, as is found for serpins from other organisms. Values for stoichiometry of inhibition (SI) range from 1.0 to 3.5 for most of the serpin–proteinase interactions tested, suggesting that these interactions might serve as good models for those that would occur in vivo.
Members of the serine, cysteine, metallo and aspartic endopeptidase groups are found in plants (as are corresponding proteinaceous inhibitors), but one of the intriguing aspects of the study of plant serpin function is that plants appear to lack genes encoding chymotrypsin-like serine proteinases (family S1 in Clan PA). Thus, in plant genomes, there are no genes homologous to those encoding elastase, thrombin, trypsin, plasminogen activator, granzyme B or other similar enzymes involved in proteolytic cascades in animals. The closest relative of these enzymes discovered in the plant kingdom is DegP, which belongs to family S1C of Clan PA. To our knowledge, no experiments have been performed to test the ability of plant serpins to inhibit this proteinase.
According to the MEROPS peptidase databaseFootnote 40, the Arabidopsis genome contains 678 putative peptidases and 148 non-peptidase homologues. These include members of almost 30 clansFootnote 41 of which several contain members known to be inhibited (at least in vitro) by serpins (plant or otherwise). The first experimental evidence identifying an endogenous target proteinase for a plant serpin was obtained recently (Vercammen et al. 2006). A yeast two-hybrid experiment screening for Arabidopsis proteins that could act as substrates or inhibitors of the Arabidopsis cysteine proteinase metacaspase 9 (AtMC9)—the product of one of nine metacaspase genes in Arabidopsis (Trobacher et al. 2006)—yielded the LR serpin ArathZx (named AtSerpin1 by the authors). Recombinant AtMC9 was shown to be inhibited by recombinant ArathZx (rArathZx) in vitro, forming 1:1 SDS-stable complexes. rArathZx was also capable of inhibiting AtMC4 and trypsin in vitro, demonstrating its ability to inhibit both cysteine and serine proteinases. Combinatorial tetrapeptide library screening in the same study identified the optimum fluorogenic substrate for AtMC9 as Ac-Val-Arg-Pro-Arg-amido-4-methyl-coumarin (Ac-VRPR-AMC), the amino acid sequence of which compared favourably with the P4–P1 residues in the RCL of ArathZx (IKLR). Evidence based on Western blots of apoplast extract from AtMC9- and ArathZx-overexpression lines indicated that both these proteins were present in the extracellular space of Arabidopsis root tips (Vercammen et al. 2006). Immunogold localization of ArathZx (which was not possible for AtMC9) suggested that the serpin was located in the endoplasmic reticulum, the Golgi apparatus, and in the cell wall. Further work will be required to establish whether ArathZx regulates the activity of AtMC9 in vivo, and whether the serpin inhibits endogenous (or exogenous) proteinases so far untested.
Cysteine proteinases are common in plants, but the functions of the great majority remain unknown. Arabidopsis contains 32 genes that encode papain-like (subfamily C1A), four that encode legumain-like (family C13)Footnote 42 and nine that encode metacaspase-type cysteine proteinases (family C14) (Trobacher et al. 2006). Classical animal caspases are absent from plants, but many of the cysteine proteinases present have been shown to be involved in various aspects of programmed cell death (PCD). Some of these cysteine proteinases act earlier in the process of PCD than others, but it is yet to be determined how specific early-acting cysteine proteinases are regulated (Trobacher et al. 2006).
Insects contain a range of digestive proteinases that might be inhibited by plant serpins. These include serine proteinases of the chymotrypsin family and cysteine proteinases. The gut of many insects is alkaline (Jongsma and Bolter 1997), providing an environment in which serpins would be expected to be active.
Possible functions of plant and algal serpins
Given the functional diversity of animal serpins, it would be reasonable to expect a range of functions for plant serpins. This diversity would be reflected in the identity of target enzymes and the timing and location of serpin expression. With only one serpin gene in the C. reinhardtii genome, serpins in unicellular green algae are unlikely to participate in a diverse range of functions. It appears more likely that a serpin would be required to regulate a specific proteolytic event in the cell. As serpin genes are absent from the fully sequenced genomes of yeast and several unicellular eukaryotes, the function of the green algal serpin may be to regulate a process that is not universal to all eukaryotes. It is possible, however, that the process is indeed universal but that an inhibitor of another protein family substitutes for the serpin in species where the serpin is absent.
In a recent review on serpins, it was suggested that those in plants may have a role in the complex pathways involved in upregulating the host immune response, as do serpins in insects, rather than directly interacting with pathogens (Law et al. 2006). This is a valid hypothesis for some plant serpins, but others are found in very high concentrations in seeds, including those of monocots such as the Poaceae cereals and eudicots such as apple (Hejgaard et al. 2005). Mature barley (Hejgaard 1982) and wheat (Rosenkrands et al. 1994) grain have been shown to contain serpins at concentrations of up to several percent total protein. It appears unlikely that these particular serpins could be acting as regulatory proteins (at least in the seeds themselves) given such high concentrations. Rather, they are probably laid down in seeds as a defensive shield to protect storage proteins from digestion by insects or fungi (Hejgaard 2001; Hejgaard and Roberts 2007; Ostergaard et al. 2000).
Defence roles for specific groups of serpins have been suggested for various animal systems, including mouse (Hill and Hastie 1987) and nematode worms including Caenorhabditis elegans (Zang and Maizels 2001). In the former case, a defence role was suggested after the ratio of synonymous to non-synonymous mutations was determined for the region of the serpin cDNA encoding the RCL. Comparison of this ratio to that determined for the whole serpin cDNA provided evidence for positive Darwinian evolution of the reactive centre residues of the serpin. This was consistent with adaption of the RCL to a range of proteinases with different proteolytic specificities, which might derive from pest or disease organisms. In the case of nematodes, a defence role was suggested due the ability of the serpins to inhibit specific mammalian proteinases such as thrombin.
The theory that at least some plant serpins are defence proteins is consistent with several studies in which insect (Manduca sexta) serpins were over-expressed in crop plants to test for their effect on resistance to pests. Claimed effects of the serpin transgene expression were a delay in the onset of thrip predation of alfalfa (Medicago sativa) (Thomas et al. 1994) and a reduction in the fecundity of sweet potato whitefly type B (Bemisia tabaci) in tobacco and cotton (Thomas et al. 1995a; Thomas et al. 1995b). To our knowledge, the transgenic plants examined in these studies are the only plants in which foreign serpin genes have been expressed for the purpose of providing a direct defence shield against pests.
Most of the serpins in mature wheat and rye grain contain reactive centres that resemble the Gln-rich repeat sequences found in prolamin storage proteins in the endosperm (Hejgaard 2001, 2005; Ostergaard et al. 2000; Rosenkrands et al. 1994). This is compelling but circumstantial evidence that the reactive centres of the grain serpins evolved to inhibit exogenous proteinases from insects and/or microbes (and possibly birds) that have adapted to breakdown endosperm storage proteins. Experimental evidence is required to demonstrate whether the complement of abundant serpins in seeds participates in the defence against pests or pathogens through inhibition of digestive enzymes. If seed serpins were acting purely as storage proteins, it is likely that they would have lost their inhibitory activity during evolution. In proteomics experiments on barley grain extracts after germination, a “vast amount” of serpins came to mask many protein spots in 2D gel experiments (Finnie et al. 2004a); however, it is unknown whether the serpins were being broken down, thereby acting as storage proteins, in addition to any defence functions. Serpins associated with tissues other than the endosperm of seeds may well carry out roles in defence; for example, those identified in the phloem of eudicot (la Cour Petersen et al. 2005; Roberts et al. 2003; Yoo et al. 2000) as well as monocot plants (Roberts et al. 2003). If these proteinases were from exogenous sources, the phloem serpins would be playing a direct role in defence.
An alternative function for serpins in the phloem may be to maintain the integrity of important signalling peptides or proteins through inhibition of destructive proteinase activity. Phloem sap is now known to contain the products of thousands of plant genes (mRNAs and proteins) and is thought to mediate a highly sophisticated long-distance signalling network in plants (Lough and Lucas 2006). The concentration of serpins in the phloem sap of pumpkin (Cucurbita maxima) was shown to increase upon challenge with the piercing-sucking aphid, Myzus persicae (Yoo et al. 2000), consistent with a role in defence signalling. Through grafting experiments using pumpkin and cucumber (Cucumis sativus) plants (both cucurbits), phloem serpins were shown to be mobile (graft transmissible) and thus to be potential signal molecules (la Cour Petersen et al. 2005) possibly involved in the regulation of programmed cell death (PCD) or defence pathways.
Plant PCD is an integral part of a large range of developmental processes, including xylem element differentiation, sculpting of leaf shape, leaf senescence, anther dehiscence, flower senescence, removal of maternal tissues during seed development and in the death of aleurone cells in cereal seeds. PCD also plays a critical role in plant responses to stress, including responses to hypoxia, shading, temperature extremes, drought and oxidative stress. The possibility that one or more of these processes may be regulated by inhibitory serpins should be investigated. Expression of the Arabidopsis serpin gene at locus At1g47710 (coding for ArathZx) was suggested by microarray experiments to be upregulated in Arabidopsis subjected to cold stress (Fowler and Thomashow 2002; Kreps et al. 2002). Cell lysis due to cold stress or wounding may release proteinases that have the potential to damage neighbouring cells. Thus, some plant serpins may act in a manner analogous to the intracellular serpins in mammals that function to protect cells from proteinases released during inflammatory responses (Silverman et al. 2004). Experimental work is required to test this hypothesis.
In a recent study, At1g47710 serpin expression was reported to be upregulated in leaves of plants in which roots had first been primed by infection with a nonpathogenic Pseudomonas strain—producing “induced systemic resistance” (ISR)—and the leaves then challenged with the pathogenic strain Pseudomonas syringae pv. tomato. Although the degree of upregulation observed was small, no serpin genes were upregulated by the priming alone, suggesting that ArathZx may have a role in the regulation of plant defence. This result is compatible with the hypothesis that ArathZx inhibits AtMC9 in vivo (as it does in vitro) (Vercammen et al. 2006), as metacaspases are likely to participate in plant defence signal transduction pathways (Trobacher et al. 2006). It is possible that further analysis of the conserved reactive centre of the Zx serpins and the specificity of the metacaspases could support a physiological role for one or more of these proteinases. For example, the conservation of P3 Lys in some branches of the plant serpin phylogram (Fig. 2; Table 4) might indicate a binding pocket with an acidic residue in the target proteinase. Similarly, a hydrophobic P3 residue—as found in the Zx serpins from all plants of the subfamily Pooideae—would most likely adapt to a hydrophobic pocket.
An EST sequence encoding a full-length Zx serpin has been assembled from Brachypodium distachyon, a temperate grass that has been suggested as a convenient “genomic bridge” between rice and the temperate cereals and forage grasses (included in Fig. 2). The genome size of B. distachyon and the genomes’ proportion of repetitive DNA are comparable to Arabidopsis and rice, its seed size is relatively large, and it is more closely related to the economically important temperate grasses than rice. B. distachyon may therefore be a worthwhile model species in which to study serpin expression and functions in the future.
Some putatively functional plant serpins contain putative N- and/or C-terminal extensions and non-RCL loop deletions (Tables 2 and 3). Although genes for some of these nonstandard serpins are associated with evidence for expression, in no case is there experimental evidence for (or against) correct protein folding to produce a functional polypeptide. To date, no N- or C-terminal sequences of plant serpins have been associated with specific properties or functions.
Often terminal extensions, surface loop insertions or deletions important for serpin function are reflected in isoelectric points (pI)Footnote 43 of the serpin proteins that differ substantially from the norm. For example, the chicken serpin MENT has a pI of 9.38Footnote 44, consistent with its function in chromatin condensation (Grigoryev et al. 1999; McGowan et al. 2006). It is possible that plant serpins with high pI values may also be involved in functions relating to binding negatively charged chemical species. Using the available 21 monocot and 32 eudicot full-length serpin sequences utilized to generate Fig. 2, pI values were calculated. There was no difference between the monocot and eudicot pI values overall: average monocot pI value = 5.79 ± 0.92 (SD); eudicot 5.81 ± 0.50. However, among the monocot serpins were two with outlying pI values, namely OrysaZxb with pI = 8.84 (compared to pI = 5.75 for OrysaZxa), and ZeamaZ9 with pI = 4.00. Markedly different pI values suggest that specific serpins may be localized to particular compartments of the cell dependent on the pH of the compartment. The Lys-rich N-terminal sequences of ArathZ5 and ArathZ2 may indicate possible interactions with negatively charged chemical species such as DNA.
Our analysis of the serpin gene complements in Arabidopsis and rice and their expression (Table 2 and 3) suggests that plants, as described for animals with several examples in this review, may contain serpins with functions that are not related to inhibitory action. For illustration, the structural basis for the binding of hormone ligand to human TBG was recently solved, showing that tetra-iodo thyroxine is carried in a surface pocket of the protein and that the binding-release mechanism is dependent on the presence of a Pro at the P8 position in the RCL (Zhou et al. 2006). It is interesting that plant brassinosteroids are structurally related to steroid hormones in animals and are now recognized as one of the six major groups of phytohormones (Haubrick and Assmann 2006); thus, although no plant serpins are known with P8 Pro, the possible existence of hormone-binding plant serpins is a suggested topic for further investigation.
Until now all plant serpins characterised were found to be inhibitory. The discovery of expressed genes encoding closely related non-inhibitory plant serpins from rice and maize, representing a distinct branch in the phylogenetic analysis (OrysaZ9 and ZeamaZ9 in Fig. 2), may provide a promising starting point for a breakthrough in this direction. These “Z9 serpins” have a conserved but unusual hinge region with P12-P9 ETSV instead of the canonical AAAA in inhibitory serpins; the subsequent sequence P7–P2—of their unusually short RCL—is highly conserved (OrysaZ9, Table 2). Research to identify putative binding partners (proteins or other molecules) is required to understand the functions of these serpins.
Concluding remarks
Viewing the serpin family of proteins as a whole, the plant kingdom has remained a blind spot for functional information for a couple of decades. Based on our current knowledge of both gene expression and protein properties (both tested and predicted), plants serpins are likely to participate in a range of biochemical pathways in distinct cell types, tissues and organs. It appears likely that the LR serpins share a common function throughout the kingdom and that some other plant serpins have evolved to fill functional niches specific for particular groups of plants. The abundant seed serpins (and possibly serpins in other organs/tissues) are likely to be involved in direct defence, while other specific serpins may be involved in the regulation of defence activation and/or programmed cell death.
One means by which plant and green algal serpin functions are likely to be discovered is through analysis of mutants in which expression of specific serpin genes is knocked out or upregulated. Serpin T-DNA mutants of Arabidopsis are beginning to reveal phenotypes distinct from those of the wild-type when plants are placed under specific stress conditions. Results of experiments of this type are likely to stimulate new research into the control of plant proteolysis and its importance for plant biology.
Notes
AGI is an acronym for the author of this article, “The Arabidopsis Genome Initiative”.
Chapters within Silverman and Lomas (eds) (2007) Molecular and Cellular Aspects of the Serpinopathies and Disorders in Serpin Activity, World Scientific (ISBN-10 981–256–963–4) are recommended reading, but many are not cited in this review because of lack of easy access for most readers.
In the cellulosome, dockerin domains are conserved noncatalytic subdomains borne by catalytic subunits. The dockerins bind to cohesion (receptor) domains of the protein CipA (Beguin and Lemaire 1996).
All numbers of this type in the review (including tables) are accession numbers.
There are many errors in the sequencing and only a few clear overlaps here, but at least two serpins appear to be expressed.
Serpins in Populus will not be considered here as annotation of the genome is at only a preliminary stage.
“IRGSP” is an acronym for the author of this article, “International Rice Genome Sequencing Project”.
TAIR7, released April 2007
The pox virus serpin crmA lacks D helix and has shortened versions of helix A and helix E (Simonovic et al. 2000).
Among these three loci, only the gene at At1g64030 can encode a true serpin (the others encode proteins with 185 and 121 amino acid residues).
The current dataset used to train WoLF PSORT contains over 12,000 animal sequences and more than 2,000 plant and fungi sequences, respectively. It was gathered mainly from Uniprot, but several hundred Arabidopsis sequences from the Gene Ontology database were also included.
Prediction scores obtained with values 8 only are mentioned here.
Location: Chlre3/scaffold_28:1327628–1332277; Protein ID: 149241
This is the subject of experiments currently underway in Roberts’ laboratory.
See http://www.tigr.org/tdb/e2k1/osa1/; May 2007
In the TIGR annotation, Os03g41419 contains four introns, and Os03g41438, Os11g12410 and Os11g12420 contain two introns, but these annotations have errors (see footnotes for Table 3). Interestingly, for Os03g41419, the TIGR-annotated exons 4 and 5 appear to be possible alternative splicing variants, each encoding the C-terminal approximately two-thirds of the protein sequence starting at identical positions; however, there is no EST evidence to suggest that alternative splicing occurs with this gene.
The alignment was performed using the Gonnet series for the substitution matrix, a gap opening penalty=10 and a gap extension penalty=0.2.
Attempts to generate a “branch and bound” tree failed due to insufficient computing power available.
In the latter clustering, reference is not to conservation of reactive centres, but rather to the whole protein; SoltuZ1a and LycesZ1 have P2-P1′ PLS and TTS, respectively, and thus would not be expected to inhibit proteinases of similar specificity.
An 8% acrylamide gel and a buffer at pH ~8 are normally used.
This layer is not present in wheat grain.
The nucellus is located between the testa and the endosperm.
Plant Expression DataBase; see http://www.plexdb.org
Matches Exemplar Sequence Barley1_01878
Matches Exemplar Sequence Barley1_01877
DAP = days after pollination
The Arabidopsis Information Resource; see http://www.arabidopsis.org
Personal communication
Using individual a priori statistical tests at the 5% level
A herbacious perennial from the family Asteraceae
In contrast to the fixed length of the N-terminal region of the RCL (P17 to P1), the length of the C-terminal region (ending at the conserved sequence DFVAD) ranges from 6 to 18 residues among the plant serpins.
The MEROPS webpage used was created 23 April 2007.
Peptidase clans found in Arabidopsis are AA, AD, CA, CD, CE, CF, MA, MC, ME, MF, MG, MH, MJ, MK, MM, MP, PA, PB, PC, SB, SC, SE, SF, SJ, SK, SP, ST, T- and U-.
The legumains are asparagine-specific cysteine proteinases that cleave peptide bonds at the carboxyl group of asparagines, or in some cases, at relatively low pH (4.0) and with reduced activity at the carbonyl group of aspartic acid. Legumain, from jackbean (Canavalia ensiformis), is the type identifier for the family (Trobacher et al. 2006).
The pH at which a protein has no net charge.
Predicted using the program ProtParam available at http://www.expasy.org
References
Aleshkov SB, Fa M, Karolin J, Strandberg L, Johansson LB, Wilczynska M, Ny T (1996) Biochemical and biophysical studies of reactive center cleaved plasminogen activator inhibitor type 1. The distance between P3 and P1’ determined by donor–donor fluorescence energy transfer. J Biol Chem 271:21231–21238
Annand RR, Dahlen JR, Sprecher CA, De Dreu P, Foster DC, Mankovich JA, Talanian RV, Kisiel W, Giegel DA (1999) Caspase-1 (interleukin-1beta-converting enzyme) is inhibited by the human serpin analogue proteinase inhibitor 9. Biochem J 342:655–665
Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
Atchley WR, Lokot T, Wollenberg K, Dress A, Ragg H (2001) Phylogenetic analyses of amino acid variation in the serpin proteins. Mol Biol Evol 18:1502–1511
Baglin TP, Carrell RW, Church FC, Esmon CT, Huntington JA (2002) Crystal structures of native and thrombin-complexed heparin cofactor II reveal a multistep allosteric mechanism. Proc Natl Acad Sci USA 99:11079–11084
Barbour KW, Goodwin RL, Guillonneau F, Wang YP, Baumann H, Berger FG (2002) Functional diversification during evolution of the murine alpha(1)-proteinase inhibitor family: role of the hypervariable reactive center loop. Mol Biol Evol 19:718–727
Becerra SP, Sagasti A, Spinella P, Notario V (1995) Pigment epithelium-derived factor behaves like a noninhibitory serpin. Neurotrophic activity does not require the serpin reactive loop. J Biol Chem 270:25992–25999
Beguin P, Lemaire M (1996) The cellulosome: an exocellular, multiprotein complex specialized in cellulose degradation. Crit Rev Biochem Mol Biol 31:201–236
Benarafa C, Remold-O’Donnell E (2005) The ovalbumin serpins revisited: perspective from the chicken genome of clade B serpin evolution in vertebrates. Proc Natl Acad Sci USA 102:11367–11372
Brandt A, Svendsen I, Hejgaard J (1990) A plant serpin gene. Structure, organization and expression of the gene encoding barley protein Z4. Eur J Biochem 194:499–505
Bruch M, Weiss V, Engel J (1988) Plasma serine proteinase inhibitors (serpins) exhibit major conformational changes and a large increase in conformational stability upon cleavage at their reactive sites. J Biol Chem 263:16626–16630
Buck MJ, Atchley WR (2005) Networks of coevolving sites in structural and functional domains of serpin proteins. Mol Biol Evol 22:1627–1634
Carrell RW, Evan DLI (1992) Serpins: mobile conformations in a family of proteinase inhibitors. Curr Opin Struct Biol 2:438–446
Carrell RW, Owen MC (1985) Plakalbumin, alpha 1-antitrypsin, antithrombin and the mechanism of inflammatory thrombosis. Nature 317:730–732
Carrell R, Travis J (1985) Alpha1-antitrypsin and the serpins: variation and countervariation. Trends Biochem Sci 10:20–24
Chauhan RS, Farman ML, Zhang HB, Leong SA (2002) Genetic and physical mapping of a rice blast resistance locus, Pi-CO39(t), that corresponds to the avirulence gene AVR1-CO39 of Magnaporthe grisea. Mol Genet Genom 267:603–612
Chen WQJ, Chang SH, Hudson ME, Kwan WK, Li JQ, Estes B, Knoll D, Shi L, Zhu T (2005) Contribution of transcriptional regulation to natural variations in Arabidopsis. Genome Biol 6:R32
Christeller JT, Farley PC, Ramsay RJ, Sullivan PA, Laing WA (1998) Purification, characterization and cloning of an aspartic proteinase inhibitor from squash phloem exudate. Eur J Biochem 254:160–167
Creighton TE, Darby NJ (1989) Functional evolutionary divergence of proteolytic enzymes and their inhibitors. Trends Biochem Sci 14:319–324
Curioni A, Pressi G, Furegon L, Peruffo ADB (1995) Major proteins of beer and their precursors in barley: electrophoretic and immunological studies. J Agric Food Chem 43:2620–2626
Dahl SW, Rasmussen SK, Hejgaard J (1996a) Heterologous expression of three plant serpins with distinct inhibitory specificities. J Biol Chem 271:25083–25088
Dahl SW, Rasmussen SK, Petersen LC, Hejgaard J (1996b) Inhibition of coagulation factors by recombinant barley serpin BSZx. FEBS Lett 394:165–168
Dahlen JR, Foster DC, Kisiel W (1997) Human proteinase inhibitor 9 (PI9) is a potent inhibitor of subtilisin A. Biochem Biophys Res Commun 238:329–333
Dahlen JR, Jean F, Thomas G, Foster DC, Kisiel W (1998) Inhibition of soluble recombinant furin by human proteinase inhibitor 8. J Biol Chem 273:1851–1854
Dementiev A, Simonovic M, Volz K, Gettins PG (2003) Canonical inhibitor-like interactions explain reactivity of alpha1-proteinase inhibitor Pittsburgh and antithrombin with proteinases. J Biol Chem 278:37881–37887
Dementiev A, Petitou M, Herbert JM, Gettins PGW (2004) The ternary complex of antithrombin-anhydrothrombin heparin reveals the basis of inhibitor specificity. Nat Struct Mol Biol 11:863–867
Druka A, Muehlbauer G, Druka I, Caldo R, Baumann U, Rostoks N, Schreiber A, Wise R, Close T, Kleinhofs A, Graner A, Schulman A, Langridge P, Sato K, Hayes P, McNicol J, Marshall D, Waugh R (2006) An atlas of gene expression from seed to seed through barley development. Funct Integr Genomics 6:202–211
Evans DE, Hejgaard J (1999) The impact of malt derived proteins on beer foam quality. Part 1. The effect of germination and kilning on the level of protein Z4, protein Z7 and LTP1. J Inst Brew 105:159–169
Evans DE, Sheehan MC (2002) Don’t be fobbed off: the substance of beer foam - A review. J Am Soc Brew Chem 60:47–57
Finer-Moore JS, Kossiakoff AA, Hurley JH, Earnest T, Stroud RM (1992) Solvent structure in crystals of trypsin determined by X-ray and neutron diffraction. Proteins 12:203–222
Finnie C, Svensson B (2003) Feasibility study of a tissue-specific approach to barley proteome analysis: aleurone layer, endosperm, embryo and single seeds. J Cer Sci 38:217–227
Finnie C, Melchior S, Roepstorff P, Svensson B (2002) Proteome analysis of grain filling and seed maturation in barley. Plant Physiol 129:1308–1319
Finnie C, Maeda K, Ostergaard O, Bak-Jensen KS, Larsen J, Svensson B (2004a) Aspects of the barley seed proteome during development and germination. Biochem Soc Trans 32:517–519
Finnie C, Steenholdt T, Roda Noguera O, Knudsen S, Larsen J, Brinch-Pedersen H, Bach Holm P, Olsen O, Svensson B (2004b) Environmental and transgene expression effects on the barley seed proteome. Phytochemistry 65:1619–1627
Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14:1675–1690
Fulton KF, Buckle AM, Cabrita LD, Irving JA, Butcher RE, Smith I, Reeve S, Lesk AM, Bottomley SP, Rossjohn J, Whisstock JC (2005) The high resolution crystal structure of a native thermostable serpin reveals the complex mechanism underpinning the stressed to relaxed transition. J Biol Chem 280:8435–8442
Gan H, Wang Y, Jiang HB, Mita K, Kanost MR (2001) A bacteria-induced, intracellular serpin in granular hemocytes of Manduca sexta. Insect Biochem Mol Biol 31:887–898
Gettins PGW (2002) Serpin structure, mechanism, and function. Chem Rev 102:4751–4803
Gettins PGW (2007) Mechanisms of serpin inhibition. In: Silverman GA, Lomas DA (eds) Molecular and cellular aspects of the serpinopathies and disorders in serpin activity. World Scientific, New Jersey, pp 67–100
Giavalisco P, Nordhoff E, Kreitler T, Kloppel KD, Lehrach H, Klose J, Gobom J (2005) Proteome analysis of Arabidopsis thaliana by two-dimensional gel electrophoresis and matrix-assisted laser desorption/ionisation-time of flight mass spectrometry. Proteomics 5:1902–1913
Giese H, Hejgaard J (1984) Synthesis of salt-soluble proteins in barley. Pulse-labeling study of grain filling in liquid-cultured detached spikes. Planta 161:172–177
Gorinstein S, Zemser M, Vargas-Albores F, Ochoa JL, Paredes-Lopez O, Scheler C, Salnikow J, Martin-Belloso O, Trakhtenberg S (1999) Proteins and amino acids in beers, their contents and relationships with other analytical data. Food Chem 67:71–78
Grigoryev SA, Bednar J, Woodcock CL (1999) MENT, a heterochromatin protein that mediates higher order chromatin folding, is a new serpin family member. J Biol Chem 274:5626–5636
Grossman AR (2005) Paths toward algal genomics. Plant Physiol 137:410–427
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723
Han J, Zhang H, Min G, Kemler D, Hashimoto C (2000) A novel Drosophila serpin that inhibits serine proteases. FEBS Lett 468:194–198
Hannah MA, Heyer AG, Hincha DK (2005) A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PloS Genet 1: e26
Harris EH (2001) Chlamydomonas as a model organism. Annu Rev Plant Physiol Plant Mol Biol 52:363–406
Haubrick LL, Assmann SM (2006) Brassinosteroids and plant function: some clues, more puzzles. Plant Cell Environ 29:446–457
Hejgaard J (1976) Free and protein-bound beta-amylases of barley grain: characterization by two-dimensional immunoelectrophoresis. Physiol Plant 38:293–299
Hejgaard J (1977) Origin of a dominant beer protein: immunochemical identity with a beta-amylase-associated protein from barley. J Inst Brew 83:94–96
Hejgaard J (1978) ‘Free’ and ‘bound’ beta-amylases during malting of barley, characterization by two-dimensional immunoelectrophoresis. J Inst Brew 84:43–46
Hejgaard J (1982) Purification and properties of protein Z—a major albumin of barley endosperm. Physiol Plant 54:174–182
Hejgaard J (2001) Inhibitory serpins from rye grain with glutamine as P-1 and P-2 residues in the reactive center. FEBS Lett 488:149–153
Hejgaard J (2005) Inhibitory plant serpins with a sequence of three glutamine residues in the reactive center. Biol Chem 386:1319–1323
Hejgaard J, Bog-Hansen TC (1974) Quantitative immunoelectrophoresis of barley and malt proteins. J Inst Brew 80:436–442
Hejgaard J, Boisen S (1980) High-lysine proteins in Hiproly barley breeding: identification, nutritional significance and new screening methods. Hereditas 93:311–320
Hejgaard J, Carlsen S (1977) Immunoelectrophoretic identification of a heterodimer beta-amylase in extracts of barley grain. J Sci Food Agric 28:900–904
Hejgaard J, Hauge S (2002) Serpins of oat (Avena sativa) grain with distinct reactive centres and inhibitory specificity. Physiol Plant 116:155–163
Hejgaard J, Kaersgaard P (1983) Purification and properties of the major antigenic beer protein of barley origin. J Inst Brew 89:402–410
Hejgaard J, Roberts TH (2007) Plant serpins. In: Silverman GA Lomas DA (eds) Molecular and cellular aspects of the serpinopathies and disorders in serpin activity. World Scientific, New Jersey, pp 279–300
Hejgaard J, Sorensen SB (1975) Characterization of a protein-rich beer fraction by two-dimensional immunoelectrophoretic techniques. Comptes Rendus des Travaux du Laboratoire Carlsberg 40:187–203
Hejgaard J, Rasmussen SK, Brandt A, Svendsen I (1985) Sequence homology between barley endosperm protein Z and protease inhibitors of the alpha-1-antitrypsin family. FEBS Lett 180:89–94
Hejgaard J, Laing WA, Marttila S, Gleave AP, Roberts TH (2005) Serpins in fruit and vegetative tissues of apple (Malus domestica): expression of four serpins with distinct reactive centres and characterisation of a major inhibitory seed form, MdZ1b. Funct Plant Biol 32:517–527
Hill RE, Hastie ND (1987) Accelerated evolution in the reactive centre regions of serine protease inhibitors. Nature 326:96–99
Hook VYH, Purviance RT, Azaryan AV, Hubbard G, Krieger TJ (1993) Purification and characterization of alpha 1-antichymotrypsin-like protease inhibitor that regulates prohormone thiol protease involved in enkephalin precursor processing. J Biol Chem 268:20570–20577
Hunt LT, Dayhoff MO (1980) A surprising new protein superfamily containing ovalbumin, antithrombin- III, and alpha 1-proteinase inhibitor. Biochem Biophys Res Commun 95:864–871
Huntington JA, Read RJ, Carrell RW (2000) Structure of a serpin–protease complex shows inhibition by deformation. Nature 407:923–926
Ibarra CA, Blouse GE, Christian TD, Shore JD (2004) The contribution of the exosite residues of plasminogen activator inhibitor-1 to proteinase inhibition. J Biol Chem 279:3643–3650
Inglis JD, Hill RE (1991) The murine Spi-2 proteinase inhibitor locus: a multigene family with a hypervariable reactive site domain. EMBO J 10:255–261
International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436:793–800
Irving JA, Pike RN, Lesk AM, Whisstock JC (2000) Phylogeny of the serpin superfamily. Implications of amino acid conservation for structure and function. Genome Res 10:1845–1864
Irving JA, Shushanov SS, Pike RN, Popova EY, Bromme D, Coetzer THT, Bottomley SP, Boulynko IA, Grigoryev SA, Whisstock JC (2002a) Inhibitory activity of a heterochromatin-associated serpin (MENT) against papain-like cysteine proteinases affects chromatin structure and blocks cell proliferation. J Biol Chem 277:13192–13201
Irving JA, Steenbakkers PJM, Lesk AM, den Camp HJMO, Pike RN, Whisstock JC (2002b) Serpins in prokaryotes. Mol Biol Evol 19:1881–1890
Irving JA, Cabrita LD, Rossjohn J, Pike RN, Bottomley SP, Whisstock JC (2003) The 1.5 angstrom crystal structure of a prokaryote serpin: controlling conformational change in a heated environment. Structure 11:387–397
Irving JA, Askew DJ, Whisstock JC (2004) Computational analysis of evolution and conservation in a protein superfamily. Methods 32:73–92
Ivanov D, Emonet C, Foata F, Affolter M, Delley M, Fisseha M, Blum-Sperisen S, Kochhar S, Arigoni F (2006) A serpin from the gut bacterium Bifidobacterium longum inhibits eukaryotic elastase-like serine proteases. J Biol Chem 281:17246–17252
Jiang H, Kanost MR (1997) Characterization and functional analysis of 12 naturally occurring reactive site variants of serpin-1 from Manduca sexta. J Biol Chem 272:1082–1087
Jiang H, Mulnix AB, Kanost MR (1995) Expression and characterization of recombinant Manduca sexta serpin-1B and site-directed mutants that change its inhibitory selectivity. Insect Biochem Mol Biol 25:1093–1100
Jiang H, Wang Y, Huang Y, Mulnix AB, Kadel J, Cole K, Kanost MR (1996) Organization of serpin gene-1 from Manduca sexta. Evolution of a family of alternate exons encoding the reactive site loop. J Biol Chem 271:28017–28023
Johnson DJ, Li W, Adams TE, Huntington JA (2006) Antithrombin-S195A factor Xa-heparin structure reveals the allosteric mechanism of antithrombin activation. EMBO J 25:2029–2037
Jongsma MA, Bolter C (1997) The adaption of insects to plant protease inhibitors. J Insect Physiol 43:885–895
Kaersgaard P, Hejgaard J (1979) Antigenic beer macromolecules: an experimental survey of purification methods. J Inst Brew 85:103–111
Kaiserman D, Bird PI (2005) Analysis of vertebrate genomes suggests a new model for clade B serpin evolution. BMC Genomics 6:167
Kang SH, Barak Y, Lamed R, Bayer EA, Morrison M (2006) The functional repertoire of prokaryote cellulosomes includes the serpin superfamily of serine proteinase inhibitors. Mol Microbiol 60:1344–1354
Kanost MR, Prasad SV, Wells MA (1989) Primary structure of a member of the serpin superfamily of proteinase inhibitors from an insect, Manduca sexta. J Biol Chem 264:965–972
Ke SH, Coombs GS, Tachias K, Navre M, Corey DR, Madison EL (1997) Distinguishing the specificities of closely related proteases. Role of P3 in substrate and inhibitor discrimination between tissue-type plasminogen activator and urokinase. J Biol Chem 272:16603–16609
Keates SE, Kostman TA, Anderson JD, Bailey BA (2003) Altered gene expression in three plant species in response to treatment with Nep1, a fungal protein that causes necrosis. Plant Physiol 132:1610–1622
Koller A, Washburn MP, Lange BM, Andon NL, Deciu C, Haynes PA, Hays L, Schieltz D, Ulaszek R, Wei J, Wolters D, Yates JR (2002) Proteomic survey of metabolic pathways in rice. Proc Natl Acad Sci USA 99:11969–11974
Komiyama T, Ray CA, Pickup DJ, Howard AD, Thornberry NA, Peterson EP, Salvesen G (1994) Inhibition of interleukin-1 beta converting enzyme by the cowpox virus serpin CrmA. An example of cross-class inhibition. J Biol Chem 269:19331–19337
Kreis M, Williamson M, Buxton B, Pywell J, Hejgaard J, Svendsen I (1987) Primary structure and differential expression of beta-amylase in normal and mutant barleys. Eur J Biochem 169:517–525
Krem MM, Di Cera E (2003) Conserved ser residues, the shutter region, and speciation in serpin evolution. J Biol Chem 278:37810–37814
Kreps JA, Wu YJ, Chang HS, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130:2129–2141
la Cour Petersen M, Hejgaard J, Thompson GA, Schulz A (2005) Cucurbit phloem serpins are graft-transmissible and appear to be resistant to turnover in the sieve element–companion cell complex. J Exp Bot 56:3111–120
Law RHP, Zhang QW, McGowan S, Buckle AM, Silverman GA, Wong W, Rosado CJ, Langendorf CG, Pike RN, Bird PI, Whisstock JC (2006) An overview of the serpin superfamily. Genome Biol 7:216
Lawrence DA, Ginsburg D, Day DE, Berkenpas MB, Verhamme IM, Kvassman JO, Shore JD (1995) Serpin-protease complexes are trapped as stable acyl-enzyme intermediates. J Biol Chem 270:25309–25312
Leicht M, Long GL, Chandra T, Kurachi K, Kidd VJ, Mace M Jr., Davie EW, Woo SL (1982) Sequence homology and structural comparison between the chromosomal human alpha 1-antitrypsin and chicken ovalbumin genes. Nature 297:655–659
Li J, Wang Z, Canagarajah B, Jiang H, Kanost M, Goldsmith EJ (1999) The structure of active serpin 1K from Manduca sexta. Struct Fold Des 7:103–109
Li W, Johnson DJ, Esmon CT, Huntington JA (2004) Structure of the antithrombin–thrombin–heparin ternary complex reveals the antithrombotic mechanism of heparin. Nat Struct Mol Biol 11:857–862
Ligoxygakis P, Roth S, Reichhart JM (2003) A serpin regulates dorsal–ventral axis formation in the Drosophila embryo. Curr Biol 13:2097–2102
Lough TJ, Lucas WJ (2006) Integrative plant biology: role of phloem long-distance macromolecular trafficking. Annu Rev Plant Biol 57:203–232
Luke CJ, Pak SC, Askew DJ, Askew YS, Smith JE, Silverman GA (2006) Selective conservation of the RSL-encoding, proteinase inhibitory-type, clade L serpins in Caenorhabditis species. Front Biosci 11:581–594
Lusk LT, Goldstein H, Ryder D (1995) Independent role of beer proteins, melanoidins and polysaccharides in foam formation. J Am Soc Brew Chem 53:93–103
Marshall CJ (1993) Evolutionary relationships among the serpins. Philos Trans Royal Soc London B Biol Sci 342:101–119
Matheson NR, van Halbeek H, Travis J (1991) Evidence for a tetrahedral intermediate complex during serpin–proteinase interactions. J Biol Chem 266:13489–13491
McGowan S, Buckle AM, Irving JA, Ong PC, Bashtannyk-Puhalovich TA, Kan WT, Henderson KN, Bulynko YA, Popova EY, Smith AI, Bottomley SP, Rossjohn J, Grigoryev SA, Pike RN, Whisstock JC (2006) X-ray crystal structure of MENT: evidence for functional loop-sheet polymers in chromatin condensation. EMBO J 25:3144–3155
Mellet P, Boudier C, Mely Y, Bieth JG (1998) Stopped flow fluorescence energy transfer measurement of the rate constants describing the reversible formation and the irreversible rearrangement of the elastase-alpha1-proteinase inhibitor complex. J Biol Chem 273:9119–9123
Mills ENC, Kauffman JA, Morgan MRA, Field JM, Hejgaard J, Proudlove MO, Onishi A (1998) Immunological study of hydrophobic polypeptides in beer. J Agric Food Chem 46:4475–4483
Murray C, Christeller JT (1995) Purification of a trypsin inhibitor (PFTI) from pumpkin fruit phloem exudate and isolation of putative trypsin and chymotrypsin inhibitor cDNA clones. Biol Chem 376:281–287
Nielsen G, Johansen H, Jensen J, Hejgaard J (1983) Localization on barley chromosome 4 of genes coding for β-amylase (Bmy1) and protein Z (Paz1). Barley Genet Newslett 13:55–57
Nixon JE, Wang A, Field J, Morrison HG, McArthur AG, Sogin ML, Loftus BJ, Samuelson J (2002) Evidence for lateral transfer of genes encoding ferredoxins, nitroreductases, NADH oxidase, and alcohol dehydrogenase 3 from anaerobic prokaryotes to Giardia lamblia and Entamoeba histolytica. Eukaryot Cell 1:181–190
O’Malley KM, Nair SA, Rubin H, Cooperman BS (1997) The kinetic mechanism of serpin-proteinase complex formation. An intermediate between the Michaelis complex and the inhibited complex. J Biol Chem 272:5354–5359
Oley M, Letzel MC, Ragg H (2004) Inhibition of furin by serpin Spn4A from Drosophila melanogaster. FEBS Lett 577:165–169
Olson ST, Swanson R, Day D, Verhamme I, Kvassman J, Shore JD (2001) Resolution of Michaelis complex, acylation, and conformational change steps in the reactions of the serpin, plasminogen activator inhibitor-1, with tissue plasminogen activator and trypsin. Biochemistry 40:11742–11756
Ostergaard H, Rasmussen SK, Roberts TH, Hejgaard J (2000) Inhibitory serpins from wheat grain with reactive centers resembling glutamine-rich repeats of prolamin storage proteins—cloning and characterization of five major molecular forms. J Biol Chem 275:33272–33279
Ostergaard O, Melchior S, Roepstorff P, Svensson B (2002) Initial proteome analysis of mature barley seeds and malt. Proteomics 2:733–739
Ostergaard O, Finnie C, Laugesen S, Roepstorff P, Svensson B (2004) Proteome analysis of barley seeds: identification of major proteins from two-dimensional gels (pl 4–7). Proteomics 4:2437–2447
Pak SC, Kumar V, Tsu C, Luke CJ, Askew YS, Askew DJ, Mills DR, Bromme D, Silverman GA (2004) SRP-2 is a cross-class inhibitor that participates in postembryonic development of the nematode Caenorhabditis elegans: initial characterization of the clade L serpins. J Biol Chem 279:15448–15459
Pak SC, Tsu C, Luke CJ, Askew YS, Silverman GA (2006) The Caenorhabditis elegans muscle specific serpin, SRP-3, neutralizes chymotrypsin-like serine peptidases. Biochemistry 45:4474–4480
Patston PA, Gettins P, Beechem J, Schapira M (1991) Mechanism of serpin action: evidence that C1 inhibitor functions as a suicide substrate. Biochemistry 30:8876–8882
Pearce MC, Pike RN, Lesk AM, Bottomley SP (2007) Serpin conformations molecular and cellular aspects of the serpinopathies and disorders in serpin activity. Molecular and cellular aspects of the serpinopathies and disorder in serpin activity. World Scientific, New Jersey, pp 35–66
Perrocheau L, Rogniaux H, Boivin P, Marion D (2005) Probing heat-stable water-soluble proteins from barley to malt and beer. Proteomics 5:2849–2858
Perron MJ, Blouse GE, Shore JD (2003) Distortion of the catalytic domain of tissue-type plasminogen activator by plasminogen activator inhibitor-1 coincides with the formation of stable serpin–proteinase complexes. J Biol Chem 278:48197–48203
Peterson FC, Gordon NC, Gettins PG (2000) Formation of a noncovalent serpin–proteinase complex involves no conformational change in the serpin. Use of 1H-15N HSQC NMR as a sensitive nonperturbing monitor of conformation. Biochemistry 39:11884–11892
Plotnick MI, Schechter NM, Wang ZM, Liu X, Rubin H (1997) Role of the P6-P3 region of the serpin reactive loop in the formation and breakdown of the inhibitory complex. Biochemistry 36:14601–14608
Potempa J, Shieh BH, Travis J (1988) Alpha-2-antiplasmin: a serpin with two separate but overlapping reactive sites. Science 241:699–700
Rasmussen SK (1993) A gene coding for a new plant serpin. Biochim Biophys Acta 1172:151–154
Rasmussen SK, Dahl SW, Norgard A, Hejgaard J (1996) A recombinant wheat serpin with inhibitory activity. Plant Mol Biol 30:673–677
Raven PH, Evert RF, Eichhorn SE (2005) Biology of plants. W.H. Freeman and Company, New York
Rawlings ND, Tolle DP, Barrett AJ (2004) Evolutionary families of peptidase inhibitors. Biochem J 378:705–716
Ray CA, Black RA, Kronheim SR, Greenstreet TA, Sleath PR, Salvesen GS, Pickup DJ (1992) Viral inhibition of inflammation: cowpox virus encodes an inhibitor of the interleukin-1 beta converting enzyme. Cell 69:597–604
Reichhart JM (2005) Tip of another iceberg: Drosophila serpins. Trends Cell Biol 15:659–665
Rezaie AR (2006) Pentasaccharide enhances the inactivation of factor xa by antithrombin by promoting the assembly of a Michaelis-type intermediate complex. Demonstration by rapid kinetic, surface plasmon resonance, and competitive binding studies. Biochemistry 45:5324–5329
Riahi Y, Siman-Tov R, Ankri S (2004) Molecular cloning, expression and characterization of a serine proteinase inhibitor gene from Entamoeba histolytica. Mol Biochem Parasitol 133:153–162
Roberts TH, Marttila S, Rasmussen SK, Hejgaard J (2003) Differential gene expression for suicide-substrate serine proteinase inhibitors (serpins) in vegetative and grain tissues of barley. J Exp Bot 54:2251–2263
Roberts TH, Hejgaard J, Saunders NFW, Cavicchioli R, Curmi PMG (2004) Serpins in unicellular Eukarya, Archaea, and Bacteria: sequence analysis and evolution. J Mol Evol 59:437–447
Robertson AS, Belorgey D, Lilley KS, Lomas DA, Gubb D, Dafforn TR (2003) Characterization of the necrotic protein that regulates the Toll-mediated immune response in Drosophila. J Biol Chem 278:6175–6180
Rollini P, Fournier RE (1997) A 370-kb cosmid contig of the serpin gene cluster on human chromosome 14q32.1: molecular linkage of the genes encoding alpha 1-antichymotrypsin, protein C inhibitor, kallistatin, alpha 1-antitrypsin, and corticosteroid-binding globulin. Genomics 46:409–415
Rosenkrands I, Hejgaard J, Rasmussen SK, Bjorn SE (1994) Serpins from wheat grain. FEBS Lett 343:75–80
Ruuska SA, Girke T, Benning C, Ohlrogge JB (2002) Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14:1191–1206
Schechter NM, Plotnick MI (2004) Measurement of the kinetic parameters mediating protease-serpin inhibition. Methods 32:159–168
Schick C, Pemberton PA, Shi GP, Kamachi Y, Cataltepe S, Bartuski AJ, Gornstein ER, Bromme D, Chapman HA, Silverman GA (1998) Cross-class inhibition of the cysteine proteinases cathepsins K, L, and S by the serpin squamous cell carcinoma antigen 1: a kinetic analysis. Biochemistry 37:5258–5266
Schwartz BS, Espana F (1999) Two distinct urokinase-serpin interactions regulate the initiation of cell surface-associated plasminogen activation. J Biol Chem 274:15278–15283
Scott FL, Eyre HJ, Lioumi M, Ragoussis J, Irving JA, Sutherland GA, Bird PI (1999) Human ovalbumin serpin evolution: phylogenic analysis, gene organization, and identification of new PI8-related genes suggest that two interchromosomal and several intrachromosomal duplications generated the gene clusters at 18q21-q23 and 6p25. Genomics 62:490–499
Shieh BH, Potempa J, Travis J (1989) The use of alpha 2-antiplasmin as a model for the demonstration of complex reversibility in serpins. J Biol Chem 264:13420–13423
Silverman GA, Lomas DA (eds) (2007) Molecular and cellular aspects of the serpinopathies and disorders in serpin activity. World Scientific, New Jersey
Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, Gettins PGW, Irving JA, Lomas DA, Luke CJ, Moyer RW, Pemberton PA, Remold-O'Donnell E, Salvesen GS, Travis J, Whisstock JC (2001) The serpins are an expanding superfamily of structurally similar but functionally diverse proteins—evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J Biol Chem 276:33293–33296
Silverman GA, Whisstock JC, Askew DJ, Pak SC, Luke CJ, Cataltepe S, Irving JA, Bird PI (2004) Human clade B serpins (ov-serpins) belong to a cohort of evolutionarily dispersed intracellular proteinase inhibitor clades that protect cells from promiscuous proteolysis. Cell Mol Life Sci 61:301–325
Simonovic M, Gettins PGW, Volz K (2000) Crystal structure of viral serpin crmA provides insights into its mechanism of cysteine proteinase inhibition. Protein Sci 9:1423–1427
Sorensen MB, Cameron-Mills V, Brandt A (1989) Transcriptional and post-transcriptional regulation of gene expression in developing barley endosperm. Mol Gen Genet 217:195–201
Sprecher CA, Morgenstern KA, Mathewes S, Dahlen JR, Schrader SK, Foster DC, Kisiel W (1995) Molecular cloning, expression, and partial characterization of two novel members of the ovalbumin family of serine proteinase inhibitors. J Biol Chem 270:29854–29861
Springhetti EM, Istomina NE, Whisstock JC, Nikitina T, Woodcock CL, Grigoryev SA (2003) Role of the M-loop and reactive center loop domains in the folding and bridging of nucleosome arrays by MENT. J Biol Chem 278:43384–43393
Stein PE, Tewkesbury DA, Carrell RW (1989) Ovalbumin and angiotensinogen lack serpin S-R conformational change. Biochem J 262:103–107
Stratikos E, Gettins PG (1997) Major proteinase movement upon stable serpin–proteinase complex formation. Proc Natl Acad Sci USA 94:453–458
Sun J, Ooms L, Bird CH, Sutton VR, Trapani JA, Bird PI (1997) A new family of 10 murine ovalbumin serpins includes two homologs of proteinase inhibitor 8 and two homologs of the granzyme B inhibitor (proteinase inhibitor 9). J Biol Chem 272:15434–15441
Sun J, Stephens R, Mirza G, Kanai H, Ragoussis J, Bird PI (1998) A serpin gene cluster on human chromosome 6p25 contains PI6, PI9 and ELANH2 which have a common structure almost identical to the 18q21 ovalbumin serpin genes. Cytogenet Cell Genet 82:273–277
Sun J, Whisstock JC, Harriott P, Walker B, Novak A, Thompson PE, Smith AI, Bird PI (2001) Importance of the P4′ residue in human granzyme B inhibitors and substrates revealed by scanning mutagenesis of the proteinase inhibitor 9 reactive center loop. J Biol Chem 276:15177–15184
Swofford DL (2002) PAUP. Phylogenetic analysis using parsimony (and other methods). Sinauer and Associates, Sunderland, Massachusetts
Tanaka N, Fujita M, Handa H, Murayama S, Uemura M, Kawamura Y, Mitsui T, Mikami S, Tozawa Y, Yoshinaga T, Komatsu S (2004) Proteomics of the rice cell: systematic identification of the protein populations in subcellular compartments. Mol Gen Genet 271:566–576
Thomas JC, Wasmann CC, Echt C, Dunn RL, Bohnert HJ, McCoy TJ (1994) Introduction and expression of an insect proteinase inhibitor in alfalfa (Medicago sativa L.). Plant Cell Rep 14:31–36
Thomas JC, Adams DG, Keppenne VD, Wasmann CC, Brown JK, Kanost MR, Bohnert HJ (1995a) Manduca sexta encoded protease inhibitors expressed in Nicotiana tabacum provide protection against insects. Plant Physiol Biochem 33:611–614
Thomas JC, Adams DG, Keppenne VD, Wasmann CC, Brown JK, Kanost MR, Bohnert HJ (1995b) Protease inhibitors of Manduca sexta expressed in transgenic cotton. Plant Cell Rep 14:758–762
Thomas L, Moore NR, Miller S, Booth NA (2007) The C-terminus of alpha2-antiplasmin interacts with endothelial cells. Br J Haematol 136:472–479
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res 25:4876–4882
Trobacher CP, Senatore A, Greenwood JS (2006) Masterminds or minions? Cysteine proteinases in plant programmed cell death. Can J Bot 84:651–667
Tsutsui Y, Liu L, Gershenson A, Wintrode PL (2006) The conformational dynamics of a metastable serpin studied by hydrogen exchange and mass spectrometry. Biochemistry 45:6561–6569
Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, Schein J, Sterck L, Aerts A, Bhalerao RR, Bhalerao RP, Blaudez D, Boerjan W, Brun A, Brunner A, Busov V, Campbell M, Carlson J, Chalot M, Chapman J, Chen GL, Cooper D, Coutinho PM, Couturier J, Covert S, Cronk Q, Cunningham R, Davis J, Degroeve S, Dejardin A, Depamphilis C, Detter J, Dirks B, Dubchak I, Duplessis S, Ehlting J, Ellis B, Gendler K, Goodstein D, Gribskov M, Grimwood J, Groover A, Gunter L, Hamberger B, Heinze B, Helariutta Y, Henrissat B, Holligan D, Holt R, Huang W, Islam-Faridi N, Jones S, Jones-Rhoades M, Jorgensen R, Joshi C, Kangasjarvi J, Karlsson J, Kelleher C, Kirkpatrick R, Kirst M, Kohler A, Kalluri U, Larimer F, Leebens-Mack J, Leple JC, Locascio P, Lou Y, Lucas S, Martin F, Montanini B, Napoli C, Nelson DR, Nelson C, Nieminen K, Nilsson O, Pereda V, Peter G, Philippe R, Pilate G, Poliakov A, Razumovskaya J, Richardson P, Rinaldi C, Ritland K, Rouze P, Ryaboy D, Schmutz J, Schrader J, Segerman B, Shin H, Siddiqui A, Sterky F, Terry A, Tsai CJ, Uberbacher E, Unneberg P, Vahala J, Wall K, Wessler S, Yang G, Yin T, Douglas C, Marra M, Sandberg G, Van de Peer Y, Rokhsar D (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313:1596–1604
Vercammen D, Belenghi B, van de Cotte B, Beunens T, Gavigan JA, De Rycke R, Brackenier A, Inze D, Harris JL, Van Breusegem F (2006) Serpin1 of Arabidopsis thaliana is a suicide inhibitor for Metacaspase 9. J Mol Biol 364:625–636
Walz C, Giavalisco P, Schad M, Juenger M, Klose J, Kehr J (2004) Proteomics of curcurbit phloem exudate reveals a network of defence proteins. Phytochemistry 65:1795–1804
Whisstock JC, Bottomley SP (2006) Molecular gymnastics: serpin structure, folding and misfolding. Curr Opin Struct Biol 16:761–768
Whisstock J, Skinner R, Lesk AM (1998) An atlas of serpin conformations. Trends Biochem Sci 23:63–67
Whisstock JC, Irving JA, Bottomley SP, Pike RN, Lesk AM (1999) Serpins in the Caenorhabditis elegans genome. Proteins 36:31–41
Wilczynska M, Fa M, Ohlsson PI, Ny T (1995) The inhibition mechanism of serpins. Evidence that the mobile reactive center loop is cleaved in the native protease–inhibitor complex. J Biol Chem 270:29652–29655
Xu X, Zhang SS, Barnstable CJ, Tombran-Tink J (2006) Molecular phylogeny of the antiangiogenic and neurotrophic serpin, pigment epithelium derived factor in vertebrates. BMC Genomics 7:248
Ye S, Cech AL, Belmares R, Bergstrom RC, Tong YR, Corey DR, Kanost MR, Goldsmith EJ (2001) The structure of a Michaelis serpin–protease complex. Nat Struct Biol 8:979–983
Yokoi S, Yamashiro K, Kunitake N, Koshino S (1994) Hydrophobic beer proteins and their function in beer foam. J Am Soc Brew Chem 52:123–126
Yoo BC, Aoki K, Xiang Y, Campbell LR, Hull RJ, Xoconostle-Cazares B, Monzer J, Lee JY, Ullman DE, Lucas WJ (2000) Characterization of Cucurbita maxima phloem serpin-1 (CmPS-1)—a developmentally regulated elastase inhibitor. J Biol Chem 275:35122–35128
Zang XX, Maizels RM (2001) Serine proteinase inhibitors from nematodes and the arms race between host and pathogen. Trends Biochem Sci 26:191–197
Zhang Q, Law R, Buckle AM, Cabrita L, McGowan S, Irving JA, Faux NG, Lesk AM, Bottomley SP, Whisstock JC (2007) Serpins in prokaryotes. In: Silverman GA, Lomas DA (eds) Molecular and cellular aspects of the serpinopathies and disorders in serpin activity. World Scientific, New Jersey, pp 131–162
Zhou Q, Snipas S, Orth K, Muzio M, Dixit VM, Salvesen GS (1997) Target protease specificity of the viral serpin CrmA. Analysis of five caspases. J Biol Chem 272:7797–7800
Zhou AW, Carrell RW, Huntington JA (2001) The serpin inhibitory mechanism is critically dependent on the length of the reactive center loop. J Biol Chem 276:27541–27547
Zhou A, Wei Z, Read RJ, Carrell RW (2006) Structural mechanism for the carriage and release of thyroxine in the blood. Proc Natl Acad Sci USA 103:13321–13326
Zhu YF, Wang Y, Gorman MJ, Jiang HB, Kanost MR (2003) Manduca sexta serpin-3 regulates prophenoloxidase activation in response to infection by inhibiting prophenoloxidase-activating proteinases. J Biol Chem 278:46556–46564
Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) Genevestigator. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136:2621–2632
Zou Z, Jiang H (2005) Manduca sexta serpin-6 regulates immune serine proteinases PAP-3 and HP8. cDNA cloning, protein expression, inhibition kinetics, and function elucidation. J Biol Chem 280:14341–14348
Acknowledgments
We thank Moreland Gibbs and David Briscoe (Macquarie University) for advice on the phylogenetic analysis; Roberts’ past and current research students Karlie Neilson, Tom Joss and Joon-Woo Ahn for helpful discussions; Robert Fluhr (Weizmann Institute) for insights into the possible roles of plant serpins; and Macquarie University for funding support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Roberts, T.H., Hejgaard, J. Serpins in plants and green algae. Funct Integr Genomics 8, 1–27 (2008). https://doi.org/10.1007/s10142-007-0059-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-007-0059-2