Abstract
Fish are the main source of the n-3 highly unsaturated fatty acids, which are crucial for human health. Their synthesis from C18 precursors is mediated by desaturases and elongases, but the activity of these enzymes has not been conclusively established in marine fish species. This study reports the cloning, tissue expression, and functional characterization of a sea bass (Dicentrarchus labrax L.) Δ6-desaturase and one of its splicing variants. Two cDNAs with open reading frames of 1,346 and 1,354 bp were cloned and named D6D and D6D-V, respectively. Both deduced protein sequences (445 and 387 amino acids, respectively) contained two transmembrane regions and the N-terminal cytochrome b5 domain with the HPGG motif characteristic of microsomal desaturases. D6D presents three histidine-rich regions, whereas in D6D-V, an insertion of eight nucleotides in the boundaries of exons 10 and 11 modified the third histidine-rich domain and led to insertion of a premature STOP codon, resulting in a shorter predicted protein. Quantitative real-time polymerase chain reaction assay of gene expression showed that D6D was highly expressed in the brain and intestine, and to a lesser extent, in muscle and liver; meanwhile, D6D-V was expressed in all tissues tested, but at level at least 200-fold lower than D6D. Functional analysis in yeast showed that sea bass D6D encodes a fully functional Δ6-desaturase with no residual Δ5-desaturase activity. This desaturase does not exhibit a clear preference for n-3 versus n-6 C18 substrates. Interestingly, D6D-V is a nonfunctional protein, suggesting that the C-terminal end is indispensable for protein activity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
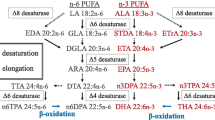
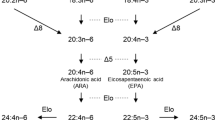
The decrease in worldwide fisheries recorded in the last years (Tidwell and Allan 2001; Pauly et al. 2005; Brunner et al. 2009), in conjunction with aquaculture expansion (FAO 2006), leads to the utilization of vegetable oils rich in C18 PUFA in aquafeeds (Bell and Waagbø 2008). However, the capacity of marine fish species to bioconvert the vegetable C18 precursors linoleic (LN; 18:2n-6) and linolenic (LNA; 18:3n-3) acids into long-chain highly unsaturated fatty acids (HUFA; chain length ≥C20 with ≥3 double bonds), eicosapentaenoic (EPA; 20:5n-3), docosahexaenoic (DHA; 22:6n-3), and arachidonic (AA; 20:4n-6) acids is controversial. In freshwater species such as Atlantic salmon (Salmo salar) or rainbow trout (Oncorhynchus mykiss), the desaturation/elongation pathway is under nutritional regulation. Thus, when these fish species are fed a diet lacking fish oil, they are capable of modulating the activity of the enzymes to produce the long-chain HUFAs (Tocher et al. 2006; Zheng et al. 2005). In contrast, most marine fish species, when deprived of long-chain HUFAs, are not capable of completing the desaturation/elongation steps that allow the synthesis of EPA, DHA, and AA. Therefore, they have a dietary requirement for long-chain HUFA. In this regard, the low level of EPA and DHA biosynthesis from C18 vegetable precursors in sea bream hepatocytes has been related to a low Δ5-desaturase activity (Tocher and Ghioni 1999), whereas in the turbot TF cell line, the HUFA elongation/desaturation pathway seems to be impaired in the C18 to C20 elongation step (Ghioni et al. 1999). In the particular case of sea bass (Dicentrarchus labrax L.), the flesh fatty acid profile is impoverished in long-chain n-3 and n-6 fatty acids when fish are fed vegetable-oil diets (Mourente et al. 2005b; Mourente and Bell 2006), with a concomitant decrease of nutritional value. However, previous studies have proved that this fish species possess the desaturase/elongase activities necessary to produce EPA, AA, and DHA from C18 fatty acids (Mourente and Dick 2002; Mourente et al. 2005a), even if the conversion rates are extremely low. In this context, it is important to better characterize the different enzymes (elongases and desaturases) involved in sea bass HUFA synthesis pathway in order to improve the use of vegetable oils lacking long-chain HUFA for marine fish aquafeeds.
Fatty acid desaturases introduce double bounds in selected positions of the acyl chains (Cook 1996). Particularly, Δ6-desaturases are the first enzymes involved in the biosynthesis of long-chain HUFA from the C18 n-3 and n-6 precursors. Over the last few years, some desaturases have been isolated and characterized from different fish species (Supplementary Table 1). Thus, full-length cDNAs encoding Δ6-desaturases from rainbow trout (O. mykiss), sea bream (Sparus aurata), Atlantic salmon (S. salar), cherry salmon (Oncorhynchus masou), carp (Cyprinus carpio), turbot (Scophthalmus maximus), Nile tilapia (Oreochromis niloticus), cod (Gadus morhua), and cobia (Rachycentron canadum) are now available. In addition, Δ5-desaturases from Atlantic salmon and cherry salmon, as well as a zebrafish (Danio rerio) desaturase with both Δ5/Δ6 activities, have been characterized (Hastings et al. 2001). These enzymes contain three histidine boxes, up to three hydrophobic domains, and an N-terminal cytochrome b5-like domain (Shanklin et al. 1994). A high percentage of identity (around 90%) between the Δ5-and Δ6-desaturase cDNAs of the same species have been reported (Zheng et al. 2005).
To get deeper insight into the desaturation capacity of sea bass (D. labrax L.), the objective of this study was to clone the Δ6-desaturase, to survey its expression profile in different tissues, and finally to functionally characterize this enzyme in order to elucidate its role in the HUFA synthesis pathway of this fish species.
Materials and Methods
Cloning of Putative Fatty Acyl Desaturases from Sea Bass
Total RNA was extracted from sea bass larvae (day 45 post hatching) fed with a fish-meal/fish-oil diet using the TRIzol® reagent (Invitrogen, Ltd., USA). Five micrograms of RNA were reverse-transcribed using iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA, USA) primed by the oligonucleotide Reverse 0 (Table 1). Available fish desaturase sequences (Supplementary Table 1) were aligned to design a degenerate primer named Forward 1 (Table 1). Primer Reverse 1 was designed after alignment of sequences contained in sea bass partial clones obtained from multi-tissue cDNA libraries described in sigenae databases (cdn22p0006m07, cdnp0002m05, and cdn24p0001j11; http://public-contigbrowser.sigenae.org:9090). These primers were used for polymerase chain reaction (PCR) amplification in a TC-512 instrument (Techne, UK) under the following conditions: initial denaturation 10 min at 94°C, 37 cycles of 1 min denaturation at 94°C, 45 s annealing at 55°C, and 1 min 30 s at 72°C for elongation, and a final extension at 72°C for 10 min. A nested PCR was then performed using primers Forward 2 and Reverse 2 (Table 1) to improve amplification specificity. Fragment at the expected length was then purified, cloned into the pCRTM II-TOPO plasmid, and used for transformation of ONE Shot™ competent cells (Invitrogen, Ltd., USA). The fragment was sequenced and found to be 845 bp long.
From this sequence, the specific primers 5′RACE1, 5′RACE2, and 5′RACE3 (Table 1) were designed and used to obtain the 5′UTR region with the 5′/3′RACE kit (Roche, USA) following the manufacturer's instructions. After cloning and sequencing the isolated DNA fragment as described above, the 5′UTR specific primer Forward 3 (Table 1) was designed. This primer was combined with the 3′UTR Reverse 3 primer, designed according to sequences contained in sea bass available clones, to obtain the full-length open reading frames (ORF) by using the Advantage® cDNA Polymerase Mix (Clontech Laboratories, Inc., USA) under the following conditions: 10 min at 94°C, 40 cycles of 1 min denaturation at 94°C, 45 s annealing at 62°C, and 1 min 45 s at 72°C for elongation, and a final extension at 72°C for 10 min. Cloning and sequencing of the PCR products revealed the presence of two highly homologous cDNAs that were named sea bass-D6D and sea bass-D6D-V, respectively. All PCR and RACE products were sequenced by Millegen (France).
For comparative analyses, the deduced amino acid sequences of Δ6-desaturases from various species were aligned using ClustalX, and sequence phylogenies were predicted using the neighbor-joining method (Saitou and Nei 1987).
Sea Bass Tissue RNA Extraction and Quantitative Real-Time PCR (qPCR)
Nine tissues (brain, pyloric caeca, anterior intestine, posterior intestine, muscle, liver, kidney, heart, and skin) of two adult fish fed with a commercial fish-meal/fish-oil diet were isolated, and total RNA extraction was immediately performed using TRIzol® reagent. Twelve micrograms of total RNA were reverse-transcribed into cDNA by using the QuantiTect Reverse Transcription Kit (QIAGEN, Germany). Expression of both sea bass-D6D and sea bass-D6D-V was studied by quantitative real-time (qPCR) using specific primers (Table 1). PCR product sizes for sea bass-D6D and D6D-V were 138 and 150 bp, respectively. Amplification from cDNA samples was performed using iQ™SYBR® Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA) under the following conditions: 1 min 30 s of initial denaturation at 95°C, 45 cycles of 30 s at 95°C, and 1 min at 65°C. This was followed by analysis of melting curve of the PCR products to confirm single PCR products. EF1 was used as a housekeeping gene for normalization of mRNA levels. Thermal cycling and fluorescence detection were conducted in a MyiQ single color real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA, USA).
Heterologous Expression of Sea Bass Desaturase cDNA in Yeast
Forward 3 and Reverse 3 primers (Table 1) were used for the amplification of the ORFs of each desaturase transcript (D6D and D6D-V) from cDNA template synthesized from larvae total RNA. PCR products were cloned into the pYES2.1 TOPO expression plasmid (Invitrogen) to obtain the recombinant plasmids pYESD6D and pYESD6D-V, which were then used to transform TOP10F′ Escherichia coli (Invitrogen). After sequence confirmation, Saccharomyces cerevisiae strain INVSc1 (Invitrogen) was transformed with one of the recombinant plasmids according to the pYES2.1TOPO®TA Expression Kit manufacturer's instructions. Yeast transformed with empty pYES2.1 plasmid (pYES2-Empty) was used as control. Transformants containing pYES2-Empty, pYESD6D, or pYESD6D-V were selected on solid minimal medium plates lacking uracil.
For functional expression, cultures were grown at 25°C in minimal medium containing 0.67% (w/v) nitrogen base, 0.19% (w/v) dropout medium, and 2% raffinose (w/v). Expression of the transgene was induced at an OD600 of 0.2 by supplementing with galactose to 2% (w/v) and tergitol to 1% (w/v) final concentration. At that time, the appropriate fatty acids were added as follows (final concentration): 0.5 mM of 18:3n-3, 0.5 mM of 18:2n-6, or 1 mM of 20:4n-3 (eicosatetraenoic acid (ETA)). Incubation was carried out at 25°C for 3 days in a shaking incubator.
Fatty Acid Analysis
Yeast cells were harvested by centrifugation (10,000 g, 15 min). Total lipid extraction was performed according to Folch et al. (1957) with dichloromethane replacing chloroform. Briefly, cells were homogenized in 2 ml of 2:1 dichloromethane:methanol (v/v) containing 0.01% butylated hydroxytoluene. Cell debris was discarded by centrifugation (14,000 g, 10 min, 4°C). Eight hundred microliters of NaCl 0.75% were added to the supernatant, and samples were decanted for 1 h at RT. Then, the bottom phase was transferred into a microcentrifugation tube for saponification (90°C, 3 min) using 0.5 ml 2 M KOH-methanol. Two milliliters of hexane were added, and after centrifugation (630 g, 10 min), the supernatant was recovered, and the hexane evaporated. Five hundred microliters of 6 N HCl and 2 ml of hexane were added, and after centrifugation (630 g, 10 min), the supernatant was recovered and evaporated. To prepare fatty acid methyl esters (FAMEs), 1 ml HCl 2.5% in MeOH was added (90°C, 3 min), and after addition of 1 ml double-distilled water and of 2 ml hexane, samples were centrifuged (630 g, 10 min), and the supernatant was recovered, evaporated, and 0.5 ml of hexane was added to the tube. FAMEs were separated by GLC (Clarus500 Perkin-Elmer with a flame ionization detector, BPX 70 capillary column: 30 m × 0.22 mm i.d. × 0.25 µm film thickness). The percentage of conversion (conversion rate) of each FA substrate was calculated as follows: 100 × (product area/(substrate area + product area)).
Results
Cloning of Putative Fatty Acyl Desaturase from Sea Bass
Starting from partial cDNA sequences obtained from several clones of multi-tissue libraries, the sequence of a putative desaturase transcript was completed by 5′ and 3′RACE. This allowed us to identify two transcripts of 1,346 and 1,354 nucleotides, respectively, named sea bass-D6D (GenBank accession no. EU647692) and sea bass-D6D-V (GenBank accession no. EU439924). The alignment of the two cDNA sequences (Fig. 1) revealed an insertion of eight nucleotides at the boundaries of exon 10 and exon 11 in the D6D-V cDNA, which interrupted the amino acid translation just after exon 10, and gave an amino acid sequence of 387 aa rather than 445 aa for D6D. The alignment of the Δ6-desaturase cDNA with the genomic sequences (obtained by 454 sequencing of the D. labrax BAC clone bassbac-102L2, GenBank accession no. FP671139) revealed 12 exons spanning 7,158 bp of genomic DNA (gDNA) as illustrated in Fig. 1. All splice sites except the donor site of intron 5 (GC..AG) exhibit the consensus splice site GT..AG. The cDNA aligned completely to the genomic DNA and resulted in only one nucleotide mismatch in the 3′UTR at position 59 (A in cDNA, G in gDNA; data not shown).
Exon/intron boundaries of sea bass D6D and D6D-V gene (exons 1–12). Location of different exons is shown by alternation of white/gray areas. 5′ and 3′ extremities of intronic sequences are indicated in lowercase letters. Hash in the boundaries of exon5/exon6 denotes a nonconsensus splice donor, and the size of each intron is indicated between brackets. Asterisks correspond to the nucleotides conserved between the two sea bass cDNA sequences. Predicted amino acid translation noted in the boundaries of exon10/exon11 shows the stop in the translation of the D6D-V cDNA
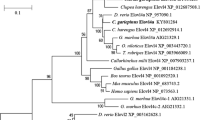
The comparison of the sea bass D6D amino acid sequences with zebrafish, mouse, and human Δ6-desaturase (Fig. 2) revealed an identity of 68%, 64%, and 65%, respectively. In addition, two transmembrane domains, the typical desaturase N-terminal cytochrome b5-like domain (HPGG), as well as the three characteristic membrane-bound histidine motifs (HDxGH, HFQHH, and QIEHH), were well conserved in the sea bass D6D amino acid sequence. In the alternative splicing transcript (D6D-V), the third histidine motif was replaced by the QIEHQ sequence (Fig. 2). A pair-wise comparison between fish and human desaturases (Supplementary Table 2) showed that sea bass amino acid sequences exhibit identity ranging from 82% to 94% with marine fish sequences, while values were lower with desaturases isolated from salmonids (around 76%) and zebrafish (68%). In addition, about half of the residues were identical between the sea bass and the human desaturases. Phylogenetic analysis comparing the two sea bass sequences with putative and characterized Δ5- and Δ6-desaturases from fish showed that sea bass D6D and D6D-V sequences cluster with cobia, sea bream, and turbot sequences rather than with salmonids, carp, and zebrafish desaturases (Fig. 3).
Alignment of the deduced amino acid sequences from sea bass D6D and D6D-V with desaturases from zebrafish (GenBank accession no. Q9DEX7), mouse (GenBank accession no. NP062673), and human (GenBank accession no. NP004256). The cytochrome b5-domain is dot-underlined. Putative transmembrane domains are shown in shaded areas, and three histidine-rich regions are framed. The asterisks indicate the heme-binding motif (HPGG), and two highly conserved histidine positions are in bold
Phylogeny of desaturase deduced amino acid sequences. The tree was constructed by the neighbor-joining method (Saitou and Nei 1987) using CLUSTALX and NJPLOT. The horizontal branch length is proportional to the amino acid substitution rate per site. The numbers represent the frequencies with which the tree topology presented here was replicated after 1,000 bootstrap iterations. Arrows highlight the location of sea bass Δ6-desaturase variants. Asterisks denote desaturase sequences that have not been functionally characterized
D6D and D6D-V Gene Expression in Several Sea Bass Tissues
D6D and D6D-V expression in sea bass tissues (brain, liver, pyloric caeca, anterior intestine, posterior intestine, muscle, kidney, heart, and skin) was monitored by quantitative PCR analysis (Fig. 4). Results show that D6D was mainly expressed in brain and intestine, followed by muscle and liver, while its expression was weak in the other tissues considered. Sea bass D6D-V expression was maximal in the anterior intestine, followed by posterior intestine, brain, and pyloric caeca. Interestingly, the absolute Ct values recorded for D6D (ranging from 19 to 22) in comparison to those obtained for D6D-V (between 28 and 30) showed that the splicing variant was expressed at a level at least 200-fold lower than D6D (data not shown).
Heterologous Expression of Sea Bass Desaturase cDNA in Yeast
To complete our analysis, we functionally characterized both newly identified sea bass desaturases by heterologous expression in S. cerevisiae (Table 2). Their activity and specificity was assessed by feeding recombinant yeast expressing pYESD6D or pYESD6D-V with potential Δ6 (18:2n-6 and 18:3n-3) and Δ5 (20:4n-3) desaturase substrates. The fatty acid composition of the yeast transformed with the empty vector showed the presence of the main fatty acids currently found in the yeast host (16:0, 16:1n-7, 18:0, and 18:1n-9), together with the exogenously fed fatty acids (see fatty acids profiles in Supplementary Fig. 1). In feeding experiments conducted with yeast containing pYESD6D, additional peaks were observed when 18:3n-3 and 18:2n-6 were added to the culture medium. These new fatty acids, stearidonic acid 18:4n-3 and gamma-linolenic acid 18:3n-6, respectively, were identified by gas chromatography analysis and based on comparison with chromatograms obtained in presence of pure standards (data not shown). The percentage of conversion ranged from 7.4 to 10.4 for the LNA and from 5.7 to 8.2 for the LN, according to different replicates of the feeding experiments (Table 2). This observation suggested that the sea bass D6D exhibits no significant preference between C18 n-6 and n-3 PUFAs. In addition, no desaturation products were observed in feeding experiments using yeast transformed with the plasmid pYESD6D-V. Moreover, no EPA was found when ETA (20:4n-3) was added in the induction medium for any of the constructs tested.
Discussion
Data presented in this work represent an important step in the understanding of the molecular basis of marine fish HUFA synthesis.
The D6D sea bass cDNA ORF encoded for a 445 amino acid protein which is more similar to sea bream (Seiliez et al. 2003), turbot (Zheng et al. 2004), and cobia (Zheng et al. 2009) than to salmonid sequences, as revealed by amino acid pair-wise comparison and phylogenetic analysis. The functional characterization of the protein in yeast confirmed the sea bass enzyme as a Δ6-desaturase, with no preference for either the n-3 substrate 18:3n-3 or the n-6 linoleate. Similar results have been observed after functional characterization of mammalian, fungal, and moss Δ6-desaturases (Aki et al. 1999; Cho et al. 1999; Kajikawa et al. 2004; Kaewsuwan et al. 2006), with the exception of the plant family Primula sp. Δ6-desaturase and of the Mantoniella squamata Δ6-desaturase that preferentially used the n-3 substrates (Sayanova et al. 2003; Hoffmann et al. 2008). In contrast, the selectivity for n-3 rather than for n-6 substrates is clear for the zebrafish bifunctional Δ5/Δ6-desaturase (Hastings et al. 2001), as well as for other fish Δ6-desaturases (O. mykiss, S. aurata, C. carpio, and Psetta maximus; Zheng et al. 2004). The percentage of conversion of 18:3n-3 to 18:4n-3 measured for the sea bass D6D ranged from 7.4 to 10.4 for the LNA and from 5.7 to 8.2 for the LN. This low desaturation capacity could explain, at least partially, sea bass impairment in the bioconversion of the C18 fatty acids provided by vegetable oil-based diets (Mourente et al. 2005a).
Some of the fish Δ6-desaturases characterized so far presented a low capacity to desaturate the Δ5 substrates 20:4n-3 and 20:3n-6, with conversion rates from 0.2% in rainbow trout (Zheng et al. 2004), sea bream (Zheng et al. 2004), or turbot (Zheng et al. 2004) to 2.3% in Atlantic salmon (Zheng et al. 2005) or cobia (Zheng et al. 2009). In our report, no Δ5 activity was detected in transgenic yeasts containing the D6D sea bass desaturase genes and cultivated in presence of 20:4n-3, indicating that like Atlantic cod (Tocher et al. 2006), the sea bass Δ6-desaturase protein was not able to desaturate ETA at the Δ5 position or that this activity was extremely low and thus was not detectable under the conditions tested.
Results obtained in this work, in conjunction with the existing literature concerning the controversial HUFA synthesis efficiency in marine fish species (Sargent et al. 2002), and especially in sea bass (Mourente and Dick 2002; Mourente et al. 2005a), allow us to hypothesize on the occurrence of Δ5-desaturase activity in this fish. Firstly, the low activity detected for sea bass liver (Mourente et al. 2005a) could be related to an unknown gene encoding for a Δ5-desaturase, and thus responsible for the trace amounts of EPA (20:5n-3) and AA (20:4n-6) detected in hepatocyte cultures in presence of C18 precursors (Mourente et al. 2005a). The existence of an enzyme with Δ5-desaturation capacity is supported by the fact that different desaturases with distinct Δ6 and Δ5 specificities have been reported for freshwater species (Hastings et al. 2005), even if the origin of the two independent desaturase genes is unclear (Zheng et al. 2004; Napier et al. 2003). However, the low capacity of marine fish species to synthesize long-chain HUFA from C18 precursors could also be related to an absence of the Δ5-desaturase gene. In this regard, to the best of our knowledge, no marine fish Δ5-desaturase proteins have been reported. Moreover, preliminary analysis of data produced by “shot gun” genome sequencing suggests the existence of a unique desaturase gene in sea bass (data not shown). If this is the real situation, it is possible to suggest that the zebrafish bifunctional desaturase might have evolved towards a unifunctional Δ6-desaturase by changes in catalytic residues, which have abolished the Δ5 activity in marine fish species. The difference in evolution of desaturase activities between freshwater and marine fish could be related to the fact that marine fish natural diet is rich in EPA and DHA, while freshwater fish diet is usually poor in long-chain HUFAs. Therefore, the freshwater species need to cover their requirements by the desaturation and elongation of C18 precursors (Sargent et al. 1995, 1999).
Recent studies in sea bass HUFA pathway have shown the occurrence of numerous splicing variants for the D6D gene. Thus, in addition to the D6D-V detected in the present work (GenBank accession no. EU439924), five other variants exist in the NCBI database (GenBank accession no. AM746707, AM746706, AM746705, AM746704, and AM746703). Despite the high homology between the two sequences identified in this study, the absence of desaturase activity for the D6D-V splicing variant indicated that the C-terminal end should be indispensable for the function of the protein. In addition, even if the physiological occurrence of all these splicing variants is uncertain, the qPCR analysis performed in our study demonstrated that D6D-V was transcribed in several tissues from juvenile sea bass (brain, intestine, muscle, liver, kidney, heart, and skin), at a much lower level than the gene encoding for the sea bass D6D. Concerning its role in regulation of Δ6-desaturase activity, it is possible that the D6D-V, like the other identified variants, decrease substrate availability by trapping C18 precursors into its preserved substrate-binding site. This would then diminish the apparent activity of the main protein. The strong expression of the D6D gene in brain is in concordance with previous results obtained for Atlantic cod (Tocher et al. 2006) and cobia (Zheng et al. 2009). In addition, the higher level of transcripts detected in intestine when compared to liver is in agreement with the level of enzymatic activity monitored in vitro by Mourente et al. (2005a), who have measured a stronger desaturation rates in enterocytes than in hepatocytes. Further studies will be needed to fully address the correlation between D6D expression levels and desaturation capacity in different sea bass organs.
In summary, we reported here the characterization of the sea bass Δ6-desaturase and showed its high level of expression in the brain and intestine. This work also demonstrates the existence of a predicted 387 amino acids nonfunctional splicing variant expressed in a wide range of sea bass tissues. The functional Δ6-desaturase did not exhibit selectivity towards n-3 or n-6 C18 substrates, and no residual Δ5-desaturase activity was detected after heterologous expression in yeast. The low conversion rate determined for sea bass Δ6-desaturase could, at least partially, explain the low HUFA biosynthesis when this species is fed vegetable-based diets. The discovery of this limited step within the HUFA synthesis process in sea bass might paves the way for further manipulation and increase of the activity of the pathway to allow efficient and effective use of vegetable oils in aquaculture. In fact, whilst the sea bass Δ6-desaturase cDNA sequence has a strong homology with those of other fish species exhibiting better conversion rates, we can not rule out the hypothesis that the low efficiency and dysfunction of this enzyme can be due to mutation(s) within the nucleotide sequence. Accordingly, since the use of transgenic fish to commercial ends is not well perceived by consumers at the present time, it is possible to consider the selection of sea bass family exhibiting better rates of Δ6-desaturase activity as soon as gene polymorphism for this enzyme will be determined. In addition, regarding other potential limiting steps in sea bass HUFA synthesis pathway, such as bioconversion of ETA into EPA, other oil resources rich in HUFAs such as microalgae may be considered in aquafeeds.
References
Aki T, Shimada Y, Inagaki K, Higashimoto H, Kawamoto S, Shiget S, Ono K, Suzuki O (1999) Molecular cloning and functional characterisation of rat Δ6 fatty acid desaturase. Biochem Biophys Res Commun 255:575–579
Bell JG, Waagbø R (2008) Safe and nutritious aquaculture produce: benefits and risks of alternative sustainable aquafeeds. In: Holmer M, Black KD, Duarte CM, Marba N, Karakassis I (eds) Aquaculture in the ecosystem. Springer Verlag, Netherlands, pp 185–225
Brunner EJ, Jones PJ, Friel S, Bartley M (2009) Fish, human health and marine ecosystem health: policies in collision. Int J Epidemiol 38:93–100
Cho HP, Nakamura MT, Clarke SD (1999) Cloning, expression and nutritional regulation of the human Δ6-desaturase. J Biol Chem 274:471–477
Cook HW (1996) Fatty acid desaturation and chain elongation in eukaryote. In: Vance DE, Vance JE (eds) Biochemistry of lipids, lipoproteins and membranes, vol 129. Elsevier, Amsterdam, p 152
Folch J, Lees M, Sloane-Stanley GH (1957) A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem 226:497–509
Food and Agricultural Organisation (FAO) (2006). State of world aquaculture 2008. Fao Fisheries Technical Paper No. 500. FAO, Rome, p 134
Ghioni C, Tocher DR, Bell MV, Dick JR, Sargent JR (1999) Low C18 to C20 fatty acid elongase activity and limited conversion of stearidonic acid, 18:4n-3, to eicosapentaenoic acid, 20:5n-3, in a cell line from the turbot, Scophthalmus maximus. Biochim Biophys Acta 1437:170–181
Hastings N, Agaba M, Tocher DR, Leaver MJ, Dick JR, Sargent JR, Teale AJ (2001) A vertebrate fatty acid desaturase with Δ5 and Δ6 activities. Proc Natl Acad Sci USA 98:14304–14309
Hastings N, Agaba MK, Tocher DR, Zheng X, Dickson CA, Dick JR, Teale AJ (2005) Molecular cloning and functional characterization of fatty acyl desaturase and elongase cDNAs involved in the production of eicosapentaenoic and docosahexaenoic acids from α-linolenic acid in Atlantic salmon (Salmo salar). Mar Biotechnol 6:463–474
Hoffmann M, Wagner M, Abbadi A, Fulda M, Feussner I (2008) Metabolic engineering of ω3-very long chain polyunsaturated fatty acid production by an exclusively acyl-CoA-dependent pathway. J Biol Chem 283:22352–22362
Kaewsuwan S, Cahoon EB, Perroud P-F, Wiwat C, Panvisavas N, Quatrano RS, Cove DJ, Bunyapraphatsara N (2006) Identification and functional characterization of the moss Physcomitrella patens Δ5-desaturase gene involved in arachidonic and eicosapentaenoic acid biosynthesis. J Biol Chem 281:21988–21997
Kajikawa M, Yamato KT, Kohzu Y, Nojiri M, Sakuradani E, Shimizu S, Sakai Y, Fukuzawa H, Ohyama K (2004) Isolation and characterization of D6-desaturase, an ELO-like enzyme and D5-desaturase from the Liverwort Marchantia polymorpha and production of arachidonic and eicosapentaenoic acids in the methylotrophic yeast Pichia pastoris. Plant Mol Biol 54:335–352
Mourente G, Bell JG (2006) Partial replacement of dietary fish oil with blends of vegetable oils (rapeseed, linseed and palm oils) in diets for European sea bass (Dicentrarchus labrax L.) over a long term growth study: effects on muscle and liver fatty acid composition and effectiveness of a fish oil finishing diet. Comp Biochem Physiol B 145:389–399
Mourente G, Dick JR (2002) Influence of partial substitution of dietary fish oil by vegetable oils on the metabolism of [1-C14]18:3n-3 in isolated hepatocytes of European sea bass (Dicentrarchus labrax L.). Fish Physiol Biochem 26:297–308
Mourente G, Dick JR, Bell JG, Tocher DR (2005a) Effect of partial substitution of dietary fish oil by vegetable oils on desaturation and beta-oxidation of [1-C-14]18:3n-3 (LNA) and [1-C-14]20:5n-3 (EPA) in hepatocytes and enterocytes of European sea bass (Dicentrarchus labrax L.). Aquaculture 248:173–186
Mourente G, Good JE, Bell JG (2005b) Partial substitution of fish oil with rapeseed, linseed and olive oils in diets for European sea bass (Dicentrarchus labrax L.): effects on flesh fatty acid composition, plasma prostaglandins E2 and F2α, immune function and effectiveness of a fish oil finishing diet. Aquac Nutr 11:25–40
Napier JA, Michaelson LV, Sayanova O (2003) The role of cytochrome b5 fusion desaturases in the synthesis of polyunsaturated fatty acids. Prost Leukot Essent Fatty Acids 68:135–143
Pauly D, Watson R, Alder J (2005) Global trends in world fisheries: impacts on marine ecosystems and food security. Philos Trans R Soc Lond B Biol Sci 360:5–12
Saitou N, Nei M (1987) The neighbor-joining method. A new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sargent JR, Bell JR, Bell MV, Henderson RJ, Tocher DR (1995) Requirement criteria for essential fatty acids. J Appl Ichthyol 11:183–198
Sargent JR, Bell JG, McEvoy L, Tocher DR, Estevez A (1999) Recent developments in the essential fatty acid nutrition of fish. Aquaculture 177:191–199
Sargent JR, Tocher DR, Bell JG (2002) The lipids. In: Halver JE, Hardy RW (eds) Fish nutrition, 3rd edn. Academic, USA, pp 181–257
Sayanova OV, Beaudoin F, Michaelson LV, Shewry PR, Napier JA (2003) Identification of Primula fatty acid Δ6-desaturases with n-3 substrate preferences. FEBS Lett 542:100–104
Seiliez I, Panserat S, Corraze G, Kaushik S, Bergot P (2003) Cloning and nutritional regulation of a Δ6-desaturase-like enzyme in the marine teleost gilthead sea bream (Sparus aurata). Comp Biochem Physiol B 135:449–460
Shanklin J, Whittle E, Fox BG (1994) Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry 33:12787–12794
Tidwell JH, Allan GL (2001) Fish as food: aquaculture's contribution. Ecological and economic impacts and contributions of fish farming and capture fisheries. Embo Reports 2:958–963
Tocher DR, Ghioni C (1999) Fatty acid metabolism in marine fish: low activity of Δ5 desaturation in gilthead sea bream (Sparus aurata) cells. Lipids 34:433–440
Tocher D, Zheng X, Schlechtriem C, Hastings N, Dick J, Teale A (2006) Highly unsaturated fatty acid synthesis in marine fish: cloning, functional characterization, and nutritional regulation of fatty acyl Δ6-desaturase of Atlantic cod (Gadus morhua L.). Lipids 41:1003–1016
Zheng X, Seiliez I, Hastings N, Tocher DR, Panserat S, Dickson CA, Bergot P, Teale AJ (2004) Characterization and comparison of fatty acyl Δ6-desaturase cDNAs from freshwater and marine teleost fish species. Comp Biochem Physiol B 139:269–279
Zheng X, Tocher DR, Dickson CA, Dick JR, Bell JG, Teale AJ (2005) Highly unsaturated fatty acid synthesis in vertebrates: new insights with the cloning and characterization of a Δ6-desaturase of Atlantic salmon. Lipids 40:13–24
Zheng X, King Z, Xu Y, Monroig O, Morais S, Tocher DR (2009) Physiological roles of fatty acyl desaturases and elongases in marine fish: characterisation of cDNAs of fatty acyl Δ6-desaturase and elovl5 elongase of cobia (Rachycentron canadum). Aquaculture 290:122–131
Acknowledgements
This work was partly supported by Marine Genomique Europe, by Axe1 “Génomique et Chimie Bleue” Europôle Mer, and by IFR 148 ScInBioS. The authors thank Erick Desmarais for his assistance in gene sequence treatment. E. Santigosa was supported by a postdoctoral fellowship from the Fundación Alfonso Martin Escudero (Spain).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Santigosa, E., Geay, F., Tonon, T. et al. Cloning, Tissue Expression Analysis, and Functional Characterization of Two Δ6-Desaturase Variants of Sea Bass (Dicentrarchus labrax L.). Mar Biotechnol 13, 22–31 (2011). https://doi.org/10.1007/s10126-010-9264-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-010-9264-4