Abstract
Variations in the mitochondrial helicase Twinkle (PEO1) gene are usually associated with autosomal dominant chronic progressive external ophthalmoplegia (PEO). We describe five patients from two unrelated Alsatian families with the new R374W variation in the Twinkle linker region who progressively developed an autosomal dominant multisystem disorder with PEO, hearing loss, myopathy, dysphagia, dysphonia, sensory neuropathy, and late-onset dementia resembling Alzheimer’s disease. These observations demonstrate that Twinkle variations in the linker domain alter cerebral function and further implicate disrupted mitochondrial DNA integrity in the pathogenesis of dementia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Twinkle protein is a helicase, which is involved in mitochondrial DNA (mtDNA) replication [1]. Variations in the nuclear PEO1 gene which encodes Twinkle cause autosomal dominant progressive external ophthalmoplegia (adPEO) with multiple mtDNA deletions in muscle (OMIM *606075). Relatively few patients with variations in PEO1 have been reported, and clinical findings ranged from isolated progressive external ophthalmoplegia (PEO) to PEO associated with parkinsonism, sensory neuropathy, cataracts, or limb weakness [2–4]. Here, we report five patients from two unrelated families who presented with a progressive multisystem disorder with PEO, hearing loss, myopathy, sensory neuropathy, and late-onset dementia and caused by a new variation in the linker region of Twinkle.
Materials and methods
Clinical and biological assessment
We studied a population of five patients (three females, two males) with Twinkle-linked autosomal dominant mitochondrial disease from two unrelated pedigrees originating from Alsace (France). The five patients were studied at age 42, 62, 64, 81, and 82, respectively (Table 1). All patients underwent standardized clinical examinations and biological tests, including serum creatine kinase and lactate levels at rest.
Neurophysiological studies
They were performed in three patients and included nerve conduction study in the four limbs and electromyography using a concentric needle electrode in at least three muscles (upper limb, lower limb, and bulbar).
Morphologic and biochemical analyses
Muscle biopsy was performed in two patients and was processed with standard methods for histology and histochemistry. Enzymatic activities of the respiratory chain complexes were measured in muscle homogenates as reported previously [5].
Molecular investigations
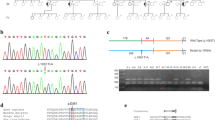
Total DNA was extracted from muscle or blood samples by standard methods. mtDNA derived from muscle was submitted to long-range polymerase chain reaction (PCR) amplification using eight combinations of primers as described previously [6]. The five exons and intron–exon boundaries of the PEO1 gene (GenBank GeneID 56652) were sequenced from genomic DNA of the five patients and seven of their relatives (Fig. 1).
a, b Pedigree of two families with the R374W Twinkle variation. Black boxes indicate the patients affected. The arrows indicate the probands. N indicates a genetically tested patient with a normal result. c Brain MRI of patient AIII-1 demonstrating mild cerebral atrophy. d Muscle biopsy of patient AIII-1 showing a ragged-red fiber (Gomori trichrome stain, ×250). e Long-range PCR analysis of muscle DNA from patient AIII-I revealing multiple mtDNA deletions. Lanes 1–7 primer pairs used for the detection of mtDNA deletions. Lane 8 primer pair used as an internal control to amplify wild-type (wt) mtDNA in a region not usually deleted. Examples of amplified fragments from wt mtDNA are indicated by arrows. M is a 1-kb-size marker. f Sequence analysis of the DNA region encompassing the heterozygous c.1120C>T variation (p.R374W) in patient AIII-1. The variation is indicated with an arrow
Other investigations
Brain magnetic resonance imaging (MRI), evaluation of Mini-mental Status Examination (MMSE) score, electrocardiogram, echocardiogram, and lung function tests were performed in three patients.
Results
Clinical and biological features
Subjects AIII-1 (proband, family A), AIII-5, BII-3 (proband, family B), BII-1, and BIII-7 all developed progressive eyelid drooping in their 40s. When examined at age 42, patient BIII-7 presented with isolated mild bilateral ptosis. In their 50s and 60s, patients AIII-1, AIII-5, BII-3, and BII-1 developed progressive hearing loss and limitation of extraocular eye movements without diplopia. When examined at age 62 and 64, respectively, patients BII-3 and BII-1 showed severe bilateral ptosis, ophthalmoparesis, mild proximal limb weakness, abolished tendon reflexes, and no obvious cognitive impairment. In their early 70s, patients AIII-1 and AIII-5 began having gait difficulty, dysphagia, and dysphonia. They also developed progressive cognitive impairment, with patient AIII-5 being diagnosed with Alzheimer’s disease (AD) at age 79. When examined at age 81 and 82, respectively, patients AIII-5 and AIII-1 showed marked bilateral ptosis, ophthalmoparesis, severe proximal limb weakness, and abolished tendon reflexes. They also had a MMSE score of 12 and 15, respectively, indicative of severe cognitive impairment. Patients AI-1, AII-1, and AII-2 were not examined but they were said to have presented in their 70s and 80s with severe cognitive impairment, bilateral ptosis, hearing loss, and gait abnormalities. Furthermore, patient BI-1 had been diagnosed with AD at age 78, when presenting with a MMSE score of 12. Neurological examination of subjects AIII-2, AIII-3, and AIII-4 was normal (Fig. 1). Serum creatine kinase and lactate at rest were within normal values in all five patients.
Neuradiological and neurophysiological features
In patients AIII-1, AIII-5, and BII-3, brain MRI demonstrated mild central and cortical cerebral atrophy and no cerebellar atrophy (Fig. 1). Electromyography revealed a myopathic pattern, and nerve conduction studies showed a reduction of sural nerves amplitudes, indicative of axonal sensory peripheral neuropathy (patients mean sural nerve distal amplitude, 1.0 μV; normal >3.5 μV).
Morphologic, biochemical, and molecular results
In patients AIII-1 and BII-3, muscle histochemistry showed cytochrome c oxidase (COX)-positive ragged-red fibers (RRF) and numerous COX-negative non-RRF (Fig. 1). Mitochondrial respiratory chain complex and citrate synthase enzyme activities were in the normal range, and long-range PCR analysis of the muscle DNA revealed multiple mtDNA deletions in both probands (Fig. 1). The novel heterozygous missense variation c.1120C>T, resulting in an amino acid change of the highly conserved Arg-374 to Trp (p.R374W), was identified in patients AIII-1, AIII-5, BII-1, BII-3, and BIII-7 (Fig. 1). Genotype analysis showed segregation of this novel allele with the disease phenotype in both families (Fig. 1).
Discussion
Twinkle is the mtDNA helicase which is essential for mtDNA maintenance and regulation of mtDNA copy number in mammals [7]. Replication of mtDNA involves a complex machinery called replisome including the mtDNA polymerase γ (polg γ) and the Twinkle helicase. The replication machinery uses double-stranded mtDNA as template to synthesize single-stranded mtDNA molecules [8]. Dominant mutations in genes encoding Twinkle (PEO1) or the two subunits of polg γ (POLG1 and POLG2) result in multiple mtDNA deletions in postmitotic tissues over time. Typical clinical features include PEO, which is sometimes associated with exercise intolerance, muscle weakness, polyneuropathy, deafness, ataxia, and cataracts [8].
Here, we report five patients from two unrelated families who presented with a progressive multisystem disorder caused by a new variation (R374W) in Twinkle. From the neuromuscular point of view, the disease in these patients fitted the classic form of PEO1 gene-linked adPEO. The symptoms the patients presented with, including PEO, sensory neuropathy, myopathy, hearing loss, dysphagia, and dysphonia, have all been previously described in patients with PEO1 gene mutations [1–4]. Some of them presented with the clinical triad of sensory ataxic neuropathy, dysarthria, and ophthalmoparesis, a phenotype previously associated with mutations in POLG1, highlighting the clinical overlap in adPEO [9]. All patients had a typical clinical course with ptosis, ophthalmoplegia, hearing loss, and polyneuropathy appearing in their 40s to 60s, but in addition developed gait abnormalities, dysphonia, dysphagia, and dementia in their 70s and 80s (Table 1). These findings demonstrate that the disorder is progressive in postmitotic tissues, thus corroborating previous clinical and mouse model studies of PEO1 mutations [3, 7].
The presence of severe dementia in our patients is unusual. Although the existence of an independent condition affecting the central nervous system (CNS) cannot be strictly ruled out, this study suggests that the PEO1 mutation may be responsible of the cognitive impairment in these patients. The putative link between the PEO1 mutation and dementia is further strengthened by the absence of dementia in all other family members without the variant R374W. On a clinical viewpoint, when using the National Institute of Neurological and Communicative Disorders and Stroke–Alzheimer’s Disease and Related Disorders Association Alzheimer’s criteria, the cognitive decline the patients presented with was undistinguishable from probable AD [10]. Strengthening this point, brain MRI did not show any of the white matter abnormalities sometimes observed in mitochondrial diseases such as mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes [11]. Interestingly, it has been shown that the R374Q change in the same codon also causes CNS abnormalities, including parkinsonism, major depression, and bipolar disorder [4]. More recently, moderate dementia was reported in two patients with the K319E Twinkle variation [9], and dementia associated with depression was described in a 71-year-old woman with the R303Q Twinkle variation [12]. Severe retarded depression without obvious dementia was described as the major clinical symptom in a female patient with PEO1 gene-linked adPEO [1, 13]. Autopsy of her brain found morphological abnormalities in neuronal mitochondria and a high percentage of deleted mtDNA molecules in basal ganglia and cerebral frontal cortex [13]. Mouse model studies of PEO1 mutations also demonstrate an accumulation of mtDNA deletions in the brain [7]. Taken together, these data strongly suggest that PEO1 variations alter cerebral function. In keeping with this concept, there is a strong evidence that mtDNA may contribute to the mitochondrial dysfunction observed in several subtypes of dementia, including AD [14]. Our report fully supports this concept and establishes for the first time a direct link between mtDNA alterations and late-onset dementia.
The new variation R374W we describe in this study is located in the linker region of Twinkle, which is of dramatic importance for the DNA helicase activity of the protein [8]. The Twinkle protein forms a stable hexamer, and the linker region between the helicase and primase-related domains is critical for protein hexamerization and helicase activity [8]. Helicase activity is coupled with an ATPase activity as Twinkle-mediated DNA unwinding is ATP dependent. In the wild-type sequence of the Twinkle linker region, residue R374 is strictly conserved from mammals to phage T7 [1, 8]. Another variation, R374Q, which has been previously described in adPEO, emphasizes the importance of codon 374 [1]. Indeed, R374Q abolishes the ability of Twinkle to form hexamers and consequently reduces by more than 90% the ATPase and DNA helicase activities compared to the wild-type protein [8]. Interestingly, the monomeric mutant R374Q can form hexamers in the presence of wild-type Twinkle [8, 15]. These data support the hypothesis that R374Q would be embryonically lethal if present in homozygous state [8, 15]. In this context, one may predict that the heterozygous R374W variant identified in our patients also provokes a dramatically reduced helicase activity. Indeed, substitution of the very basic residue arginine (R) with the hydrophobic aromatic tryptophan (W) induces a severe change in the structure of the linker region, probably more drastic than the arginine (R) to glutamine (Q) change. Consequently, as W has no positive charge on the side chain like R, the R374W variation may compromise normal specific interactions between Twinkle monomers like R374Q [8]. In addition, R374W may directly cause replication pausing or stalling, leading to widespread accumulation of multiple mtDNA deletions and gradual mitochondrial respiratory dysfunction, especially in skeletal muscle and brain [15].
Conclusion
Here, we report for the first time a direct link between late-onset dementia and mtDNA damage. Our observations also show that late-onset dementia may be a feature of adPEO and emphasize the central role of the Twinkle linker region in the mtDNA replication machinery. Further clinical, molecular and functional expression studies are now needed to better characterize the consequences of dominant Twinkle variations on brain function.
References
Spelbrink JN, Li FY, Tiranti V et al (2001) Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat Genet 28:223–231
Virgilio R, Ronchi D, Hadjigeorgiou GM et al (2008) Novel Twinkle (PEO1) gene mutations in mendelian progressive external ophthalmoplegia. J Neurol 255:1384–1391
Jeppesen TD, Schwartz M, Colding-Jorgensen E, Krag T, Hauerslev S, Vissing J (2008) Phenotype and clinical course in a family with a new de novo Twinkle gene mutation. Neuromuscul Disord 18:306–309
Baloh RH, Salavaggione E, Milbrandt J, Pestronk A (2007) Familial parkinsonism and ophthalmoplegia from a mutation in the mitochondrial DNA helicase Twinkle. Arch Neurol 64:998–1000
de Camaret Mousson B, Taanman J-W, Padet S et al (2007) Kinetic properties of mutant deoxyguanosine kinase in a case of reversible hepatic mtDNA depletion. Biochem J 402:377–385
Ferlin T, Guironnet G, Barnoux MC, Dumoulin R, Stepien G, Mousson B (1997) Detection of mitochondrial DNA deletions by a screening procedure using the polymerase chain reaction. Mol Cell Biochem 174:221–225
Tyynismaa H, Mjosund KP, Wanrooij S et al (2005) Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proc Nat Acad Sci U S A 102:17687–17692
Korhonen JA, Pande V, Holmlund T et al (2008) Structure-function defects of the TWINKLE linker region in progressive external ophthalmoplegia. J Mol Biol 377:691–705
Hudson G, Deschauer M, Busse K, Zierz S, Chinnery PF (2005) Sensory ataxic neuropathy due to a novel C10Orf2 mutation with probable germline mosaicism. Neurology 64:371–373
McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM (1984) Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 34:939–944
Pavlakis SG, Phillips PC, DiMauro S, De Vivo DC, Rowland LP (1984) Mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes: a distinctive clinical syndrome. Annals of Neurology 16:481–488
Van Hove JL, Cunningham V, Rice C et al (2009) Finding Twinkle in the eyes of a 71-year-old lady: a case report and review of the genotypic and phenotypic spectrum of Twinkle-related dominant disease. Am J Med Genet 149A:861–867
Suomalainen A, Majander A, Haltia M et al (1992) Multiple deletions of mitochondrial DNA in several tissues of a patient with severe retarded depression and familial progressive external ophthalmoplegia. J Clin Invest 90:61–66
Lin MT, Beal MF (2006) Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443:787–795
Goffart S, Cooper HM, Tyynismaa H, Wanrooij S, Suomalainen A, Spelbrink JN (2009) Twinkle mutations associated with autosomal dominant progressive external ophthalmoplegia lead to impaired helicase function and in vivo replication stalling. Hum Mol Genet 18:328–340
Author information
Authors and Affiliations
Corresponding author
Additional information
The authors (Andoni Echaniz-Laguna, Jean-Baptiste Chanson, Jean-Marie Wilhelm, François Sellal, Martine Mayençon, Michel Mohr, Christine Tranchant, Bénédicte Mousson de Camaret) disclose all financial support for their work and other potential conflicts of interest.
Rights and permissions
About this article
Cite this article
Echaniz-Laguna, A., Chanson, JB., Wilhelm, JM. et al. A novel variation in the Twinkle linker region causing late-onset dementia. Neurogenetics 11, 21–25 (2010). https://doi.org/10.1007/s10048-009-0202-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-009-0202-4