Abstract
Understanding and predicting patterns arising from the dynamics of marine food webs is central to trophic and community ecology and numerical models of food webs constitute a primary tool to simulate these dynamics. Food web simulation models are often highly complex while at the same time often too constrained to reproduce the level of variability observed in real systems. The recently developed non-deterministic network dynamics (NDND) modelling framework has been suggested as a simulation alternative, which can generate multiple patterns of food web variability despite great structural simplicity. Two important aspects of the NDND modelling framework remain unexplored: first the derivation of model input parameters from empirical or theoretical studies and second the evaluation of the model simulations against observations. We provide a methodology for the derivation of model parameters based on empirical observations, the metabolic theory of ecology and life-history theory and apply it to the specific case of the Barents Sea food web. We then evaluate the ability of the NDND simulations to reproduce a wide range of patterns of food web dynamics against observations collected in the Barents Sea during 28 years. Patterns emerging from the simulations include trends and cycles in biomass, trophic levels and transfer efficiency, density-dependent growth, top-down vs bottom-up oscillations, ecosystem level stability and synchrony and trophic functional responses. The ability of the NDND to generate so many patterns observed empirically in the Barents Sea is remarkable given that it is based only on random trophic interactions operating within few constraints set by ecological rules. Our results show that investigations of food web dynamics in marine ecosystems, including the definition of reference states and responses to climate and exploitation pressures, may be achieved with models that are structurally simple and based on few well-established assumptions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ecological systems are intrinsically complex; they consist of many components (for example, species, habitats), without a central control of the system dynamics. Such systems are self-organised (interactions generate the system’s organisation) and display complex and non-linear feedback mechanisms. Furthermore, ecological systems are usually characterised by being capable of innovation and operate at multiple scales (for example, from viruses to whales and micrometres to ocean basin scales), and offer a limited window of observability. In addition, stochastic phenomena play an important role in real-world physical and biological systems, and thus in shaping the dynamics of such systems. As a result, ecosystems are difficult to analyse, understand and, not the least, model.
Marine ecosystems are known to undergo major structural and functional changes (regime shifts) with possible long-lasting consequences (Blenckner and Niiranen 2013). These events, which are often caused by interactions among multiple drivers (climate, overfishing, eutrophication and species invasions) (Lees and others 2006), may have major social–ecological consequences and are difficult to predict (Scheffer and Carpenter 2003; Lade and others 2013). Interpreting the state and dynamics of ecosystems requires a reference of the potential variability of ecosystems (Clark and others 2001). Retrospective data can inform about the past variability of the system and help in resolving underlying causes and rates of ecological change. Most temporal series of ecological observations of marine systems go back 3–5 decades at most, a time frame that is too short to inform about system dynamics given the long lifespans of many species and the temporal dynamics of historic environmental drivers (Lotze and Worm 2008). Constructing reference baselines from past observations is often difficult as it involves a wide range of information sources, such as paleontological, archaeological, historical and fisheries records (Lotze and Worm 2008), as well as many assumptions which can result in large uncertainties (Longhurst 2010). A complementary approach is to derive references for the variability of ecosystems from numerical simulation models of ecosystem dynamics (Landres and others 1999). Ideally, a numerical model should be parsimonious and it should be possible to evaluate model outputs against observations. Parsimony implies that the model be based on a limited number of principles, each derived from well-defined underlying assumptions, that the number of model entities be restricted to the minimum necessary, and that input data and parameters are few and derived from observations or theoretical considerations.
Food web models, which represent a specific class of ecosystem models that describe trophic interactions between species or groups of species in terms of energy or biomass fluxes, are commonly used to simulate ecosystem dynamics (Christensen and Walters 2004). A recurrent difficulty, in building food web models, is to ensure that the dynamics represented and simulated in a mathematical model bares some degree of isomorphism with the dynamics in real ecosystems (Ruiz and Kuikka 2012). Most operational food web models today are highly complex, with structures that rely on large numbers of parameters and state variables, and with many parameter values not derived from direct observations or lacking theoretical support. Despite their complexity, food web models are usually too constrained to reproduce the variability patterns observed in real systems, and they are rarely capable of simulating regime shifts or other ‘ecological surprises’ that are commonly observed (Blenckner and Niiranen 2013; Möllmann and others 2015).
A novel approach to food web modelling, introduced by Mullon and others (2009), was evaluated by Planque and others (2014a) who termed it “non-deterministic network dynamics” (NDND) modelling. The NDND model, with a basis in the concept of “viability” in mathematics (Aubin 1991; Aubin and others 2011), does not rely on optimal solutions but rather on sets of possible viable system states and trajectories. It is well known that the dynamic behaviour of modelled food webs is highly sensitive to the shape of the consumer–resource functional responses (Williams and Martinez 2004; Koen-Alonso and Yodzis 2005; Kearney and others 2012), which are difficult to specify and even more so to parameterise (Hunsicker and others 2011). In contrast to conventional food web models, which rely on numerous consumer–resource functional responses, the trophic fluxes in the NDND model are simulated as stochastic (non-deterministic) processes whilst the system state and trajectory are constrained by few simple rules. Treating this process as a constrained random process, or an emergent property, seems appropriate given the considerable uncertainty and variability of the consumer–resource functional responses (Englund and Leonardsen 2008).
In the present study, we develop a parsimonious food web dynamic simulation model for the Barents Sea, using the NDND modelling framework. A crucial step in model development is the setting of input parameter values, and we show how these can be derived from metabolic theory (Brown and others 2004) and life-history theory (Hoenig 1983). Another critical step is model evaluation through comparison of model simulation outputs against empirical observations. Because the NDND model is a stochastic simulation model, it is not possible to directly match simulated trajectories to those observed in nature. Instead, we use summary statistics of specific ecosystem properties (food web biomass and fluxes, biomass variability, growth and density dependence, trophic controls, temporal stability, synchrony and consumer–resource functional responses) to evaluate model performance by comparison with empirical observations, following the pattern-oriented modelling approach advocated by Grimm and Railsback (2012). Finally, we show that many of the ecosystem variability patterns empirically observed in the Barents Sea can emerge from the simulations of this simple model.
Materials and Methods
Barents Sea Ecosystem
The Barents Sea is a high-latitude, seasonally ice-covered, Arctic shelf sea ecosystem that is strongly influenced by the advection of relatively warm North Atlantic water from the Norwegian Sea (Dalpadado and others 2014). Four decades of observations have revealed major fluctuations in species abundance, spatial distribution and trophic interactions (Johannesen and others 2012; Dalpadado and others 2012). Climate warming has been accompanied by changes in fish community structure, particularly in northern areas where the Arctic fish community has been gradually replaced by an Atlantic community (Fossheim and others 2015). Analyses of historical records (Johannesen and others 2012) have revealed patterns of variability such as regime shifts, decadal variations in trophic interactions or multi-annual community changes. The degree to which these result from climate pressures and exploitation patterns or simply reflect the natural variability of marine ecosystems remains unclear. Information on year-to-year fluctuations in environmental and biotic conditions, synthesised in Johannesen and others (2012) and ICES (2014a, b), are used in this study as the primary sources of data to evaluate the results of model simulations against empirical observations.
Modelling Principles
Here we provide a brief summary of model concepts, structure and parameters followed by the specific details of the model implementation for the Barents Sea food web.
We use the NDND modelling framework and define the food web as a network consisting of nodes and links. Nodes are trophospecies (trophic groups) and can refer to taxonomic species, groups of species or specific developmental stages of single species. Links represent trophic interactions between trophospecies. The topology of the food web is invariant during a simulation. Contrary to other food web models, such as Ecopath with Ecosim and Atlantis (Christensen and Walters 2004; Fulton 2001), trophic interactions are not deterministically specified in the NDND framework. Instead, the key principle is that at each time step, trophic flows are drawn randomly within a restricted set defined by a few physical and biological constraints. The physical constraints are determined by the law of conservation of mass, which entails that fluctuations in the biomass of individual species solely result from the balance between gains (feeding, import) and losses (predation, metabolic losses, export). Trophic flows from prey to predators cannot be negative. Naturally, biomasses are constrained to remain positive, each with a minimum threshold value called refuge biomass. Species-specific biological constraints also include satiation: the maximum consumption rate, and inertia: the maximum population growth and mortality rates. The latter are derived from life-history theory and the metabolic theory of ecology. Additional input parameters for potential assimilation efficiency (the proportion of prey biomass that can potentially be assimilated) and for other losses (the rate of metabolic losses and losses not explicitly accounted for in the model) are also required. In the original NDND formulation, biomass is used as a surrogate for energy, so variations in energy content per unit biomass in different organisms are ignored. This can be an inappropriate assumption when various prey organisms display different energy concentrations (for example, echinoderms vs. fish). Therefore, an additional species-specific input parameter was introduced in the present version of the model to account for variations in energy content: digestibility correction factor. The product of the potential assimilation efficiency by the digestibility correction factor is the absorption efficiency (the proportion of prey biomass digested and absorbed).
In summary, the NDND is a mass balance model in which food web topology is fixed, trophic flows are drawn randomly, food intake does not exceed satiation, biomass variations are bounded between a maximum growth rate and a maximum mortality rate (inertia) and biomass does not fall below a minimum level (refuge biomass).
The dynamics of species in the food web is defined by the general equation:
where ∑ j F ji represents the biomass fluxes to species i (consumption), ∑ j F ij represents the biomass fluxes from species i (predatory loss), γ i is the potential assimilation efficiency of species i, κ j is the digestibility correction factor for species j, I i is import (immigration), E i is export (fishing, emigration), μ i is “other losses” (metabolic losses and other mortality not accounted for explicitly in the model) and B i is the biomass of species i. At each time step, a set of trophic flows (F) is drawn randomly, given that mass balance and physical and biological constraints are satisfied. A detailed description of the model structure is given in Planque and others (2014a) and the method for sampling trophic flows is detailed in Subbey and others (2016).
Configuration of the Barents Sea Model
The temporal resolution of the model is annual and the spatial scale is the Barents Sea shelf (1.6 million km2). The topology—who eats whom—is presented in Figure 1, and the list of taxa included in each trophospecies is detailed in Appendix S1. Simulations start in 1986 and starting biomass values are derived from the literature as described in Appendix S2. Biological input parameters inertia, other losses and satiation were derived from metabolic theory (Yodzis and Innes 1992; Brown and others 2004), life-history theory (Hoenig 1983) and previously reported values (Blanchard and others 2002; Makarieva and others 2008). The derivation of biological input parameters is detailed in Supplementary Material (S3). Simulations where run for 200 years. The first 28 years (1986–2013) are used for comparison against empirical observations.
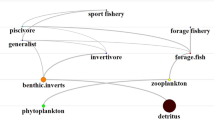
Trophospecies (circles), trophic interactions (black arrows) and imports (green arrows). The size and thickness of the circles and lines are indicative of species biomass trophic and imports flows, respectively (median of 1000 simulations of 28 year). Trophic flows are pointing to predators. Looped flows denote cannibalism (Color figure online).
Food Web Topology
Three mass balance food web models have previously been developed for the Barents Sea (Blanchard and others 2002; Dommasnes and others 2001; Skaret and Pitcher unpublished). These comprise 41, 30 and 58 functional groups and 267, 169 and 545 trophic links respectively. The use of food web topologies with such a level of complexity within the NDND framework is unrealistic and inappropriate since the NDND model is designed to simulate the variability of whole-ecosystem properties, rather than detailed population dynamics. Food web complexity can be substantially reduced; typically to less than 14 species and 40 trophic links while conserving essential properties at the ecosystem level (Ulanowicz and others 2014). The topology used in the present study is a simplification into eight functional groups covering all taxa included in earlier food web models, but excluding detritus. The eight trophospecies are phytoplankton, herbivorous and omnivorous zooplankton, benthos, pelagic and demersal fish, marine mammals and birds (Figure 1).
Biomasses, Import and export
Starting biomasses were derived from literature and, when possible, several estimates from different sources were used for each species. To avoid bias toward a particular source, we used the geometric mean of the estimates as the input value. Refuge biomasses were set arbitrarily to 1% of starting biomasses. The annual net primary production (modelled as import of phytoplankton) and the import of herbivorous and omnivorous zooplankton from the Norwegian Sea were derived from published estimates. As for starting biomasses, when several estimates were available from different sources, the geometric mean of these was used. The imports of benthos, pelagic and demersal fish, marine mammals and birds were set to zero. The imports are assumed to be constant throughout the simulation period. The export term represents processes such as fishing and emigration and was set to zero for all species. Supplementary simulations were conducted to explore the possible effects of fishing (Supplementary Material S4). A detailed description of the derivation of input data is given in the Supplementary Material (S2).
Assimilation Efficiency and Digestibility
The absorption efficiency, defined as the ratio of absorbed to ingested food, can vary markedly depending on the quality and quantity of prey, the consumer species and the stage/age of the consumer (Valiela 1984). As stated in Eq. 1, the absorption of a species is the product of the potential assimilation efficiency (γ) of a consumer and the prey digestibility correction factor (κ). These coefficients were selected so that the absorption efficiencies (γ(predator) · κ(prey)) were similar to the values reported in the literature, ranging from 0.56 (absorption efficiency of demersal fish feeding on benthos) to 0.90 (absorption efficiency of marine mammals feeding on zooplankton and pelagic fish). These values are similar to the absorption efficiencies reported by Blanchard and others (2002): 0.65–0.82 for benthos, 0.80 for pelagic fish, 0.82 for zooplankton and 0.89 for sea mammals, birds and demersal fish. Note that the digestibility correction factor (κ) is irrelevant for mammals and birds because they are not preyed upon in this model.
Inertia, Other Losses and Satiation
In the NDND, the inertia coefficient (ρ) accounts both for maximum mortality and maximum growth rates (using −ρ and +ρ). Maximum mortality and growth rate can be derived from life-history theory (Hoenig 1983; Hewitt and Hoenig 2005), metabolic theory of ecology (Savage and others 2004) or from allometric relationships (Yodzis and Innes 1992).
The term “other losses” (μ) includes metabolic losses and losses not explicitly accounted for in the model, such as disease-induced mortality. The rate of metabolic losses is approximated by the field metabolic rate (FMR) of a species, which is assumed to be three times the basal metabolic rate (BMR, Speakman and Selman 2003). This estimate of μ is a minimum because it only accounts for metabolic losses but not for other types of losses. Estimates of μ were derived from three different sources (Yodzis and Innes 1992; Gillooly and others 2001; Makarieva and others 2008), and the arithmetic mean of these estimates was used as the input value to the NDND model. In contrast, to inertia (ρ) and other losses (μ), estimates of satiation (σ) were derived from one source (Yodzis and Innes 1992), based on allometric scaling relationships. The detailed derivations of inertia, other losses and satiation coefficients are provided in Supplementary Material (S3). The set of input parameters and starting values for the Barents Sea NDND model is summarised in Table 1.
Model Evaluation
The outputs of the NDND model cannot be evaluated by measuring the likelihood of observed versus simulated biomass trajectories because modelled trajectories are stochastic and are not aimed at fitting observational data directly. Because the NDND model is designed to generate sets of possible viable trajectories—rather than to reconstruct observed past trajectories—comparison between observed and simulated time series should be conducted by checking whether empirical trajectories lie within the simulated envelopes or not. The model is considered valid if the ensemble of simulations contains the empirical trajectories even when the latter deviate noticeable from the median trajectories.
One the other hand, the NDND model can be evaluated using summary statistics that capture key properties of the system. These summary statistics, also called second-order properties, can be calculated from model simulations and observational data (Hartig and others 2011). The model evaluation approach follows the general principles of ‘pattern-oriented modelling’ (POM, Grimm and Railsback 2005, 2012) that uses multiple criteria for design, selection and calibration of models of complex systems. Using repeated simulations, it is possible to derive empirical frequency distribution of summary statistics in the model. These distributions can then be used to empirically estimate the probability that the value of summary statistics in the observed data be obtained in the simulations. This corresponds to an empirical null hypothesis testing approach, in which the NDND is the null model. In this study, model evaluation is conducted by constructing frequency distributions of selected summary statistics (see list below) from 1000 food web trajectories. We examined the probability of observing the value of empirical summary statistics in the null model, that is, the NDND simulations. In addition, we present some summary statistics for which there are no empirical data available for direct comparison, but for which there are expectations based on expert knowledge or previous modelling attempts. The NDND model evaluation is based on the following properties: food web biomass and fluxes, biomass variability patterns, growth and density dependence, trophic controls, temporal stability synchrony and consumer–resource functional responses. For this purpose, we used the following set of summary statistics:
-
Average individual biomasses and trophic fluxes estimates based upon 1000 simulation over the period 1986–2013;
-
Trophic level (TL) diet weighted by the trophic level of its prey (TL i = 1 + ∑ j D i,j · TL j , where D i,j is the fraction of prey j in the diet of species i and TL j is the trophic level of prey j) (Pauly and others 2000);
-
Transfer efficiency (TE) the ratio between sum of predatory fluxes plus export fluxes and the throughput (sum of consumption and predatory fluxes) (Pauly and others 2000);
-
Envelope of biomass trajectories log-transformed biomass envelopes (50, 95 and 99%) of individual trophospecies for the period 1986–2013;
-
Biomass distribution of individual trophospecies;
-
Density dependence the slope of the growth rate versus biomass relationship,;
-
Trophic control (top-down and bottom-up) measured as the correlation between predator biomass and prey biomass in the previous year over sliding 15-year time periods (Johannesen and others 2012);
-
Stability of the food web the ratio of mean biomass over temporal variance in biomass for all species combined (Lehman and Tilman 2000);
-
Synchrony of the food web the ratio of temporal variance in the biomass of individual species over the temporal variance for all species (Loreau and de Mazancourt 2008);
-
Trophic functional response the consumption rate of a trophospecies as a function of prey biomass.
When possible, these statistics were derived from empirical data for the purpose of model evaluation. When empirical data were not available, model evaluation was done against theoretical expectations and literature.
Results
Average Biomasses, Trophic Flows, Trophic Levels and Transfer Efficiencies
Average biomasses and trophic flows are illustrated in Figure 1. The most important trophic fluxes are between trophospecies situated in the lower trophic levels, with flows between phytoplankton, herbivorous and omnivorous zooplankton and benthos accounting for 85% of the flows in the food web (when cannibalistic flows are excluded). The distribution of biomass in the food web shows a maximum at intermediate trophic levels, a feature that is consistent with patterns observed in many marine systems (Pranovi and others 2012).
Comparison of simulated biomasses and trophic flows with empirical observations from the Barents Sea should be viewed with caution, in particular for lower trophic levels, since the new harvestable production (defined as the net primary production or new production) is not explicitly modelled in the NDND and vertical flux of organic matter from the surface to the benthos is modelled as direct predation on phytoplankton by benthos. Despite these limitations, the NDND model generates phytoplankton–benthos biomass fluxes (95% CI 105–305 t km−2 y−1) that are within the range of export production estimates (75–550 t km−2 y−1; Klages and others 2004).
The fluxes were used to derive both the trophic level (TL) and transfer efficiency (TE) of trophospecies which are given in Table 2. The estimated TL ranges from 2 (herbivorous zooplankton) to 4.7 (mammals), whereas the mean TE ranges from 1.2 in benthos to 28.1 in herbivorous zooplankton. With the exception of benthos, the transfer efficiencies decline as trophic levels increase (Table 2).
Biomass Variability Patterns
Empirical biomass trajectories for species in the Barents over 28 years lie within the simulated 99% envelopes (Figure 2), indicating that the NDND model captures a range of biomass variations equal to or greater than what has been observed in the recent past. Biomasses of herbivorous and omnivorous zooplankton in 1994 and 1989–1991 are extreme cases which lie outside the 99% limits, but these remain within the complete simulation envelope (100% limits). The median estimates, based on 1000 simulations, are slightly lower than the observed trajectories with respect to herbivorous zooplankton, pelagic and demersal fishes and the opposite with respect to omnivorous zooplankton. Pelagic fish and, to a lesser extent, zooplankton groups frequently reach low densities, close or equal to the refuge biomass (Figures 2, 3). Patterns of inter-annual variability in the biomass of pelagic and demersal fish appear to be well simulated, while simulations tend to generate inter-annual patterns for zooplankton biomass that are more variable than empirical observations would suggest (Figure 2). This is confirmed by the distribution of biomasses of the observed and simulated 28-year biomass time series (Figure 3). The observed ranges of biomass for zooplankton and fish groups are narrower than the simulated ranges, indicating that the model generate greater biomass variability than has been empirically observed in the recent decades.
Biomass time series of herbivorous and omnivorous zooplankton, benthos, pelagic and demersal fishes, mammals and birds. The light, medium light and dark grey area denote 99, 95 and 50% simulation envelopes, respectively. The red and black solid lines denote the biomass trajectory in the empirical observations and the median of 1000 biomass simulations, respectively. The horizontal hatched blue lines mark the refuge biomasses and the two tilted hatched blue lines denote the lower and upper inertia boundaries. The dashed black lines denote three random simulations (Color figure online).
Biomass distribution (log-scale) of herbivorous and omnivorous zooplankton, benthos, pelagic and demersal fishes, mammals and birds (from 1000 simulations of 28 year). Bars represent the simulated data, the red lines denote the median empirical estimates for each group and the pink region indicates the range of empirical observations (when available). Vertical dotted blue lines denote the refuge biomass for each trophospecies (Color figure online).
Density Dependence and Trophic Control
The model simulations revealed high variability in population growth rates for all species (Figure 4). For high trophic levels, simulated growth rates often cover the entire range allowed by model constraints and there was no evidence of density dependence. For lower trophic levels, variations in growth rates were substantial but within model limits and there appeared to be an inverse relation between maximum growth rates and biomass. All but two empirical observations (one pelagic and one demersal not illustrated in the figure) were within the 95% envelope indicating that the model encompasses these growth values.
Growth rate versus biomass for herbivorous and omnivorous zooplankton, benthos, pelagic and demersal fishes, mammals and birds. Simulated and empirical data are represented by black and red dots, respectively. The red lines are fitted to empirical data (second-order polynom). The green lines highlight how the maximum population growth rate declines with increasing population biomass for herbivorous and omnivorous zooplankton, benthos and pelagic fishes (Color figure online).
Negative and positive correlations between predator and prey biomass have been interpreted as top-down and bottom-up controls, respectively (Cury and others 2003), and tracking how correlations between predators and prey vary with time has been used to reveal interdecadal changes in trophic controls in the Barents Sea (Johannesen and others 2012). Time series of correlation derived from simulations can resemble those observed from empirical data (Figure 5). Typically, stochastic simulations lead to interdecadal oscillations between apparent top-down and bottom-up controls. This feature is particularly salient for demersal versus pelagic fishes (Figure 5, upper right panel). The aggregated results (Figure 5, lower panel) show that the observed trophic correlations lie within the 95% confidence intervals of the modelled data, thereby confirming that the model covers the range of situations observed in the wild. Correlations derived from empirical observations are more negative, on average, than those derived from simulations, suggesting a dominance of top-down control in Barents Sea during the period 1986–2013.
Correlations, calculated on 15-year centred sliding windows, between trophic groups (herbivorous and omnivorous zooplankton, pelagic and demersal fish) in empirical and simulated data. Top: correlations from 1993 to 2006 for empirical data (left) and one simulation (right). Bottom: boxplot of correlation from 1000 simulations shown with empirical observations (red dots). The boxes and error bars of the boxes correspond to 50 and 95% confidence intervals of the modelled data, respectively, and the horizontal solid black lines indicate the median. HZP, OZP, PEL and DEM denote herbivorous zooplankton, omnivorous zooplankton, pelagic and demersal fish, respectively (Color figure online).
Stability and Synchrony
In contrast to the evaluation criteria presented above, stability and synchrony indices provide information about the dynamics of the food web as a whole, rather than on individual trophospecies.
Simulated and empirical stability and synchrony indices, as well as indices of stability and synchrony derived by Bell and others (2014) for 19 marine ecosystems, are presented in Figure 6. Simulation-based stability and synchrony envelopes display a wide range of possible situations. Empirical observations of both indices, with the exception of the synchrony index in 1993–1997, lie within the simulated 99% envelopes. The overall stability of the food web derived from empirical observations is close to that in simulated food webs. On the other hand, synchrony derived from empirical observations is generally greater than that from the simulations. Empirical data suggest that low stability is associated with high synchrony and vice versa (R = −0.77). In contrast, there is no relationship between the two indices in the simulations or in Bell’s data. Most (95%) of the simulated stability–synchrony correlations are found between −0.8 and 0.86 (median R = 0.1, Figure 6), which are not significantly different from zero. This is similar to the stability–synchrony correlation found in Bell’s data (R = −0.12).
Food web stability (upper left panel) and synchrony (upper right panel). The dark, medium and light grey areas correspond to 50, 95 and 99% simulation envelopes, respectively (1000 simulations). Black solid lines denote the median of simulations and red solid lines denote empirical estimates. Dashed black lines denote three random simulations. The green dots show the stability and synchrony values estimated for several ecosystems by Bell and others (2014). The histogram (lower left panel) displays 1000 simulated 14-year stability–synchronicity correlations. The red and green solid lines denote the mean stability–synchronicity correlations of the empirical data for the Barents Sea and Bell’s data, respectively (Color figure online).
Trophic functional responses
Consumption-resource envelopes for four selected consumer–resource pairs are illustrated in Figure 7. Simulated consumer–resource relationships vary considerably, particularly for trophospecies feeding on lower trophic levels such as omnivorous zooplankton and pelagic fish. It appears that the simulated consumption rates are often far from the satiation boundaries (horizontal hatched lines in Figure 7), suggesting that resource availability, rather than satiation, is limiting consumption. Although trophic interactions are modelled as random processes in the NDND framework, the consumer–resource functional responses that emerge from the simulations resemble Holling-type II or III patterns (Holling 1965).
Four consumer–resource functional responses: phytoplankton–omnivorous zooplankton, omnivorous zooplankton–pelagics, pelagics–demersals and pelagic–mammals. The dark, medium and light grey areas correspond to 50, 95 and 99% simulation envelopes, respectively. The black solid lines denote the median of the simulations. The blue tilted and horizontal hatched lines denote the 1:1 (prey consumption = available prey) and satiation (consumption = satiation) constraints, respectively. The scatters correspond to 2000 randomly selected consumer–resource relationships (Color figure online).
Discussion
Rosen (1969) termed biology “the physics of intractable systems”, partly due to our inability to identify and specify the causal relationships behind the dynamics of the system. In numerical modelling, Occam’s razor law (also known as the law of parsimony) prescribes that, when faced with alternatives, the simplest explanation of a pattern (or phenomenon) is to be preferred (Blumer and others 1987). This principle is standard in statistics when seeking model variance-bias tradeoff, a compromise between model complexity and fitting accuracy (Burnham and Anderson 2002). For ecosystem models, recent trends have gone rather the opposite way, with process-based models becoming more and more complex and often integrating hundreds of functions and even larger numbers of parameters (for example, Holt and others 2014). The approach adopted in this manuscript is underpinned by Occam’s razor and attempts to explain as many patterns as possible using a simple model structure with a minimal number of parameters and assumptions.
The NDND framework is not advocated as a panacea to model the dynamics of complex ecosystems, but rather it is a simple approach primarily aimed at understanding how patterns in food web dynamics may emerge from the combination of chance and necessity (that is, stochastic interactions operating within ecological constraints). The power of the NDND model for the Barents Sea does not lie in its efficacy to best reproduce a single pattern (for example, a biomass trajectory for a single trophospecies) but rather in its ability to capture simultaneously multiple patterns of different nature while remaining simple. To the best of our knowledge, this is the first time a food web simulation model is evaluated against data for such a wide range of patterns. Simulations with the NDND model reveal that a limited set of rules and constraints is sufficient to simulate a multitude of patterns of the Barents Sea food web dynamics. Beside the food web synchrony, which is overall lower than that observed in the Barents Sea and other areas, the simulated dynamics of the other food web properties resemble empirical observations and results from other modelling studies.
Emergent Patterns
Basic food web properties, such as TE and TL of trophospecies, may be used to inform limits in food web productivity (Libralato and others 2008), which can be used to estimate fisheries harvest potential, impact of fishing (Pauly and others 1998; Shannon and others 2014) and changes in ecosystem structure over time (Pranovi and others 2014). Previous modelling studies have shown that these food web properties as well as trophic functional responses may greatly vary in time and space as a result of changes in life history and prey availability (Jennings and others 2002; Koen-Alonso and Yodzis 2005; Libralato and others 2008). Not accounting for this variability can strongly alter simulated food web dynamics and bias the estimates of food web properties.
The consumer–resource functional relationship is perhaps the most critical process in food web models and it is one of the most difficult to parameterise (Koen-Alonso 2007). Although the dynamics of modelled food webs are known to be highly sensitive to the choice of the functional responses, these are generally poorly known for wild marine populations due to the lack of large-scale consumer–resource data. Treating the functional response as an emergent property rather than as a pre-defined input, can allow for a wide range of food web dynamics and possible trajectories. There are no appropriate field observations to evaluate the functional responses emerging from the simulations, but field and modelling studies report both type II and III functional responses (Koen-Alonso and Yodzis 2005; Smout and Lindstrøm 2007), thus supporting the trophic functional relationships emerging from the NDND model.
Patterns of inter-annual variability in trophospecies biomass are of particular interest, and the NDND is capable of generating trends and pseudo-cycles that resemble those observed in the wild, as well as decadal variations in trophic controls that are similar to those empirically described for the Barents Sea. Given the stochastic nature of the model, it is likely that the simulated densities of individual trophospecies are more variable than those in the wild. This is in contrast with deterministic food web models such as EwE or Atlantis (Fulton 2001) that are often too smooth to capture natural patterns of year-to-year variability or pseudo-cycles effectively. Other deterministic ecosystem models may produce similar levels of variability by including detailed biological processes (Kishi and others 2011; Kearney and others 2012). Many of these processes are in general poorly understood and hard to be parameterised with precision and simulated dynamics can be highly sensitive to biological assumptions in these models (Gentleman and others 2003; Subbey and others 2014). The NDND offers a parsimonious complement to—either simple or complex—deterministic models for the simulation of inter-annual variability in food webs.
In comparison to the simulated data, the empirical observations suggest strong density dependence, in particular for omnivorous zooplankton and pelagic and demersal fishes. This can be a statistical artefact resulting from the small number of observations (the slopes of the growth-density functions being non-significant) or a true feature. Whether density dependence reflects compensation (growth declining towards zero with increasing abundance), overcompensation (negative growth at high abundance) or depensation (low growth at low abundance) remains a key issue in population dynamics studies and in marine fish populations (Rose and others 2001). Determining the presence and exact shape of the growth–abundance relationship is problematic for many species, and slight variations in the degree of compensation can greatly affect the modelled dynamics of animal populations (Ginzburg and others 1990; Turchin 2003; Sibly and others 2005). Aubin and others (2011) have shown that regardless of the exact shape of the population growth curve, there exists a region in the growth–biomass space where population trajectories can remain viable. The NDND simulations show great variability in species growth and abundance and appear to explore the viability space of each trophospecies, rather than to depict clear functional forms that could be interpreted unambiguously. For species with potentially high growth rates—that are also at lower trophic levels—maximum growth rates decline with species abundance indicating compensatory (herbivorous zooplankton) and overcompensatory (omnivorous zooplankton, benthos and pelagic fishes) dynamics. For birds and mammals, there is no apparent density dependence, and this likely results from the fact that their biomasses rarely approach carrying capacity in the 28-year simulation period because the biomasses for these species at the start of the simulation period are low.
Temporal changes in trophic control are well known (Hunt and McKinnell 2006), and may be indicative of changes in ecosystem states (Frank and others 2006). Thus, understanding the underlying causes behind these shifts can be beneficial to the management of marine resources. Fishing and climate are arguably the most important drivers of change in ecosystem state (Jackson and others 2001; Frank and others 2006; Lehodey and others 2006), but we show here that the observed decadal swings in trophic control may also simply emerge from random predatory interactions. The dynamic consequences of top-down control on the food web as a whole are not known, and our results indicate no apparent relationship between trophic control and food web stability. This is contrary to empirical data which indicate slightly lower food web stability (see below) when the system is top-down controlled. Whether there is a causal relationship between food web dynamics and trophic control, as indicated by the empirical data, remains to be resolved.
Given the stochastic nature of the NDND model and the lack of clear compensatory dynamics in the simulations, it is not surprising that model simulations are less synchronous than observed in the wild. This suggests that processes that favour synchronous dynamics in real systems, such as dispersal and environmental fluctuations (Gouhier and others 2010), and which are lacking in the NDND, might be required to reproduce appropriate levels of synchrony. In contrast, the simulated stability is similar to observations from the Barents Sea and other areas. This is, to some extent, in line with theory (Allesina and Pascual 2008) that suggests that the food web stability is robust to perturbations of interaction strength. It is assumed that factors that synchronise population fluctuations within and between communities are expected to be destabilising (Gouhier and others 2010), implying that synchrony and stability should be inversely related. This feature is evident in the empirical data but not in the simulations. The lack of correlation between food web synchrony and stability in the simulations is however in line with other studies (Bell and others 2014), thus the empirical food web dynamics in the Barents Sea may represent an extreme case.
Climate Variability and Fishing
Ocean climate variations and fishing were not explicitly incorporated in the model simulations, although these have been described as strong drivers of the Barents Sea ecosystem dynamics (Dalpadado and others 2012, 2014; Johannesen and others 2012). Although this could be a major shortcoming in a standard modelling approach designed to reproduce the past trajectory of ecosystem states, the NDND framework is compatible with extreme simplifications, even of the external pressures. The random sampling of trophic flows is somehow analogous to incorporating extreme variability in external controls such as climate, affecting all parts of the ecosystem. Therefore, current simulations implicitly assume random ocean climate variations, rather than climate stability. A more explicit representation of climate impact on ecosystem within the model is technically possible, but this would require in-depth understanding and quantification of the mechanisms by which ocean climate variability affects all parts of the ecosystem. As it stands, the NDND model constitutes a ‘null’ model against which the performance of more complex models that explicitly include ocean climate forcing can be evaluated.
During the 28 years investigated in this study, fishing has been significant in the Barents Sea. Removal of biomass through fishing is a systematic pressure and it affects well-defined components of the ecosystem (commercial fish and shellfish), although indirect effects on non-exploited species have also been reported. An in-depth investigation of how fishing dynamics may alter the patterns of ecosystem variability in the Barents Sea is beyond the scope of this study. However, we explored the systematic effects of fishing through sustained removal of key species groups by running supplementary simulations in which fishing export was included for the pelagic and demersal groups (Supplementary Material S4). Results from these simulations show that in the presence of fishing the biomass of demersals tends to be lower (−6.2%) at the end of the 28-year simulation period, than when fishing is not included. Conversely, the biomass of the pelagic group tends to increase (+18.8%), possibly as a result of predation release from the demersal group. These are relatively minor changes in comparison with variability in fish biomass between simulations with identical setups (1 and 3 orders of magnitude for demersals and pelagics, respectively). For other species groups, there are no visible changes in biomass in the simulations that incorporate fishing. In these simulations, other ecosystem properties (mean trophic flows, density dependence, top-down and bottom-up trophic controls, stability and synchrony) remained virtually unchanged (Supplementary Material S4). These results do not show how actual fishing that has taken place in the Barents Sea has affected the dynamics of the ecosystem, but they illustrate that the intrinsic variability generated in the NDND model simulation vastly exceeds the effect of fishing when averaged over several decades.
Limitations and Future Developments
Like other modelling approaches, the NDND relies on a set of assumptions, mathematical formulation and parameterisation, which, if violated or wrongly specified, will affect the results and conclusions. Model outputs are expected to be sensitive to the level of complexity of the food web structure, parameter values and transition probabilities.
In the NDND, the food web topology is set a priori and remains fixed during simulations. The present version of the Barents Sea application consists of a simple food web topology, inspired from Ulanowicz’s suggestion that food web complexity can be set to few species and trophic links and still conserve essential properties at ecosystem level (Ulanowicz and others 2014). Yet, alterations in the complexity of the representation of the food web can affect model behaviour. The structural food web properties (link density, connectance and so on) of the current food web topology deviate much from those derived from more complex food webs (Link 2002; Planque and others 2014b). The dynamic consequences of simplifying the topological complexity are difficult to anticipate, but the structural properties of the food web may affect the stability and, thus, the synchrony of food webs (Allesina and Pascual 2008). Few analyses of the kind have been conducted so far (Pinnegar and others 2005; Giricheva 2015), and it is unclear how general their results might be. A dedicated analysis of the influence of food web topological choices on emerging patterns in Barents Sea ecosystem simulations remains to be conducted.
We used life-history theory and metabolic theory of ecology to derive input parameter values for each individual trophospecies. These estimates are imprecise and traits are known to vary within and between species and with time (Rosa and Seibel 2008). For example, the assumption that “other losses” (μ) is equal to three times the basal metabolic rate (BMR) may be reasonable for fish and mammals (Winberg 1956, Speakman and Selman 2003) but less so for other trophospecies. Dedicated sensitivity analysis is needed to determine which parameters the model is most sensitive to and what range of dynamics may be expected given the natural variability and uncertainties in input parameter values. The realism of the NDND model may be improved using model parameters and food web structure that can dynamically vary on evolutionary time scales (for example, McKane 2004; Fussman and others 2007; Loreau 2010).
An important assumption in the NDND is that all possible transitions, from a food web state at time t to the subsequent food web state at time t + 1, are equiprobable. This assumption is consistent with Occam’s razor principle of using the simplest possible representation of the stochastic process (here a uniform distribution). While a model with mechanistic trophic functional responses constitutes a caricature of a fully known and deterministic system, the NDND model with random trophic interactions drawn from uniform distributions constitutes a caricature at the other end of the spectrum, of a modestly known, non-deterministic and highly variable system. Real food webs are likely to lie somewhere between these two extremes. Dedicated analyses are necessary to evaluate the effect of changing the probability distribution (between the uniform distribution of the current NDND and the Dirac delta function of deterministic trophic functional responses) on the model emerging patterns. Empirical support is equally needed to test whether the uniform distribution is the most appropriate representation of transition probabilities.
Reference Model
Given that the NDND model is both parsimonious and capable of simulating a multitude of patterns of food web dynamics, we suggest that the NDND model can serve as a reference or null model for the variability of marine ecosystem. The analysis of additional patterns would serve to highlight which patterns can be explained by a minimal set of rules and constraints, and which patterns may require the consideration of additional processes. Multiple simulations can be used to explore how the range of possible food web configurations may expand into the future, as exemplified in Figure 2. Projecting future ecosystem states and dynamics may not be possible with current models beyond short time horizons (Beckage and others 2011; Petchey and others 2015; Planque 2016); however, given its constrained stochastic nature, the NDND model may be used to evaluate the forecast horizon of different ecosystem properties. Complex ecosystem models are generally not designed to perform predictions, but are rather used to evaluate tradeoffs in ecosystem configurations or to investigate possible responses to specific pressures such as climate warming or exploitation (Fulton and others 2011). The NDND model can be used for similar purpose, for example, to investigate the effects of changes in environmental or fishing conditions on the food web dynamics.
Conclusions
We show here how to parameterise a simple NDND food web model using empirical observations, metabolic theory and life-history theory. Using a range of patterns to evaluate model performance against observations, we show that this parsimonious simulation model can reproduce many of the patterns observed in a real complex biological system (The Barents Sea food web) without requiring a complex model structure or deterministic trophic functional responses. Rather, this simple approach provides a reference model for expected patterns of ecosystem dynamics. Many of the patterns observed empirically in the Barents Sea can be interpreted as resulting from the interplay of random interactions with few ecological constraints.
References
Aubin JP. 1991. Viability theory. Berlin: Springer.
Aubin JP, Bayen AM, Saint-Pierre P. 2011. Viability theory: new directions. Berlin: Springer.
Allesina S, Pascual M. 2008. Network structure, predator–prey modules, and stability in large food webs. Theor Ecol 1:55–64.
Beckage B, Gross LJ, Kauffman S. 2011. The limits to prediction in ecological systems. Ecosphere. doi:10.1890/ES11-00211.1.
Bell RJ, Fogarty MJ, Collie JS. 2014. Stability in marine fish communities. Mar Ecol Prog Ser 504:221–39.
Blanchard JL, Pinnegar JK, Mackinson S. 2002. Exploring marine mammal-fishery interactions using ‘Ecopath with Ecosim’: modelling the Barents Sea Ecosystem. Sci Ser Tech Rep CEFAS Lowestoft 117:1–52.
Blenckner T, Niiranen S. 2013. Biodiversity-marine food-web structure, stability, and regime shifts. In: Roger P, Ed. Climate vulnerability. Oxford: Oxford Academic Press. p 203–12.
Blumer A, Ehrenfeucht A, Haussler D, Warmuth MK. 1987. Occam’s razor. Inform Process Lett 24:377–80.
Brown JH, Gillooly JF, Allen AP, Savage VM, West GB. 2004. Toward a metabolic theory of ecology. Ecology 85:1771–89.
Burnham KP, Anderson DR. 2002. Model selection and multimodel inference: a practical information-theoretic approach. New York: Springer.
Christensen V, Walters CJ. 2004. Ecopath with Ecosim: methods, capabilities and limitations. Ecol Mod 172:109–39.
Clark JS, Carpenter SR, Barber M, Collins S, Dobson A, Foley JA, Lodge DM, Pascual M, Pielke R, Pizer W, Pringle C, Reid WV, Rose KA, Sala O, Schlesinger WH, Wall DH, Wear D. 2001. Ecological forecasts: an emerging imperative. Science 293:657–60.
Cury P, Shannon L, Shin YJ. 2003. The functioning of marine ecosystems: a fisheries perspective. In: Sinclair M, Valdimarson G, Eds. Responsible fisheries in the marine ecosystem. FAO Fisheries Technical Paper No 400. pp 103–123.
Dalpadado P, Ingvaldsen RB, Stige LC, Bogstad B, Knutsen T, Ottersen G, Ellertsen B. 2012. Climate effects on Barents Sea ecosystem dynamics. ICES J Mar Sci 69:1303–16.
Dalpadado P, Arrigo KR, Hjøllo SS, Rey F, Ingvaldsen RB, Sperfeld E, van Dijken GL, Stige LC, Olsen A, Ottersen G. 2014. Productivity in the Barents Sea—response to recent climate variability. PLoS One. doi:10.1371/journal.pone.0095273.
Dommasnes A, Christensen V, Ellertsen B, Kvamme C, Melle W, Nøttestad L, Pedersen T, Tjelmeland S, Zeller D. 2001. An Ecopath model for the Norwegian Sea and Barents Sea. Fish Cen Res Rep 9:213–39.
Englund G, Leonardsen K. 2008. Scaling up the functional response for spatially heterogeneous systems. Ecol Lett 11:440–9.
Fossheim M, Primicerio R, Johannesen E, Ingvaldsen RB, Aschan MM, Dolgov AV. 2015. Recent warming leads to a rapid borealization of fish communities in the Arctic. Nat Clim Chang. doi:10.1038/NCLIMATE2647.
Frank KT, Petrie B, Shackell NL, Choi JS. 2006. Reconciling differences in trophic control in mid-latitude marine ecosystems. Ecol Lett 9:1096–105.
Fulton EA. 2001. The effects of model structure and complexity on the behaviour and performance of marine ecosystem models. Tasmania: Ph.D. thesis, University of Tasmania, Hobart. pp 1–429.
Fulton EA, Link JS, Kaplan IC, Savina-Rolland M, Johnson P, Ainsworth C, Horne P, Gorton R, Gamble RJ, Smith ADM, Smith DC. 2011. Lessons in modelling and management of marine ecosystems: the Atlantis experience. Fish Fish 12:171–88.
Fussman GF, Loreau M, Abrams P. 2007. Eco-evolutionary dynamics of communities and ecosystems. Funct Ecol 21:465–77.
Gentleman W, Leising A, Frost B, Strom S, Murray J. 2003. Functional responses for zooplankton feeding on multiple resources: a review of assumptions and biological dynamics. Deep-Sea Res II 50:2847–75.
Gillooly JF, Brown JH, West GB, Savage VM, Charnov EL. 2001. Effects of size and temperature on metabolic rate. Science 293:2248–51.
Ginzburg LR, Ferson S, Akcakaya HR. 1990. Reconstructibility of density dependence and the conservative assessment of extinction risks. Conserv Biol 4:63–70.
Giricheva E. 2015. Aggregation in ecosystem models and model stability. Prog Oceanogr 134:190–6.
Gouhier TC, Guichard F, Gonzalez A. 2010. Synchrony and stability of food webs in metacommunities. Am Nat 175:E16–34.
Grimm V, Railsback SF. 2005. Individual-based modelling and ecology. Princeton: Princeton University Press.
Grimm V, Railsback SF. 2012. Pattern-oriented modelling: a ‘multi-scope’ for predictive systems ecology. Philos Trans R Soc Lond B 367:298–310.
Hartig F, Calabrese JM, Reineking B, Wiegand T, Huth A. 2011. Statistical inference for stochastic simulation models-theory and application. Ecol Lett 14:816–27.
Hewitt DA, Hoenig JM. 2005. Comparison of two approaches for estimating natural mortality based on longevity. Fish Bull 103:433–7.
Hoenig JM. 1983. Empirical use of longevity data to estimate mortality rates. Fish Bull 82:898–903.
Holling CS. 1965. The functional response of predators to prey density and its role in mimicry and population regulation. Mem Entomol Soc Can 45:5–60.
Holt J, Icarus AJ, Anderson TR, Brewin R, Butenschön M, Harle J, Huse G, Lehodey P, Lindemann C, Memery L, Salihoglu B, Senina I, Yool A. 2014. Challenges in integrative approaches to modelling the marine ecosystems of the North Atlantic: physics to fish and coasts to ocean. Prog Oceanogr 129:285–313.
Hunsicker ME, Ciannelli L, Bailey KM, Buckel JA, White WJ, Link JS, Essington TE, Gaichas S, Anderson TW, Brodeur RD, Chan KS, Chen K, Englund G, Frank KT, Freitas V, Hixon MA, Hurst T, Johnson DW, Kitchell JF, Reese D, Rose GA, Sjodin H, Sydeman WJ, van der Veer HW, Vollset K, Zado S. 2011. Functional responses and scaling in predator–prey interactions of marine fishes: contemporary issues and emerging concepts. Ecol Lett 14:1288–99.
Hunt GL, McKinnell S. 2006. Interplay between top-down, bottom-up, and wasp-waist control in marine ecosystems. Prog Oceanogr 68:115–24.
ICES. 2014a. Report of the Arctic fisheries working group (AFWG). ICES CM2014/ACOM: 05. pp 1–656.
ICES. 2014b. First interim report of the working group on integrated assessment of the Barents Sea. ICES CM 2014/SSGRSP: 04. pp 1–49.
Jackson JBC, Kirby MX, Berger WH, Bjorndal KA, Botsford LW, Bourque BJ, Bradbury RH, Cooke R, Erlandson J, Estes JA, Hughes TP, Kidwell S, Lange CB, Lenihan HS, Pandolfi JM, Peterson CH, Steneck RS, Tegner MJ, Warner RR. 2001. Historical overfishing and the recent collapse of coastal ecosystems. Science 293:629–37.
Jennings S, Greenstreet SPR, Hill L, Piet GJ, Pinnegar JK, Warr KJ. 2002. Long-term trends in the trophic structure of the North Sea fish community: evidence from stable-isotope analysis, size-spectra and community metrics. Mar Biol 141:1085–97.
Johannesen E, Ingvaldsen RB, Bogstad B, Dalpadado P, Eriksen E, Gjøsæter H, Knutsen T, Skern-Mauritzen M, Stiansen JE. 2012. Changes in Barents Sea ecosystem state, 1970–2009: climate fluctuations, human impact, and trophic interactions. ICES J Mar Sci 69:880–9.
Kearney K, Stock C, Aydin K, Sarmiento JL. 2012. Coupling planktonic ecosystem and fisheries food web models for a pelagic ecosystem: description and validation for the subarctic Pacific. Ecol Mod 237–238:43–62.
Kishi MJ, S-i Ito, Megrey BA, Rose KA, Werner FE. 2011. A review of the NEMURO and NEMURO.FISH models and their application to marine ecosystem investigations. J Oceanogr 67:3–16.
Klages M, Boetius A, Christensen JP, Deubel H, Piepenburg D, Schewe I, Soltwedel T. 2004. The benthos of Arctic Seas and its role for the organic carbon cycle at the seafloor. In: Stein R, Macdonald RW, Eds. The organic carbon cycle in the Arctic Ocean. Berlin: Springer. p 139–67.
Koen-Alonso M, Yodzis P. 2005. Multispecies modelling of some components of the marine community of northern and central Patagonia, Argentina. Can J Fish Aquat Sci 62:1490–512.
Koen-Alonso M. 2007. A process-oriented approach to the multispecies functional response. In: Rooney N, McCann KS, Noakes DLG, Eds. From energetics to ecosystems: the dynamics and structure of ecological systems. Dordrecht: Springer. p 1–36.
Lade SJ, Tavoni A, Levin SA, Schlüter M. 2013. Regime shifts in a social-ecological system. Theor Ecol 6:359–72.
Landres PB, Morgan P, Swanson FJ. 1999. Overview of the use of natural variability concepts in managing ecological systems. Ecol Appl 9:1179–88.
Lehman C, Tilman D. 2000. Biodiversity, stability and productivity in competitive communities. Am Nat 156:534–52.
Lees K, Pitois S, Scott C, Frid C, Mackinson S. 2006. Characterizing regime shifts in the marine environment. Fish Fish 7:104–27.
Lehodey PJ, Alheit J, Barange M, Baumgartner T, Beaugrand G, Drinkwater K, Fromentin J-M, Hare SR, Ottersen G, Perry RI, Roy C, van der Lingen CD, Werner F. 2006. Climate variability, fish and fisheries. J Climate 19:5009–30.
Libralato S, Coll M, Tudela S, Palomera I, Pranovi F. 2008. Novel index for quantification of ecosystem effects of fishing as removal of secondary production. Mar Ecol Prog Ser 355:107–29.
Link J. 2002. Does food web theory work for marine ecosystems? Mar Ecol Prog Ser 230:1–9.
Longhurst A. 2010. Mismanagement of marine fisheries. Cambridge: Cambridge University Press.
Loreau M, de Mazancourt C. 2008. Species synchrony and its drivers: neutral and nonneutral community dynamics in fluctuating environments. Am Nat 172:E48–66.
Loreau M. 2010. Linking biodiversity and ecosystems: towards a unifying ecological theory. Philos Trans R Soc Lond B 365:49–60.
Lotze HK, Worm B. 2008. Historical baselines for large marine animals. Trends Ecol Evol 24:254–62.
Makarieva AS, Gorshkov VG, Li Bai-Lian, Chown SL, Reich PB, Gavrilov VM. 2008. Mean mass-specific metabolic rates are strikingly similar across life’s major domains: evidence for life’s metabolic optimum. Proc Natl Acad Sci USA 105:16994–9.
McKane AJ. 2004. Evolving complex food webs. Eur Phys J B 38:287–95.
Möllmann C, Folke C, Edwards M, Conversi A. 2015. Marine regime shifts around the globe: theory, drivers and impact. Philos Trans R Soc Lond B. doi:10.1098/rstb.2013.0260.
Mullon C, Fréon P, Cury P, Shannon L, Roy C. 2009. A minimal model of the variability of marine ecosystems. Fish Fish 10:115–31.
Pauly D, Christensen V, Dalsgaard J, Froese R, Torres F Jr. 1998. Fishing down marine food webs. Science 279:860–3.
Pauly D, Christensen V, Froese R, Palomares M. 2000. Fishing down aquatic food webs. Am Sci 88:46–51.
Petchey OL, Pontarp M, Massie TM, Kéfi S, Arpat O, Weilenmann M, Palamara GM, Altermatt F, Matthews B, Levine JM, Childs DZ, McGill BJ, Schaepman ME, Schmid B, Spaak P, Beckerman AP, Pennekamp F, Pearse IS. 2015. The ecological forecast horizon, and examples of its uses and determinants. Ecol Lett 18:597–611.
Pinnegar JK, Blanchard JL, Mackinson S, Scott RD, Duplisea DE. 2005. Aggregation and removal of weak-links in food web models: system stability and recovery from disturbance. Ecol Mod 184:229–48.
Planque B. 2016. Projecting the future state of marine ecosystems, ‘la grande illusion’? ICES J Mar Sci 73:204–8.
Planque B, Lindstrøm U, Subbey S. 2014a. Non-deterministic modelling of food web dynamics. PLoS One. doi:10.1371/journal.pone.0108243.
Planque B, Primicerio R, Michalsen K, Aschan M, Certain G, Dalpadado P, Gjøsæter H, Hansen C, Johannesen E, Jørgensen LL, Kolsum I, Kortsch S, Leclerc L-M, Omli L, Skern-Mauritzen M, Wiedmann M. 2014b. Who eats whom in the Barents Sea: a food web topology from plankton to whales. Ecology 95:E95–124.
Pranovi F, Link J, Fu C, Cook AM, Liu H, Gaichas S, Friedland KD, Utne KR, Benoit HP. 2012. Trophic-level determinants of biomass accumulation in marine ecosystems. Mar Ecol Prog Ser 459:185–201.
Pranovi F, Libralato S, Zucchetta M, Link J. 2014. Biomass accumulation across trophic levels: analysis of landings for the Mediterranean Sea. Mar Ecol Prog Ser 512:201–16.
Rosa R, Seibel BA. 2008. Synergistic effects of climate-related variables suggest future physiological impairment in a top oceanic predator. Proc Natl Acad Sci USA 105:20776–80.
Rose K, Cowan JH Jr, Winemiller KO, Myers RA, Hilborn GR. 2001. Compensatory density dependence in fish populations: importance, controversy, understanding and prognosis. Fish Fish 2:293–327.
Rosen R. 1969. On mathematics and biology. In: Saaty TL, Weyl FJ, Eds. The spirit and use of mathematical sciences. New York: McGraw-Hill Press. p 203–18.
Ruiz J, Kuikka S. 2012. Tangled ecosystem models: the temptations of siren songs and silence. Sci Mar 76:191–4.
Savage VM, Gillooly JF, Brown JH, West GB, Charnov EL. 2004. Effects of body size and temperature on population growth. Am Nat 163:429–41.
Scheffer M, Carpenter SR. 2003. Catastrophic regime shifts in ecosystems: linking theory to observation. Trends Ecol Evol 18:648–56.
Shannon L, Osman W, Jarre A. 2014. Trophic level-based indicators to track fishing impacts across marine ecosystems. Mar Ecol Prog Ser 512:115–40.
Sibly RM, Barker D, Denham MC, Hone J, Pagel M. 2005. On the regulation of populations of mammals, birds, fish, and insects. Science 309:607–10.
Smout S, Lindstrøm U. 2007. Multispecies functional response of the minke whale Balaenoptera acutorostrata based on small-scale foraging studies. Mar Ecol Prog Ser 341:277–91.
Speakman JR, Selman C. 2003. Physical activity and resting metabolic rate. P Nutr Soc 62:621–34.
Subbey S, Devine JA, Schaarschmidt U, Nash RDM. 2014. Modelling and forecasting stock–recruitment: current and future perspectives. ICES J Mar Sci 71:2307–22.
Subbey S, Planque B, Lindstrøm U. 2016. Exploring stochasticity and imprecise knowledge based on linear inequality constraints. J Math Biol 73:575–95.
Turchin P. 2003. Complex population dynamics. Princeton: Princeton University Press.
Ulanowicz RE, Holt RD, Barfield M. 2014. Limits on ecosystem trophic complexity: insights from ecological network analysis. Ecol Lett 17:127–36.
Valiela I. 1984. Marine ecological processes. Chicago: Springer.
Williams RJ, Martinez ND. 2004. Stabilization of chaotic and non-permanent food-web dynamics. Eur Phys J B 38:297–303.
Winberg GG. 1956. Rate of metabolism and food requirements of fishes. Fish Res Bd Can Transl Ser 194:1–253.
Yodzis P, Innes S. 1992. Body size and consumer-resource dynamics. Am Nat 139:1151–75.
Acknowledgments
The authors wish to thank the editor and two anonymous reviewers for their critical, inspiring and constructive inputs to earlier versions of the manuscript. This work was supported by the Marine Processes program of the Institute of Marine Research, Norway.
Author information
Authors and Affiliations
Corresponding author
Additional information
Ulf Lindstrøm and Benjamin Planque have contributed equally to the manuscript.
Author Contributions
Benjamin Planque conceived the study; Ulf Lindstrøm and Benjamin Planque performed research; Ulf Lindstrøm and Benjamin Planque analyzed data; Ulf Lindstrøm, Benjamin Planque and Sam Subbey have contributed new methods or models; Ulf Lindstrøm, Benjamin Planque and Sam Subbey have written the article.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lindstrøm, U., Planque, B. & Subbey, S. Multiple Patterns of Food Web Dynamics Revealed by a Minimal Non-deterministic Model. Ecosystems 20, 163–182 (2017). https://doi.org/10.1007/s10021-016-0022-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10021-016-0022-y