Abstract
Acidithiobacillus ferrooxidans is a heavy metal-tolerant acidophilic chemolithotroph found in acidic mine effluent and is used commercially in the bioleaching of sulfide ores. In this work, we investigated the interplay between divalent cadmium (Cd2+) resistance and expression of genes involved in the sulfur assimilation pathway (SAP). We also investigated the response of the thiol-containing metal-chelating metabolites, cysteine and glutathione(GSH), to increasing Cd2+ concentrations. During growth in the presence of 30 mM Cd2+, the concentrations of mRNA for 5 genes in the SAP pathway increased more than fourfold: these encode ATP sulfurylase (ATPS), adenosine 5′-phosphosulfate (APS) reductase, sulfite reductase (SiR), serine acetyltransferase (SAT) and O-acetylserine (thiol) lyase (OAS-TL). Increased transcription was also reflected in increased enzyme activities: those of SAT and adenosylphosphosulfate reductase (APR) reached a peak of 26- and 15.8-fold, respectively, compared to the control culture in the presence of 15 mM Cd2+. In contrast, the activity of OAS-TL, which is responsible for the biosynthesis of cysteine, was diminished. At the metabolite level, the intracellular cysteine and GSH contents nearly doubled. These results suggested that Cd2+ induced transcription of SAP genes, while directly inhibiting the activities of some enzymes (e.g., OAS-TL). Overall, these results are consistent with a detoxification/resistance mechanism involving enhanced sulfur uptake and sequestration of Cd2+ by cysteine and glutathione.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Acidithiobacillus ferrooxidans is a Gram-negative, chemolithotrophic, acidophilic Proteobacterium that plays a critical economic role in bioleaching of sulfide metal ores. Members of the genus Acidithiobacillus derive energy from metabolizing sulfur, sulfide metal ores, and ferrous iron and can be isolated from acidic environments such as mine drainage effluent and acidified sulfate-containing soils. A. ferrooxidans is the most studied species within the genera and is widely used for bioleaching, where it obtains energy by oxidation of sulfur and ferrous iron and uses molecular oxygen as the terminal electron acceptor (Vera et al. 2013). These bacteria are also notable for their resistance to high concentrations of heavy metals. A. ferrooxidans can grow in the presence of exceptionally high divalent cadmium (Cd2+) concentrations. This property is important for its use in bioleaching processes, where Cd2+ and other metal levels can be present in the 5–100 mM range (Baillet et al. 1997). Cadmium is a nonessential metal that is toxic to most organisms even at low concentrations, disrupting pathways involved in carbon, nitrogen, and sulfur metabolism, as well as cellular redox chemistry (Toppi and Gabbrielli. 1999; Blazhenko et al. 2006). Acidophilic microorganisms found in acidic mine drainage (optimal growth pH < 3) are often highly resistant to heavy metals such as cadmium, with resistance being mediated by multiple mechanisms which are generally more efficient than those found in neutrophiles.
Prokaryotes have evolved a series of general mechanisms to control the cellular levels of heavy metals that are activated when concentrations exceed acceptable levels (Nies 1999), conferring in most organisms a low resistance to Cd2+; for example, Escherichia coli has a minimum inhibitory concentration (MIC) of 0.7 mM for Cd2+ in Luria–Bertani (LB) medium, while Ralstonia sp. CH34 has an MIC of 0.8 mM (Nies 2003, 2000). In Gram-negative bacteria, resistance-nodulation cell division (RND)-driven tripartite protein complexes pump out a wide array of substrates, with the metal transporter CzcA from Cupriavidus metallidurans being a typical example (Shamim et al. 2014; Kim et al. 2011; Scherer and Nies 2009). However, these mechanisms only confer resistance at concentrations of approximately 1 mM or less and do not account for the high resistance observed in organisms found in acidic mine drainage effluent, where the concentrations of heavy metals such as cadmium are typically 1 to 2 orders of magnitude higher. A critical question is: what are the mechanisms of heavy metal resistance in highly resistant organisms such as A. ferrooxidans? Answering this question will have a profound impact on acidic mine effluent detoxification and human health (Dopson et al. 2003, 2014).
Eukaryotes have evolved distinct resistance mechanisms that enable survival in the presence of toxic heavy metals. One of the most common mechanisms found in yeast, algae, photosynthetic protists, and plants involves intracellular metal chelation by glutathione (GSH), phytochelatins (PCs), and other low molecular weight sulfur-containing peptides derived from cysteine (Cozatl et al. 2005). These peptides bind a variety of metals in the cytosol. Depending on the organism, the metal–PC or metal–GSH complexes are actively transported into vacuoles. Whether such chelating or sequestration mechanisms play a role in A. ferrooxidans Cd2+ resistance is unknown. Interestingly, all known heavy metal-chelating agents contain sulfur and are therefore products of the sulfur assimilation pathway (SAP) that is present in photosynthetic organisms, fungi, and many bacteria. Thus, the SAP may play a critical role in conferring resistance to heavy metals.
The SAP is present in photosynthetic organisms, fungi, and many bacteria. Sulfate is transported into the cytosol, where it undergoes reduction prior to incorporation into cysteine, GSH, methionine, and a variety of other metabolites (Fig. 1). According to bioinformatic analysis, a metabolic model was reconstructed for the uptake and assimilation of sulfur in A. ferrooxidans (Valdés et al. 2003). In this pathway, ATP sulfurylase (ATPS, encoded by the cysD and cysN genes) catalyzes the first step of inorganic sulfate assimilation, activating sulfate via an ATP-dependent reaction leading to the formation of adenosine 5′-phosphosulfate (APS) and pyrophosphate. APS is directly reduced to sulfite by APS reductase (encoded by the cysH gene) and then the sulfite is reduced to sulfide by NADPH-SiR (encoded by the cysIJ gene). The first step in cysteine biosynthesis involves serine acetyltransferase (SAT), which is encoded by cysE. CysE catalyzes the reaction of serine and acetyl-coenzyme A to produce OAS. The second step of cysteine synthesis involves an OAS-sulfhydrylase, also called OAS-TL (encoded by the cysM gene). OAS-TL converts O-acetylserine (OAS) and sulfide into cysteine and acetate (Fig. 1). Although a SAP has not been characterized in A. ferrooxidans, the pathway shown in Fig. 1 is a reasonable framework upon which an understanding of sulfur uptake can be built.
Changes in relative transcript levels of sulfate assimilation biosynthesis enzymes in A. ferrooxidans under Cd2+ stress, in comparison to an unchallenged control (5 mM Cd2+, 15 mM Cd2+, 30 mM Cd2+). ATPS ATP sulfurylase, APR adenosylphosphosulfate reductase, SiR sulfite reductase, SAT serine acetyltransferase, OAS o-acetylserine, OAS-TL, and o-acetylserine (thiol) lyase. The induction ratios were determined by comparing the relative transcript cDNA levels from A. ferrooxidans grown in the Cd2+ concentrations indicated versus those obtained in control cells grown in the absence of the metal and normalized by 16S rRNA expression in each case. Values of standard deviations were obtained with three biological replicates. CT, PCR cycle at which fluorescence first rises above a threshold background fluorescence. The transcript levels were final values (p < 0.05)
Previous work has shown that in yeast, protists, and plants, expression of SAP genes is up-regulated by exposure to Cd2+ (Cozatl et al. 2005). For example, the ATPS gene was induced 13-fold in the model plant species Arabidopsis thaliana upon Cd2+ (200 μM) exposure (Harada et al. 2002); Cd2+ (1 mM) stress also promoted a 6-fold increase in the ATPS protein level in the yeast Saccharomyces cerevisiae (Vido et al. 2001). RNA blot analysis of transcript amounts indicated that in 6-week-old Brassica juncea plants, the genes for ATPS and APSR were strongly induced by exposure to Cd2+ (25 μM) (Heiss et al. 1999). In addition, a DNA microarray analysis of S. cerevisiae showed that the GSH synthesis gene (gsh1) and almost all transcripts of the enzymes involved in sulfur amino acid metabolism, especially MET14 (APK) and MET17 (PAPSR), were induced at 21- and sixfold, respectively, after exposure to Cd2+ (Momose and Iwahashi 2003). Proteomic analysis of PAPSR shows that its concentration is increased fivefold in response to 1 mM Cd2+ exposure (Vido et al. 2001). Similarly, the bryophyte Physcomitrella patens reacts to elevated Cd2+ concentrations by increasing the cellular concentrations of cysteine and GSH (Koprivova et al. 2002). In contrast, recent investigations of Cd2+-stressed bryophytes showed no increase in PC biosynthesis, but instead there was a significant increase in the GSH pool (Helbig K et al. 2008a, b).
Although the genetics of the SAP are well known, little attention has been paid to cytosolic concentrations of cysteine and GSH in response to heavy metal exposure in prokaryotes. Our goal is to elucidate the mechanism of Cd2+ resistance in A. ferrooxidans. In this work, we investigate gene regulation of the SAP by Cd2+ stress in A. ferrooxidans and demonstrate a connection between sulfur assimilation and Cd2+ tolerance.
Materials and methods
Strains, media, and culture conditions
A. ferrooxidans, ATCC 23270, was obtained from the American Type Culture Collection and was routinely grown at 30 °C with shaking (170 rpm) in 9 K medium, containing (NH4)2SO4 3.0 g/L, MgSO4 0.5 g/L, K2HPO4 0.5 g/L, KCl 0.1 g/L, and Ca(NO3)2 0.01 g/L with ferrous sulfate (10 g Fe2+/L) as the energy source. The medium was adjusted to pH 2.3 using sulfuric acid before FeSO4∙7H2O was added and then filter sterilized. Bacterial growth was assessed by measuring ferrous iron oxidation rates. The ferrous iron concentration in solution was determined volumetrically by titrating against potassium dichromate (K2Cr2O7) using sodium diphenylamine sulfonate (0.2 % wt/vol) as the indicator.
Total RNA extraction and complementary DNA (cDNA) synthesis
Total RNA was isolated using TRIzol® (Invitrogen Corporation, Carlsbad, USA) and purified with the RNeasy® mini kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instructions. On-column DNase digestion was performed with RNase-free DNase (Qiagen GmbH, Hilden, Germany) to remove genomic DNA. RNA integrity was checked by 1.5 % agarose gel electrophoresis and ethidium bromide staining. Total RNA was quantified at OD260 and OD280 with a NanoDrop® ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, USA) and then served as the template to synthesize cDNA.
Primer design for real-time PCR
Real-time PCR primers were designed using Primer Premier 5.0 and then synthesized by Sagon Biotech (Sagon, Shanghai, China). Annotations of A. ferrooxidans ATCC 23270 genes and their putative cellular functions were obtained from the Institute for Genomic Research website at http://cmr.tigr.org/tigr-scripts/CMRGenomePage, and the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/nuccore/cp001219.1) (Accession: CP001219.1; GI: 218517237). All primer pairs for selected genes that encode putative SAP components are listed in Table 1. Specific fragments were amplified and purified, and were then visualized by 1.5 % agarose gel electrophoresis and ethidium bromide staining to detect whether PCR resulted in single products of the expected sizes.
Real-time PCR detection
The real-time PCR mixture (final volume 50 μL) contained 25 μL of SYBR® Green Real-time PCR Master Mix (Toyobo Co., LTD., Osaka, Japan), 2 μL of a 10 mM solution of sense/anti-sense primer, 10 μL of cDNA template, and 11 μL of nuclease-free water. Real-time PCR was carried out with an iCycler iQ Real-time PCR detection system (Bio-Rad Laboratories, Inc., Irvine, CA, USA): 1 cycle of 95 °C for 30 s, and then 40 cycles of 95 °C for 15 s, 55 °C for 30 s, and 72 °C for 30 s. At the completion of each run, melting curves for the amplicons were measured by raising the temperature at 0.5 °C intervals from 55 to 95 °C while monitoring fluorescence. The specificity of the PCR amplification was evaluated by examining the derivatized T m melting curve for symmetry and a lack of non-specific peaks. Amplification of gene transcript amounts for samples from cultures grown in the presence of Cd2+ (5, 15, 30 mM) compared to the amounts from control samples was determined by analyses of the respective amounts of PCR products. The results were normalized against 16S rRNA to correct sample-to-sample variation. Each assay was run in triplicate.
Measurements of enzyme activities
Harvested bacterial cells were lysed by sonication (22 kHz) for 5 min with 1 min intervals of cooling and cell debris was removed by centrifugation at 10,000g for 10 min. Protein contents of the crude cell-free extracts were measured by the Folin method (Lowry et al. 1951), using bovine serum albumin as the standard. The activity of APS reductase in the extracts was evaluated by measuring the reduction of ferricyanide in the presence of sulfite and adenosine monophosphate (AMP) (Truper and Rogers 1971). The activity of SAT was estimated by the formation of OAS in the presence of 5,5-dithiobis-2-nitrobenzoic acid (DTNB) (Stiller et al. 2007). The activity of SiR was determined by measuring the reduction of NADPH (340 nm) in the presence of sulfite (Ostrowski et al. 1989). The activity of OAS-TL was estimated by measuring the formation of cysteine according to the method of Chronis (Schnell et al. 2007). Assays were performed with two samples for each treatment and each assay was run in triplicate, for a total of six replicates for each sample.
Measurement of cysteine and GSH by colorimetry
The cellular content of cysteine was assayed, by measuring the ability of cell lysates to form a Co2+–cysteine complex which has an optical absorbance maximum at 358 nm. The complex formation reaction was started by adding 3.0 mL NH4Cl–NH4OH buffer solution, 1.5 mL CoCl2 solution (100 mg/L Co2+) and 1.0 mL cysteine-containing solution sequentially into 10 mL colorimetric tubes. After 5 min incubation, measure the reactants by a UV spectrophotometer. The cysteine content was estimated by comparison with the absorption of samples to which a standard cysteine solution had been added.
The GSH content in cell lysates was measured by GSH formation of 5-phosphorothioate-2-nitrobenzoic acid with 5,5-dithiobis (2-nitrobenzoic acid) (DTNB). The reaction started by mixing 2.0 mL GSH-containing solution, 2.5 mL of Na2HPO4 buffer solution (0.32 mmol/L), and 0.5 mL of DTNB solution in a 10 mL colorimetric tubes. Absorbance was measured at 416 nm within 5 min, at which 5-phosphorothioate-2-nitrobenzoic acid exhibited maximum absorption. The GSH content of cell lysates was estimated by comparison with samples containing a known concentration of GSH.
Assays were performed with two samples for each treatment and each assay was run in triplicate, a total of six replicates for each sample.
Statistical analyses
Student’s t test (independent sample t test) was used to determine the statistical significance. The p values were calculated using the null hypothesis. A p value of less than 0.05 indicates that the two sets are statistically different.
Results
Vitality of A. ferrooxidans upon Cd2+ exposure
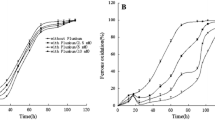
To estimate the vitality of A. ferrooxidans under Cd2+ stress, the effect of increasing Cd2+ concentrations on the iron-oxidizing activity of cultures was investigated (Fig. 2). Cd2+ inhibits iron-oxidizing activity, as judged by its effect on the rate of iron oxidation in the liquid medium. In the presence of 5 mM Cd2+, A. ferrooxidans cell extracts took a slightly longer time to oxidize 50 % of the iron compared with the control, while in the presence of higher Cd2+ concentrations the time for 50 % iron oxidation was longer by 6 and 8 min for 15 and 30 mM Cd2+, respectively, compared with the control. In addition, cell densities at the early stationary phase in the presence of 15 and 30 mM Cd2+ were smaller than those seen in the absence of the metal or in the presence of 5 mM Cd2+ (data not shown). Although A. ferrooxidans cells were still able to grow in the presence of 100 mM Cd2+, the culture cell density was very low.
Transcription patterns of genes involved in the SAP upon Cd2+ exposure
The transcript concentrations of sulfur assimilation-related genes were measured by real-time PCR to assess how Cd2+ affected gene transcription in A. ferrooxidans using total RNA extracted from A. ferrooxidans cells grown in the presence or absence of Cd2+. Treatment with Cd2+ affected transcription of genes coding for enzymes involved in sulfate reduction in a concentration-dependent manner (Fig. 1). In the presence of 5 mM Cd2+, transcript levels of the sulfate assimilation genes of A. ferrooxidans were not significantly changed, exhibiting natural fluctuations under the slightly unfavorable growth condition. In the presence of 15 mM Cd2+, most sulfate assimilation genes were up-regulated 1.2- to 3.0-fold, yet there was no observed effect on cysNC, a subunit of ATPS, suggesting that the metal concentration may have affected these sulfate assimilation genes differently. In the presence of 30 mM Cd2+, transcription levels of most sulfur assimilation genes were up-regulated, the exception being cysJ, which was slightly down-regulated. The increases in mRNA levels for ATPS (cysD-1), APR (cysH), SiR (cysI), SAT (cysE), and OAS-TL (cysM) were 7.29-, 6.58-, 4.4-, 4.0-, and 4.2-fold, respectively. Overall, exposure to high concentrations of Cd2+ had a significant effect on sulfate assimilation gene expression in A. ferrooxidans.
Thiol pools are increased upon Cd2+ stress
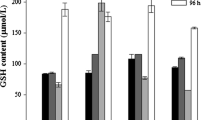
Thiol profiling was carried out to assess whether the observed increases in transcription result in changes in the concentrations of sulfur-containing metabolites. These analyses showed that thiol metabolite concentrations increased in a Cd2+ concentration-dependent manner (Table 2). GSH levels varied with Cd2+ concentration and exposure time in a more complicated fashion. Prior to 24 h of incubation, Cd2+ stress resulted in higher GSH levels compared to the control. At 72 h, GSH levels for most samples were lower than that at 12 h, with the exception being the culture incubated in the presence of 5 mM Cd2+, wherein a maximum concentration of GSH was observed. Finally, at 96 h, all the samples showed very high level of GSH, including the control.
After 12 h, Cd2+-stressed cells contained higher concentrations of cysteine and GSH than control cultures, as determined by UV spectrophotometry. In contrast, only slight changes in cysteine and GSH levels were observed 72 h after Cd2+ exposure. After 96 h these metabolites had clearly increased. Most notably, cells exposed to 15 mM Cd2+ exhibited significantly enhanced levels of both cysteine and GSH after 96 h of exposure.
Activities of cysteine-synthesizing enzymes were differentially affected by Cd2+
To examine whether altered transcription by Cd2+ stress is accompanied by changes in enzymatic activities, assays were carried out for key A. ferrooxidans SAP enzymes. When cell extracts were analyzed, enzymatic activity levels for various enzymes differed in response to Cd2+ concentrations (Table 3). The levels of SAT and APR activities were elevated with increasing Cd2+ concentration, showing peaks at 15 mM; however, the activities of SiR and OAS-TL were depressed at high metal concentrations, in spite of the observation that transcript levels of their respective genes were increased. These observations are consistent with direct Cd2+ inhibition of SiR and OAS-TL.
Discussion
Previous studies have indicated that the five metal resistance mechanisms identified in neutrophiles are also present in acidophiles, in some cases utilizing homologous proteins. However, a remarkable feature of acidophiles such as A. ferrooxidans is their tolerance of much higher concentrations of heavy metals, as judged by observed MIC values (Dopson et al. 2003). Eukaryotes provide a clue that resistance to heavy metals can be mediated by mechanisms involving chelation/sequestration by cysteine or GSH (Figueira et al. 2005; Gharieb 2004). If such a mechanism is responsible for Cd2+ resistance in A. ferrooxidans, the SAP would be predicted to be up-regulated by Cd2+, resulting in higher intracellular concentrations of GSH and cysteine (Helbig et al. 2008a, b; Hochgrafe 2007; Latinwo et al. 1998). In this study, we analyzed the effect of Cd2+ on the SAP gene transcription, enzymatic activities, and the intracellular levels of GSH and cysteine.
Real-time PCR showed that transcription of most genes in the SAP was up-regulated following exposure to elevated Cd2+. In addition, the thiol pool concentrations, including cysteine and GSH, were increased in the presence of Cd2+. Our findings are thus in agreement with published results for eukaryotes (Cozatl et al. 2005; Dominguez-Solis et al. 2001).
A surprising finding from our study was that elevated transcript levels do not necessarily result in enhanced enzymes activity levels. This phenomenon was observed for cysI (coding for SiR subunit) and cysM (coding for OAS-TL). The difference between transcription and enzymatic activity for those genes is likely due to Cd2+ binding to the enzymes, directly inhibiting their activities, a phenomenon observed for many metal irons (Wang et al. 1996; Bandyopadhyay et al. 1997). The precise mechanism of this inhibition is currently being investigated in our laboratories.
Another interesting observation made in this study was that two key enzymes in the SAP, SiR and OAS-TL, for synthesis of cysteine were depressed while the level of cysteine was elevated under Cd2+ stress. This observation suggests that A. ferrooxidans possesses an alternative biosynthetic pathway generating cysteine and GSH to that presented in Fig. 1 (Brzywczy et al. 2002; Marzluf 1997; Ono et al. 1999; Thomas and Surdin-Kerjan 1997).
Conclusion
Overall, the results presented herein clearly demonstrate that exposure to Cd2+ results in elevated expression of A. ferrooxidans SAP genes, with a concomitant increase in most of the respective enzyme activities. Critically, we show that Cd2+ exposure increases cellular concentrations of thiol-containing GSH and cysteine, providing evidence that resistance might be mediated by a mechanism involving heavy metal sequestration. A critical next step will be delineation of the fate of the thiol–metal complexes, and efforts to achieve this are ongoing in our laboratories.
References
Alonso A, Martinez JL (2000) Cloning and characterization of SmeDEF, a novel multidrug efflux pump from Stenotrophomonas maltophilia. Antimicrob Agents Chemother 44:3079–3086. doi:10.1128/AAC.44.11.3079-3086.2000
Baillet F, Magnin JP, Cheruy A, Ozil P (1997) Cadmium tolerance and uptake by a Thiobacillus ferrooxidans biomass. Environ Technol 18:631–637. doi:10.1080/09593331808616581
Bandyopadhyay D, Chatterjee AK, Datta AG (1997) Effect of cadmium, mercury and copper on partially purified hepatic flavokinase of rat. Mol Cell Biochem 167(1–2):73–80. doi:10.1023/A:1006815504302
Blazhenko OV, Zimmermann M, Kang HA, Bartosz G, Penninckx MJ, Ubiyvovk VM, Sibirny AA (2006) Accumulation of cadmium ions in the methylotrophic yeast Hansenula polymorpha. Biometals 19:593–599. doi:10.1007/s10534-006-0005-0
Brzywczy J, Sienko M, Kucharska A, Paszewski A (2002) Sulphur amino acid synthesis in Schizosaccharomyces pombe represents a specific variant of sulphur metabolism in fungi. Yeast 19:29–35. doi:10.1002/yea.798
Cozatl MD, Tavera LH, Navarro AH, Navarro HA, Sanchez RM (2005) Sulfur assimilation and glutathione metabolism under cadmium stress in yeast, protists and plants. FEMS Microbiol 29:653–671. doi:10.1016/j.femsre.09.004
Dominguez-Solis JR, Gutierrez-Alcala G, Vega JM, Romero LC, Gotor C (2001) The cytosolic O-acetylserine(thiol)lyase gene is regulated by heavy metals and can function in cadmium tolerance. J Biol Chem 276:9297–9302. doi:10.1074/jbc.M009574200
Dopson M, Austin BC, Koppineedi PR, Bond PL (2003) Growth in sulfidic mineral environments: metal resistance mechanisms in acidophilic micro-organisms. Microbiology 149:1959–1970. doi:10.1099/mic.0.26296-0
Dopson M, Ossandon FJ, Lövgren Land David SH (2014) Metal resistance or tolerance? Acidophiles confront high metal loads via both abiotic and biotic mechanisms. Front microbiol 5:1–4. doi:10.3389/fmicb.2014.00157
Figueira E, Lima IG, Pereira SIA (2005) Cadmium tolerance plasticity in Rhizobium leguminosarum bv. viciae: glutathione as a detoxifying agent. Can J Microbiol 51:7–14. doi:10.1139/W04-101
Gharieb MM, Gadd GM (2004) Role of glutathione in detoxification of metal(loid)s by Saccharomyces cerevisiae. Biometals 17:183–188. doi:10.1023/B:BIOM.0000018402.22057.62
Harada E, Yamaguchi Y, Koizumi N, Hiroshi S (2002) Cadmium stress induces production of thiol compounds and transcripts for enzymes involved in sulfur assimilation pathways in Arabidopsis. J Plant Physiol 159:445–448. doi:10.1078/0176-1617-00733
Heiss S, Schafer HJ, Haag-Kerwer A, Rausch T (1999) Cloning sulfur assimilation genes of Brassica juncea L.: cadmium differentially affects the expression of a putative low-affinity sulfate transporter and isoforms of ATP sulfurylase and APS reductase. Plant Mol Biol 39:847–857. doi:10.1023/A:1006169717355
Helbig K, Bleuel C, Krauss GJ, Nies DH (2008a) Glutathione and transition-metal homeostasis in Escherichia coli. J Bacteriol 190(15):5431–5438. doi:10.1128/JB.00271-08
Helbig K, Grosse C, Nies DH (2008b) Cadmium toxicity in glutathione mutants of Escherichia coli. J Bacteriol 190:5439–5454. doi:10.1128/JB.00272-08
Hochgrafe F, Mostertz J, Pother DC, Becher D, Helmann JD, Hecker M (2007) S-Cysteinylation is a general mechanism for thiol protection of Bacillus subtilis proteins after oxidative stress. J Biol Chem 282:25981–25985. doi:10.1074/jbc.C700105200
Kim EH, Nies DH, McEvoy MM, Rensing C (2011) Switch or funnel: how RND-type transport systems control periplasmic metal homeostasis. J Bacteriol 10(193):2381–2387. doi:10.1128/JB.01323-10
Koprivova A, Meyer AJ, Schween G, Herschbach C, Reski R, Kopriva S (2002) Functional knockout of the adenosine 5′-phosphosulfate reductase gene in Physcomitrella patens revives an old route of sulfate assimilation. J Biol Chem 277:32195–32201. doi:10.1074/jbc.M204971200
Latinwo LM, Donald C, Ikediobi C, Silver S (1998) Effects of intracellular glutathione on sensitivity of Escherichia coli to mercury and arsenite. Biochem Biophys Res Commun 242:67–70. doi:10.1006/bbrc.1997.7911
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275
Marzluf GA (1997) Molecular genetics of sulfur assimilation in filamentous fungi and yeast. Annu Rev Microbiol 51:73–96. doi:10.1146/annurev.micro.51.1.73
Momose Y, Iwahashi H (2003) Bioassay of cadmium using a DNA microarray: genome-wide expression patterns of Saccharomyces cerevisiae response to cadmium. Environ Toxicol Chem 20:2353-2360
Nies DH (1999) Microbial heavy-metal resistance. Appl Microbiol Biotechnol 51:730–750
Nies DH (2000) Heavy metal-resistant bacteria as extremophile: molecular physiology and biotechnological use of Ralstonia sp.CH34. Extremophile 4:77–82. doi:10.1007/s007920050140
Nies DH (2003) Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev 27:313–339. doi:10.1016/S0168-6445(03)00048-2
Ono BI, Hazu T, Yoshida S, Kawato T, Shinoda S, Brzvwczy J and Paszewski A (1999) Cysteine biosynthesis in Saccharomyces cerevisiae: a new outlook on pathway and regulation. Yeast 15: 1365-1375. doi:10.1002/(SICI)1097-0061(19990930)15:13<1365:AID-YEA468>3.3.CO;2-L
Ostrowski J, Barber MJ, Rueger DC, Miller BE, Siegel LM, andKredich NM (1989) Characterization of the flavoprotein moieties of NADPH-sulfite reductase from Salmonella typhimurium and Escherichia coli: physicochemical and catalytic properties, amino acid sequence deduced from the DNA sequence of CysJ and comparison with NADPH-cytochrome P-450 reductase. J Biol Chem 264:15726–15737
Scherer J, Nies DH (2009) CzcP is a novel efflux system contributing to transition metal resistance in Cupriavidus metallidurans CH34. Mol Microbiol 73:601–621. doi:10.1111/j.1365-2958.2009.06792.x
Schnell R, Oehlmann W, Singh M, Schneider G (2007) Structural insights into catalysis and inhibition of O-acetylserine sulfhydrylase from Mycobacterium tuberculosis: crystal structures of the enzyme α-aminoacrylate intermediate and an enzyme-inhibitor complex. J Biol Chem 282:23473–23481. doi:10.1074/jbc.M703518200
Shamim S, Rehman A, Qazi MH (2014) Cadmium-resistance mechanism in the bacteria cupriavidus metallidurans CH34 and pseudomonas putida mt2. Arch Environ Contam Toxicol 67:149–157. doi:10.1007/s00244-014-0009-7
Stiller I, Dancs G, Hesse H, Hoefgen R, Bánfalvi Z (2007) Improving the nutritive value of tubers: elevation of cysteine and glutathione contents in the potato cultivar White Lady by marker-free transformation. J Biotechnol 128:335–343. doi:10.1016/j.jbiotec.2006.10.015
Thomas D, Surdin-Kerjan Y (1997) Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 61:503–532
Toppi LSD, Gabbrielli R (1999) Response to cadmium in higher plants. Environ Exp Bot 41:105–130. doi:10.1016/S0098-8472(98)00058-6
Truper HG, Rogers LA (1971) Purification and properties of adenylyl sulfate reductase from the phototrophic sulfur bacterium, Thiocapsa roseopersicina. J Bacteriol 108:1112–1121
Valdés J, Veloso F, Jedlicki E, Holmes D (2003) Metabolic reconstruction of sulfur assimilation in the extremophile Acidithiobacillus ferrooxidans based on genome analysis. BMC Genom 4(51):1471–2164. doi:10.1186/1471-2164-4-51
Vera M, Schippers A, Sand W (2013) Progress in bioleaching: fundamentals and mechanisms of bacterial metal sulfide oxidation—part A. Appl Microbiol Biotechnol 97:7529–7541. doi:10.1007/s00253-013-4954-2
Vido K, Spector D, Lagniel G, Lopez S, Toledano MB, Labarre J (2001) A proteome analysis of the cadmium response in Saccharomyces cerevisiae. J Biol Chem 276:8469–8474. doi:10.1074/jbc.M008708200
Wang JY, Qiu L, Wu ED, Drlica K (1996) RNases involved in ribozyme degradation in Escherichia coli. J Bacteriol 178:1640–1645
Acknowledgments
The project is supported by National Natural Science Foundation of China (51264029), Environmental Public Welfare Scientific Research Project (201309005), Inner Mongolia Natural Science Foundation of China (2013MS0604), Program for Young Talents of Science and Technology in Universities of Inner Mongolia Autonomous Region (NJYT-14-B12), and Innovation Foundation of Inner Mongolia University of Science and Technology (2014QNGG05).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Albers.
Rights and permissions
About this article
Cite this article
Zheng, C., Chen, M., Tao, Z. et al. Differential expression of sulfur assimilation pathway genes in Acidithiobacillus ferrooxidans under Cd2+ stress: evidence from transcriptional, enzymatic, and metabolic profiles. Extremophiles 19, 429–436 (2015). https://doi.org/10.1007/s00792-014-0728-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-014-0728-8