Abstract
The enzyme 5-phosphoribosyl-1-α-diphosphate (PRPP) synthase (EC 2.7.6.1) catalyses the Mg2+-dependent transfer of a diphosphoryl group from ATP to the C1 hydroxyl group of ribose 5-phosphate resulting in the production of PRPP and AMP. A nucleotide sequence specifying Sulfolobus solfataricus PRPP synthase was synthesised in vitro with optimised codon usage for expression in Escherichia coli. Following expression of the gene in E. coli PRPP synthase was purified by heat treatment and ammonium sulphate precipitation and the structure of S. solfataricus PRPP synthase was determined at 2.8 Å resolution. A bent dimer oligomerisation was revealed, which seems to be an abundant feature among PRPP synthases for defining the adenine specificity of the substrate ATP. Molecular replacement was used to determine the S. solfataricus PRPP synthase structure with a monomer subunit of Methanocaldococcus jannaschii PRPP synthase as a search model. The two amino acid sequences share 35 % identity. The resulting asymmetric unit consists of three separated dimers. The protein was co-crystallised in the presence of AMP and ribose 5-phosphate, but in the electron density map of the active site only AMP and a sulphate ion were observed. Sulphate ion, reminiscent of the ammonium sulphate precipitation step of the purification, seems to bind tightly and, therefore, presumably occupies and blocks the ribose 5-phosphate binding site. The activity of S. solfataricus PRPP synthase is independent of phosphate ion.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The compound 5-phospho-d-ribosyl 1-α-diphosphate (PRPP) is a precursor in the biosynthesis of purine and pyrimidine nucleotides, the amino acids histidine and tryptophan as well as the co-factor NAD (Hove-Jensen 1988, 1989). In most of these reactions nitrogen of various aromatic bases or ammonia functions as a nucleophile, which attacks the anomeric carbon of PRPP with the formation of N-glycosidic bonds and with inversion of configuration (Lieberman et al. 1955). Variants of this scheme are decaprenyl-phosphate phosphoribosyltransferase and a phosphoribosyltransferase of methanopterin biosynthesis, where the nucleophiles are oxygens resulting in the formation of O-glycosidic bonds (Huang et al. 2005; White 1996), and 4-aminobenzoate phosphoribosyltransferase, where the nucleophile is the decarboxylated carbanion of 4-aminobenzoate resulting in the formation of a C-glycosidic bond (Rasche and White 1998). In all cases diphosphate is the leaving group, which is subsequently hydrolysed to Pi by diphosphatase. Microorganisms such as Escherichia coli contain 10 enzymes, designated phosphoribosyltransferases, which utilise PRPP as a substrate (Jensen et al. 2008). The synthesis of PRPP is catalysed by PRPP synthase (ATP: d-ribose 5-phosphate pyrophosphotransferase, EC 2.7.6.1) in a reaction, where the β,γ-diphosphoryl group of ATP is transferred to the C1 hydroxyl of ribose 5-phosphate: ribose 5-phosphate + ATP → PRPP + AMP (Kornberg et al. 1955). The enzyme requires a Mg2+-ion as an activator in addition to that complexed with ATP (Switzer 1969). PRPP synthase, encoded by a prs gene in most organisms, is ubiquitously found among free-living organisms, lacking only in certain specialised intracellular parasites such as chlamydiae (Xie et al. 2002).
Several classes of PRPP synthases have been identified (Krath and Hove-Jensen 2001a). Class I constitutes the “classical” PRPP synthase represented by the enzymes from organisms such as the bacterial species Bacillus subtilis (Arnvig et al. 1990), Bacillus caldolyticus (Krath and Hove-Jensen 1996), E. coli (Hove-Jensen et al. 1986), Salmonella enterica serovar Typhimurium (Switzer 1969), the three isoforms of mammals such as man and rat (Taira et al. 1987), as well as isozymes 1, 2 and 5 of Arabidopsis thaliana and isozymes 1 and 2 of spinach (Krath et al. 1999; Krath and Hove-Jensen 1999). Class I PRPP synthases have hexameric quaternary structure; diphosphoryl donor capacity is limited to ATP and, in some instances, dATP. The enzymes are allosterically inhibited by either of the ribonucleoside diphosphates ADP or GDP or both. Finally, they are activated by phosphate ion (Pi), which may be regarded as an allosteric activator, as Pi-binding competes with ribonucleoside diphosphate-binding at the allosteric site (Willemoës et al. 2000). The crystal structure of some class I enzymes have been determined, namely B. subtilis PRPP synthase and human PRPP synthase isoform 1 (Eriksen et al. 2000; Li et al. 2007). Class II PRPP synthases show broad diphosphoryl donorspecificity by accepting GTP, CTP or UTP (in addition to ATP and dATP). Allosteric regulation has not been detected among these enzymes, and, finally, their activity is independent of Pi (Krath and Hove-Jensen 2001a, b). A three-dimensional structure of a class II PRPP synthase has not been published. Following the characterisation of Methanocaldococcus jannaschii PRPP synthase the existence of class III PRPP synthases was postulated. M. jannaschii PRPP synthase revealed limited diphosphoryl donor capacity, lack of allosteric regulation by ribonucleoside diphosphate and activation by Pi, whereas a tetrameric quaternary structure was determined (Kadziola et al. 2005). Thus, the characteristics of M. jannaschii PRPP synthase appeared to be a mixture of those of class I and II. PRPP synthase from Mycobacterium tuberculosis appears to belong to class I. The quaternary structure is hexameric (Alderwick et al. 2011; Breda et al. 2012; Lucarelli et al. 2010), and the activity is allosterically inhibited by ribonucleoside diphosphate (Breda et al. 2012). Contradictory to this, it has been reported that M. turberculosis PRPP synthase activity is independent of Pi and that GTP, CTP and UTP (in addition to ATP) could be used as diphosphoryl donors (Breda et al. 2012), i.e. class II. Additionally, the structure of PRPP synthase from the thermophilic, acidophilic archaeon Thermoplasma volcanium revealed properties different from class I, II and III PRPP synthases with the enzyme’s quaternary structure being a dimer. Results of kinetic analysis of this enzyme have not been published (Cherney et al. 2011).
In the present study, the structure of PRPP synthase from the acidophilic, thermophilic arachaeon Sulfolobus solfataricus was determined revealing a bent dimer oligomerization, which seems to be a common feature among PRPP synthases in defining the adenine specificity of the substrate ATP. Implications of oligomeric structure on regulatory properties of PRPP synthase are discussed.
Materials and methods
General
Restriction endonucleases and T4 DNA ligase were purchased from New England Biolabs (Boston, MA). Materials for the isolation of plasmid DNA was obtained from Qiagen (Hilden, Germany). Molecular mass standards were obtained from Biorad (Hercules, CA). Carrier-free 32Pi (Nex-53) was purchased from Perkin Elmer (Boston, MA).
Gene synthesis
The recombinant S. solfataricus PRPP synthase-specifying gene was synthesised chemically and inserted into the pGS4 vector (Geneart, Regensburg, Germany). The sequence was designed to match the optimal codon usage of E. coli. In addition, the AT-content was reduced from 65 to 51 %, and internal, putative ribosome-binding sites and chi sites were avoided. The designed sequence upstream of the translation initiation codon was GAATTCATTAAAGAGGAGAAATTAACTATG, where a restriction endonuclease EcoRI recognition site is indicated in italics and the AUG-specifying sequence is indicated in bold. The open reading frame was followed by TAATAA CCATGG, i.e. a sequence specifying two translation stop codons, which are indicated in bold, and a restriction endonuclease NcoI recognition site, which is indicated in italics. The designed sequence was excised by digestion with EcoRI and NcoI and ligated to DNA of the expression vector pUHE23-2 (H. Bujard, University of Heidelberg, personal communication), which had been previously digested by the same restriction endonucleases. This procedure resulted in the generation of pHO565. Nucleotide sequencing revealed the expected sequence of the insert of pHO565 of which the coding sequence is ATGATTATTATTGGCGGCAGCGCGACCAACGGCATTGATGAAAGCCTGAGCAAAATTCTGAGCATTCCGCTGGTGAAAGTGGAAAACAAAATCTTTCCGGATGGCGAAAGCTATATTCGTGTGCCGAGCAGCATTCGTGATGAAGAAGTGCTGCTGGTTCAGACCACCGATTATCCGCAGGATAAACATCTGATCGAACTGTTTCTGATTGCGGAAACCATTCGTGATCTGGGCGCGAAAAAACTGACCGCGATTGTGCCGTATCTGGCCTATAGCCGTCAGGATCGTCGTTTTAAAGATGGCGAAGCGATTAGCATTAAAACCATCCTGCATATTCTGAGCGAAGTGGGCGTGAACACCCTGGTGGTGGTGGAACCGCATAAACCGGAAGAACTGAGCTATTTTAAAGGCGAACTGAAAATCGTGCATCCGTATCATCAGATTGCGCGCAAAATCAAAGAAATCATCGAAGATCCGTTTATTCTGGCCCCGGATCGTGGTGCGCTGGATCGTGCGCGTAAAATTGCGGAAGAAATCAACGCGCCGTATAGCTATATTGAAAAAGAACGTGATCGTACCACCGGCGAAGTGCGTATTAAAGAAGCGCCGAACATTAACCTGAAAGGCAAAGATGTGGTGATCATCGACGATATTATTAGCACCGGCGGCACCATTGTTCAGGCGACCCGTCTGGCCTACAGCCTGGGTGCGAAAAGCGTTACCGCAGCAGCCATTCACCTGCTGCTGGTTGGCGGTGCGAAAGAACGTCTGCGTGAAGTTGGTGTTAAAACGCTGATTGGCACCAACACCATCAACGTGAACGATAAAGATATCATCACCATCGATGTGAGCCAGAGCATTGCCCTGAGCCTG. The gene containing this open reading frame is designated prs Ss .
Cell growth and prs Ss gene expression
E. coli strain HO1088 (Δprs-4::KanR/F´lacIq TetR), which lacks endogenous PRPP synthase activity (Krath and Hove-Jensen 1999, 2001a), was transformed with pHO565 by selecting for ampicillin resistance. The prs Ss gene furthermore complemented the Δprs-4::KanR allele, indicating the synthesis in E. coli of active PRPP synthase. Cells were grown at 37 °C in NZY medium supplemented with ampicillin (100 mg L−1) and tetracycline (10 mg L−1) with aeration by shaking (Hove-Jensen and Maigaard 1993). Cell growth was monitored as optical density (OD) at 436 nm. An OD436 of 1.0 (1 cm path-length) corresponds to approximately 3 × 1011 cells L−1. At an OD436 of approximately 1.5, isopropyl 1-thio-β-d-galactopyranoside was added to a concentration of 0.50 mM to induce expression of prs Ss . Incubation continued for additional 6 h. Cells were collected by centrifugation. Cell pellets were stored at −20 °C if not used immediately.
Purification of PRPP synthase
Purification was performed at 4 °C unless otherwise stated. Cells were resuspended in 50 mM Tris/HCl, pH 7.6 and broken by ultrasonic treatment in a Soniprep ultrasonic disintegrator (model 150, Measuring and Scientific Equipment). Cell debris was removed by centrifugation and to the supernatant fluid, 0.1 volume of streptomycin sulphate [11 % (w/v) in 50 mM potassium phosphate buffer, pH 7.6] was added. Following centrifugation the supernatant fluid, the “crude” extract, was heated to 75 °C for 15 min. Denatured protein was removed by centrifugation, and the supernatant fluid, the “heat” fraction, was 40 % saturated with solid ammonium sulphate. The precipitated protein was removed by centrifugation, and the supernatant was saturated to 60 % with solid ammonium sulphate. Precipitated protein was removed by centrifugation, dissolved in 50 mM Tris/HCl, pH 7.6, and dialysed against the same buffer to generate the “40/60” fraction. The purity was evaluated by SDS-PAGE and Coomassie Brilliant Blue staining (Laemmli 1970). Protein content was determined by the bicinchoninic acid procedure with bovine serum albumin as the standard (Smith et al. 1985).
Assay of PRPP synthase activity
The standard assay of PRPP synthase activity at 60 °C was performed as follows. Extract or enzyme (10 μL), appropriately diluted in 50 mM Tris/HCl, pH 7.6 containing bovine serum albumin (1 g L−1), was mixed with 90 μL of a reaction cocktail (both prewarmed at 60 °C) containing 2.0 mM [γ-32P]ATP (approximately 1 kBq per assay) prepared as previously described (Jensen et al. 1979), 5.0 mM ribose 5-phosphate, 4.0 mM magnesium chloride, 20 mM sodium fluoride (omitted in assays of the purified enzyme), 50 mM Tris/HCl, pH 7.6. Samples (10 μL) were removed at intervals (usually 10, 30 and 60 s.) and mixed with 5 μL of 0.33 M formic acid. This 15 μL was applied to polyethyleneimine-cellulose plates (Baker-flex, J. T. Baker). After drying, PRPP and ATP were separated by thin-layer chromatography with 0.85 M potassium dihydrogen phosphate, which had been previously adjusted to pH 3.4 with 0.85 M phosphoric acid, as solvent. Radioactivity was quantified with the Cyclone Storage Phosphor System (Perkin Elmer, Wellesley, MA). Specific activity is expressed as μmol (min × mg of protein)−1.
Crystallisation
All crystallisation experiments were performed with AMP and ribose 5-phosphate added to the protein solution (12.7 mg/mL in 50 mM Tris/HCl pH 7.6) to a concentration of 10 mM of both compounds. Crystallisation conditions were tested by vapour diffusion as sitting or hanging drops. An initial JCSG+ screen (Qiagen) (Newman et al. 2005) was set up by a robot (Oryx, Douglas Instruments) in a 96-well plate with reservoir volumes of 100 μL and drop sizes of 0.4 μL. The drop composition was of 25 % reservoir and 75 % protein solution. Optimisation experiments were done in 24-well plates with 1 mL reservoirs, and total size drops of 4 μL were set up manually with ratios 2:2 or 3:1 of protein and reservoir, respectively. Crystals used for data collection were grown from reservoirs composed of 14 % PEG10000 with 0.2 M diammonium citrate, pH 5.0 and grew to a size of 350 × 100 × 100 μm3.
Data collection
Diffraction data were obtained at beamline 911–2 of the MAX-lab synchrotron source facility in Lund, Sweden. Data were collected at 120 K using crystals with no added cryo-protection. Images were collected over 180 degrees with 1° of oscillation per image. Single images were evaluated with the program Xdisp (Otwinowski and Minor 1997). Data were later indexed, integrated and scaled using the programs Denzo and Scalepack (Otwinowski and Minor 1997, 2001). Data were also analysed with HKLview from the CCP4 program suite to determine the space group (Collaborative Computational Projekt Number 4 (1994). The CCP4 suite: programs for protein crystallography). The space group was determined as P21 with cell dimensions a = 80.2 Å, b = 92.8 Å, c = 138.3 Å and β = 90.91°. The crystal diffracted to 2.8 Å resolution. Statistics are shown in Table 1.
Structure determination
S. solfataricus PRPP synthase structure was solved by molecular replacement using Phaser with M. jannaschii PRPP synthase as search model (McCoy et al. 2005). The amino acid sequences of S. solfataricus and M. jannaschii PRPP synthases share 36 % identity. Six copies of the search model were placed in the asymmetric unit displaying consecutive translational function Z scores of 3.6, 20.8, 11.0, 17.2, 17.9 and 18.8.
Refinement
S. solfataricus PRPP synthase structure was refined using simulated annealing and NCS restraints with Phenix.refine (Afonine et al. 2005). The structure was manually corrected between rounds of refinement at the graphics using Coot (Emsley and Cowtan 2004). Refinement statistics are shown in Table 1.
Amino acid sequence analysis
Amino acid sequences were aligned with the Multalin software (Corpet 1988).
Protein data bank accession codes
The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB code 4TWB), Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org).
Results
Cloning and expression of the S. solfataricus prs gene and purification of PRPP synthase
The native S. solfataricus prs gene was isolated by PCR with S. solfataricus DNA as the template. This gene, when harboured in the expression vector pUHE23-2, readily complemented the E. coli Δprs-4 allele. Activity of PRPP synthase, although low, was detectable in crude extracts of cells expressing the S. solfataricus prs gene. A gene product of the expected molecular mass (32 kDa), however, was undetectable by SDS-PAGE followed by staining with Comassie Brilliant Blue. To circumvent these problems, the gene was synthesised chemically as described in “Materials and methods” and designated prs Ss . In general, the codon usage was changed to match that of highly expressed E. coli genes. Specifically, 15 arginine-specifying AGA or AGG triplets were changed to CGT, and 27 isoleucine-specifying ATA triplets were changed to ATT or ATC. The codon adaption index of the prs Ss gene was determined as 0.589, whereas a value of 0.129 was determined for the native S. solfataricus prs gene, and a value of 0.553 for the E. coli prs gene (Sharp and Li 1987). Induction of prs Ss gene expression resulted in the appearance of a polypeptide of appropriate molecular mass by SDS-PAGE. The amount of this polypeptide reached a maximum 6–8 h after onset of induction. After overnight incubation (18–20 h) the presumptive PRPP synthase subunit band had disappeared, indicating the simultaneous synthesis and degradation (data not shown).
PRPP synthase was purified by a three-step procedure as described in “Materials and methods”. The procedure resulted in 7-fold purification, a yield of 62 % and an enzyme with a specific activity of 67 μmol (min × mg protein)−1 determined by the standard assay described in “Materials and methods”. The activity was determined in the absence of Pi, and, thus, S. solfataricus PRPP synthase activity is independent of Pi. SDS–polyacrylamide gel electrophoresis established the molecular mass of the band representing PRPP synthase as 34.0 kDa, which is comparable to the molecular mass of the deduced amino acid sequence, 32.2 kDa.
Crystal structure and active site
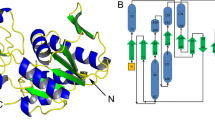
The subunit of S. solfataricus PRPP synthase comprises 291 residues. Residues 1–190 and 205–291 could be modelled into the electron density at a resolution of 2.8 Å, while the remaining stretch 191–204 was disordered. The enzyme is organised as a two-domain polypeptide. The two domains have a very similar fold indicating gene duplication at some point in evolution. This similarity is, however, not detectable at the amino acid sequence level. The active site is situated at the interface between the two domains, in this structure marked by the binding of AMP and sulphate. We assigned the following secondary structure elements. β1: Ile2-Gly6, α2: Gly11-Ser21, β3: Pro23-Val27, β3’: Glu28-Phe32, β4: Glu36-Arg40, β5: Glu48-Thr56, α6: Pro59-Leu77, β7: Lys81-Tyr88, α8: Ile104-Glu115, β9: Thr120-Val124, 310: Lys128-Phe135, β11: Glu138-Val142, α12: Pro144-Ile156, β13: Asp158-Leu162, α14: Ala168-Ile179, β15: Pro182-Ile186, β17: Lys210-Asp216, α18: Gly222-Ser234, β19: Ser239-Ala244, α20: Ala253-Gly261, β21: Thr264-Asn269, β22: Ile279-Ile281, α23: Val283-Leu291, Fig. 1. In the N-terminal domain there is a six-stranded parallel β-sheet formed by strands β3, β1, β5, β7, β9 and β11 which is sandwiched between helices α6, α8 and 310 on one side and helices α2 and α23 on the other side. An anti-parallel hairpin composed of β3’ and β4 protrudes between β3 and β5. In the C-terminal domain there is also a six-stranded parallel β-sheet formed by strands β15, β13, β17, β19, β21 and β22, which is sandwiched between helices α12 and α14 on one side and helices α18 and α20 on the other side. As for the N-terminal domain there is a protrusion between strands β15 and β17, which consists of the catalytic flexible loop with missing electron density.
Alignment of amino acid sequences of PRPP synthases. The amino acid sequences are: S.s., S. solfataricus; T.v., T. volcanium; M.j., M. jannaschii; and B.s., B. subtilis PRPP synthases; S.o., Spinacea oleracea PRPP synthase isozyme 4. Secondary structural elements of S. solfataricus PRPP synthase are indicated above the sequences. Beta sheets are indicated by arrows, helices by bars. Secondary structure elements are designated similarly to those of M. jannaschii PRPP synthase (Kadziola et al. 2005). Amino acid residues identical in all five sequences are indicated by asterisks below the spinach PRPP synthase amino acid sequence, whereas conserved amino acid residues are indicated by a dot. Conserved amino acids are valine, leucine and isoleucine; phenylalanine and tyrosine; aspartic and glutamic acid; arginine and lysine; serine and threonine; glycine and alanine. B. subtilis PRPP synthase amino acid residues, which are located at the active site are typed in red, those which are located at the allosteric, regulatory site are typed in blue, whereas those involved in subunit–subunit interactions are typed in green. Amino acid residues involved in formation of the bent dimer are typed in bold, whereas those involved in formation of the parallel dimer are typed in italic. Whenever amino acid residues of PRPP synthase of S. solfataricus, T. volcanium, M. jannaschii or spinach are identical or conserved relative to those of the B. subtilis enzyme, the colour code of the latter sequence is applied to the S. solfataricus, T. volcanium, M. jannaschii or spinach amino acid residues as well. Relevant amino acid residues of B. subtilis PRPP synthase were selected on the basis of the three-dimensional structures previously published (Eriksen et al. 2000, 2002; Nygaard 2001)
Although ribose 5-phosphate was present in the crystallisation mixture, the compound was not found in the density. Rather sulphate, which was used during purification, was seen. The sulphate oxygen atoms are accepting hydrogen bonding from Ser220-N, Thr221-N, Thr221-Oγ1, Gly222-N, Thr224-N and Thr224-Oγ1, Fig. 2. Additionally, AMP is bound to the active site. Here the adenine ring is stacked between residues Arg93 and BPhe32 (superscript B denotes a neighbouring subunit generated by non-crystallographic twofold symmetry). Adenine-N6 is donating hydrogen bonding to BAsp34-Oδ2 and BGlu36-Oε2. His127-Nε2 and Arg93-Nη1 each donate hydrogen bonding to an α-phosphate oxygen of AMP and, moreover, Arg93-Nη2 bridges to a sulphate oxygen. Two more residues are in van der Waal contact with AMP: Gln94 with the adenine ring and Phe98 with the ribose ring.
Binding of the adenylate moiety of ATP and sulphate to the active site of PRPP synthase. Residues with atoms within 4 Å from the ligands AMP and sulphate are shown. Amino acid residues of two subunits contribute to the active site. Amino acid residues from one subunit are shown in colour, whereas residues from the second subunit are shown in grey and indicated by superscript “B” to the left of the amino acid name
The quaternary structure of S. solfataricus PRPP synthase is dimeric as found also for T. volcanium PRPP synthase (Cherney et al. 2011). The interface of the bent dimer is formed between the N-terminal domains, Fig. 3. The predominant interactions at the dimer interface are the salt bridges: AAsp34–BArg93, AArg40–BAsp100, AGlu72–BHis111, AGlu72–BLys107, AAsp76–BLys107 and AAsp76–BArg97, which, with their symmetry partners, result in 12 strong interactions. These salt bridges are not conserved among all PRPP synthases.
Quaternary structures of PRPP synthases. a Propellar, hexameric PRPP synthase of B. subtilis (Eriksen et al. 2000) or of human PRPP synthase 1 (Li et al. 2007); b tetrameric PRPP synthase of M. jannaschii (Kadziola et al. 2005); c dimeric PRPP synthase of S. solfataricus (this study) or of T. volcanium (Cherney et al. 2011). The bent dimer (red and cyan) is incorporated in all of the structures
Deductions from amino acid sequence alignments
Amino acid sequence alignment of S. solfataricus PRPP synthase with those of the closely related species Sulfolobus islandicus and Sulfolobus acidocaldarius revealed identities of 93 and 57 %, respectively. Sequence identity of S. solfataricus PRPP synthase with other archaeal PRPP synthases is lower, 42, 41, 35 and 29 % with the PRPP synthases of Pyrococcus furiosus, Pyrococcus abyssi. M. jannaschii and T. volcanium, respectively, whereas the identity of S. solfataricus PRPP synthase with class I PRPP synthase of B. subtilis and class II spinach PRPP synthase isozyme 4 is 25 and 17 %, respectively. Figure 1 shows an amino acid sequence alignment of PRPP synthase from the three archaeal species S. solfataricus, T. volcanium and M. jannaschii, class I PRPP synthase of B. subtilis as well as class II spinach PRPP synthase isozyme 4. Overall only 18 amino acid residues (6 %) are identical with additional 29 conserved amino acid residues (10 %).
A colour code is applied to amino acid residues of the sequences of Fig. 1. This colour code is based on the assignment of structural and functional properties of individual amino acid residues of B. subtilis PRPP synthase as evaluated from three-dimensional structure determinations (Eriksen et al. 2000, 2002; Nygaard 2001). Amino acid residues involved in the formation of the active site are typed in red in Fig. 1. These amino acid residues include the S. sulfolobus PRPP synthase amino acid residues mentioned above or shown in Fig. 2 (Phe32, Asp34, Arg 93, Gln94, His127, Ser220, Thr224), and it is evident that these residues are highly conserved among the sequences of the five species. In contrast, amino acid residues typed in blue and involved in the formation of the allosteric, regulatory site of B. subtilis PRPP synthase are poorly conserved if at all, consistent with the lack of allosteric regulation of S. sulfolobus, T. volcanium and M. jannaschii PRPP synthases as well as spinach PRPP synthase isozyme 4. The partial conservation of B. subtilis Leu134, Gln138, Ile139 and Phe142 may indicate additional functional properties of these amino acid residues. Amino acid residues involved in the formation of subunit–subunit interactions of B. subtilis PRPP synthase are shown in green in Fig. 1. Conservation is particularly evident among residues involved in the formation of the bent dimer, i.e. B. subtilis PRPP synthase amino acid residues Glu44-Arg84. Of these 41 residues, 21 (51 %) are conserved in the S. solfataricus PRPP synthase amino acid sequence.
Discussion
The quaternary structure of S. solfataricus PRPP synthase is dimeric, and, thus, resembles that of the archaeon T. volcanium (Cherney et al. 2011). In contrast, the quaternary structure of PRPP synthase of the thermophilic, methanogenic archaeon M. jannaschii is tetrameric and appears to be built by two dimers (Kadziola et al. 2005). The significance, if any, of this structural difference for enzymatic properties remains to be established. Oligomerization in PRPP synthase is important for the enzyme’s regulatory properties. Although no biochemical evidence is available for PRPP synthase of S. sulfolobus and T. volcanium, except that the activity of the former enzyme is independent of Pi, the enzyme of M. jannaschii has been shown to bind ADP only at the active site, and that the enzyme apparently does not contain an allosteric site for ribonucleoside diphosphate inhibition (Kadziola et al. 2005). The class I PRPP synthases are allosterically inhibited by purine ribonucleoside diphosphates, ADP and/or GDP. The enzymes from B. subtilis (Eriksen et al. 2000), man (Li et al. 2007), and the Gram-negative Betaproteobacterium Burkholderia (Pseudomonas) pseudomallei (PDB code 3DAH) have hexameric quaternary structures. The allosteric site of B. subtilis PRPP synthase has been studied in detail. It consists of amino acid residues provided from three subunits, i.e. residues located at the interphase of the bent dimer as well as at the interphase of the so-called parallel dimer (Eriksen et al. 2000). The archaeal PRPP synthases do not possess these subunit–subunit interactions. In addition, as shown in Fig. 1 the specific amino acid residues involved in the formation of the allosteric site of B. subtilis PRPP synthase are far from conserved in the archaeal enzymes, and, moreover, the archaeal PRPP synthases are shorter than the class I enzymes, and specifically lack the important amino acid residues close to the carboxy-terminal end of B. subtilis PRPP synthase. Altogether, it is evident that an evolutionary effect exists, with lack of allosteric regulation of PRPP synthase of archaeal organisms and with allosteric regulation present only in class I PRPP synthases of bacteria, and eukaryotic organisms. Consistent with the apparent lack of an allosteric site in S. solfataricus PRPP synthase, the activity of the enzyme is independent of the presence of Pi. In allosterically regulated PRPP synthases Pi, an allosteric activator, and ADP, an allosteric inhibitor, bind to the same site (Willemoës et al. 2000).
In general, the specificity of the binding of the adenyl moiety of the substrate ATP in PRPP synthase is created at the interface of two subunits. In the simplest case the quaternary structure is dimeric as seen for S. solfataricus and T. volcanium PRPP synthases (Cherney et al. 2011). The tetrameric structure of M. jannaschii PRPP synthase may be regarded as built of two dimers, whereas the hexameric structures of B. subtilis (Eriksen et al. 2000), human (Li et al. 2007), and B. pseudomallei PRPP synthases may be regarded as built of three dimers (Fig. 3). Also in the cases with the tetrameric and hexameric enzymes the binding of ATP occurs at the interface of two subunits, and more specifically at the interface of the bent dimers. In the active site around AMP, residues Phe32, Asp34 and His127 of S. solfataricus PRPP synthase are fully conserved among PRPP synthases. Residues Tyr91, Arg93 and Gln94 are conserved in all class I PRPP synthases. At the ribose 5-phosphate site, residues Gly223, Thr224 as well as Asp216 and Asp217 are fully conserved while Thr221 is conserved only in class I PRPP synthases. The side chains of Asp216 and Asp217 face the ribose moiety of ribose 5-phosphate and Asp217 hydrogen-binds to His127. Two other regions of the sequence contain fully conserved residues. Lys190 and Arg192 are situated in the flexible loop and have presumably catalytic function. Asp165 and Gly167 are situated between β13 and α14 with the side chain of Asp165 hydrogen bonding to the ribose ring of ribose 5-phosphate (Cherney et al. 2011; Kadziola et al. 2005).
As mentioned in the “Introduction”, kinetic characterization defined class I and class II PRPP synthases, and a class III PRPP synthase was postulated on the basis of characterization of M. jannaschii PRPP synthase. The kinetic properties of this latter enzyme were a mixture of those of class I and class II PRPP synthases. Although only a few archaeal PRPP synthases have been characterised, the heterogeneity in physical and chemical properties suggest that the definition of a true class III is superfluous and that class I, class II and archaeal PRPP synthases may be a better definition.
Abbreviations
- PRPP:
-
5-Phospho-d-ribosyl 1-α-diphosphate
References
Afonine PV, Grosse-Kunstleve RW, Adams PD (2005) The phenix refinement framework. CCP4 Newslett 42:8
Alderwick LJ, Lloyd GS, Lloyd AJ, Lovering AL, Eggeling L, Besra GS (2011) Biochemical characterization of the Mycobacterium tuberculosis phosphoribosyl-1-pyrophosphate synthetase. Glycobiology 21:410–425. doi:10.1093/glycob/cwq173
Arnvig K, Hove-Jensen B, Switzer RL (1990) Purification and properties of phosphoribosyl-diphosphate synthetase from Bacillus subtilis. Eur J Biochem 192:195–200
Breda A, Martinelli LK, Bizarro CV, Rosado LA, Borges CB, Santos DS, Basso LA (2012) Wild-type phosphoribosylpyrophosphate synthase (PRS) from Mycobacterium tuberculosis: a bacterial class II PRS? PLoS One 7:e39245. doi:10.1371/journal.pone.0039245
Cherney MM, Cherney LT, Garen CR, James MN (2011) The structures of Thermoplasma volcanium phosphoribosyl pyrophosphate synthetase bound to ribose-5-phosphate and ATP analogs. J Mol Biol 413:844–856. doi:10.1016/j.jmb.2011.09.007
Collaborative Computational Projekt Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50:760–763
Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16:10881–10890
Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi:10.1107/S0907444904019158
Eriksen TA, Kadziola A, Bentsen AK, Harlow KW, Larsen S (2000) Structural basis for the function of Bacillus subtilis phosphoribosyl-pyrophosphate synthetase. Nat Struct Biol 7:303–308. doi:10.1038/74069
Eriksen TA, Kadziola A, Larsen S (2002) Binding of cations in Bacillus subtilis phosphoribosyldiphosphate synthetase and their role in catalysis. Protein Sci 11:271–279. doi:10.1110/ps.28502
Hove-Jensen B (1988) Mutation in the phosphoribosylpyrophosphate synthetase gene (prs) that results in simultaneous requirements for purine and pyrimidine nucleosides, nicotinamide nucleotide, histidine, and tryptophan in Escherichia coli. J Bacteriol 170:1148–1152
Hove-Jensen B (1989) Phosphoribosylpyrophosphate (PRPP)-less mutants of Escherichia coli. Mol Microbiol 3:1487–1492
Hove-Jensen B, Maigaard M (1993) Escherichia coli rpiA gene encoding ribose phosphate isomerase A. J Bacteriol 175:5628–5635
Hove-Jensen B, Harlow KW, King CJ, Switzer RL (1986) Phosphoribosylpyrophosphate synthetase of Escherichia coli. Properties of the purified enzyme and primary structure of the prs gene. J Biol Chem 261:6765–6771
Huang H, Scherman MS, D’Haeze W, Vereecke D, Holsters M, Crick DC, McNeil MR (2005) Identification and active expression of the Mycobacterium tuberculosis gene encoding 5-phospho-α-d-ribose-1-diphosphate: decaprenyl-phosphate 5-phosphoribosyltransferase, the first enzyme committed to decaprenylphosphoryl-d-arabinose synthesis. J Biol Chem 280:24539–24543. doi:10.1074/jbc.M504068200
Jensen KF, Houlberg U, Nygaard P (1979) Thin-layer chromatographic methods to isolate 32P-labeled 5-phosphoribosyl-α-1-pyrophosphate (PRPP): determination of cellular PRPP pools and assay of PRPP synthetase activity. Anal Biochem 98:254–263
Jensen KF, Dandanell G, Hove-Jensen B, Willemoës M (2008) Chapter 3.6.2, Nucleotides, nucleosides, and nucleobases. ASM Press. Accessed 18 August
Kadziola A, Jepsen CH, Johansson E, McGuire J, Larsen S, Hove-Jensen B (2005) Novel class III phosphoribosyl diphosphate synthase: structure and properties of the tetrameric, phosphate-activated, non-allosterically inhibited enzyme from Methanocaldococcus jannaschii. J Mol Biol 354:815–828
Kornberg A, Lieberman I, Simms ES (1955) Enzymatic synthesis and properties of 5-phosphoribosylpyrophosphate. J Biol Chem 215:389–402
Krath BN, Hove-Jensen B (1996) Bacillus caldolyticus prs gene encoding phosphoribosyl-diphosphate synthase. Gene 176:73–79
Krath BN, Hove-Jensen B (1999) Organellar and cytosolic localization of four phosphoribosyl diphosphate synthase isozymes in spinach. Plant Physiol 119:497–506
Krath BN, Hove-Jensen B (2001a) Class II recombinant phosphoribosyl diphosphate synthase from spinach. Phosphate independence and diphosphoryl donor specificity. J Biol Chem 276:17851–17856
Krath BN, Hove-Jensen B (2001b) Implications of secondary structure prediction and amino acid sequence comparison of class I and class II phosphoribosyl diphosphate synthases on catalysis, regulation, and quaternary structure. Protein Sci 10:2317–2324. doi:10.1110/ps.11801
Krath BN, Eriksen TA, Poulsen TS, Hove-Jensen B (1999) Cloning and sequencing of cDNAs specifying a novel class of phosphoribosyl diphosphate synthase in Arabidopsis thaliana. Biochim Biophys Acta 1430:403–408
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Li S, Lu Y, Peng B, Ding J (2007) Crystal structure of human phosphoribosylpyrophosphate synthetase 1 reveals a novel allosteric site. Biochem J 401:39–47. doi:10.1042/BJ20061066
Lieberman I, Kornberg A, Simms ES (1955) Enzymatic synthesis of pyrimidine nucleotides; orotidine-5′-phosphate and uridine-5′-phosphate. J Biol Chem 215:403–451
Lucarelli AP et al (2010) Mycobacterium tuberculosis phosphoribosylpyrophosphate synthetase: biochemical features of a crucial enzyme for mycobacterial cell wall biosynthesis. PLoS One 5:e15494. doi:10.1371/journal.pone.0015494
McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr 61:458–464. doi:10.1107/S0907444905001617
Newman J et al (2005) Towards rationalization of crystallization screening for small- to medium-sized academic laboratories: the PACT/JCSG+ strategy. Acta Crystallogr D Biol Crystallogr 61:1426–1431. doi:10.1107/S0907444905024984
Nygaard FB (2001) The molecular mechanism of catalysis and allosteric regulation in the phosphoribosyldiphosphate synthase from Bacillus subtilis. Thesis, University of Copenhagen
Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326
Otwinowski Z, Minor W (2001) DENZO and SCALEPACK volume F: crystallography of biological macromolecules. International tables for crystallography. Springer, New York
Rasche ME, White RH (1998) Mechanism for the enzymatic formation of 4-(β-d-ribofuranosyl)aminobenzene 5′-phosphate during the biosynthesis of methanopterin. Biochemistry 37:11343–11351. doi:10.1021/bi973086q
Sharp PM, Li WH (1987) The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res 15:1281–1295
Smith PK et al (1985) Measurement of protein using bicinchoninic acid. Anal Biochem 150:76–85
Switzer RL (1969) Regulation and mechanism of phosphoribosylpyrophosphate synthetase. I. Purification and properties of the enzyme from Salmonella typhimurium. J Biol Chem 244:2854–2863
Taira M, Ishijima S, Kita K, Yamada K, Iizasa T, Tatibana M (1987) Nucleotide and deduced amino acid sequences of two distinct cDNAs for rat phosphoribosylpyrophosphate synthetase. J Biol Chem 262:14867–14870
White RH (1996) Biosynthesis of methanopterin. Biochemistry 35:3447–3456. doi:10.1021/bi952308m
Willemoës M, Hove-Jensen B, Larsen S (2000) Steady state kinetic model for the binding of substrates and allosteric effectors to Escherichia coli phosphoribosyl-diphosphate synthase. J Biol Chem 275:35408–35412
Xie G, Bonner CA, Jensen RA (2002) Dynamic diversity of the tryptophan pathway in chlamydiae: reductive evolution and a novel operon for tryptophan recapture. Genome Biol 3:research0051
Acknowledgments
We thank Qunxin She (University of Copenhagen) for kindly providing DNA of S. solfataricus, Tonny D. Hansen, Jens-Christian N. Poulsen and Dorthe Boelskifte for pertinent technical assistance. The MAXLAB and DANSCATT are gratefully acknowledged for providing the infrastructure facilitating collection of synchrotron data.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The experiments performed in this study comply with the current laws of Denmark.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by S. Albers.
The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB code 4TWB), Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org).
Rights and permissions
About this article
Cite this article
Andersen, R.W., Leggio, L.L., Hove-Jensen, B. et al. Structure of dimeric, recombinant Sulfolobus solfataricus phosphoribosyl diphosphate synthase: a bent dimer defining the adenine specificity of the substrate ATP. Extremophiles 19, 407–415 (2015). https://doi.org/10.1007/s00792-014-0726-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-014-0726-x