Abstract
Two obligately anaerobic sulfidogenic bacterial strains were isolated from the full-scale Thiopaq bioreactor in Lelystad (The Netherlands) removing H2S from biogas under oxygen-limiting and moderately haloalkaline conditions. Strain HSRB-L represents a dominant culturable sulfate-reducing bacterium in the reactor. It utilizes formate, H2 (with acetate as C-source) and lactate as e-donors, and sulfate, thiosulfate and sulfite as e-acceptors. It is haloalkalitolerant, with a pH range for lithotrophic growth from 7.5 to 9.7 (optimum at 8.5–9) and a salt range from 0.1 to 1.75 M total Na+ (optimum at 0.6 M). The strain is a member of the genus Desulfonatronum and is proposed as a novel species D. alkalitolerans. The second strain, strain HTRB-L1, represents a dominant thiosulfate/sulfur reducer in the reactor. It is an obligate anaerobe utilizing formate and H2 (with acetate as C-source), lactate, pyruvate and fumarate as e-donors, and thiosulfate (incomplete reduction), sulfur, arsenate and fumarate as e-acceptors. With lactate as e-donor it also grows as an ammonifyer in the presence of nitrate and nitrite. HTRB-L1 is haloalkalitolerant, with a pH range for lithotrophic growth from 7.1 to 9.7 (optimum at 8.5) and a salt range from 0.6 to 1.5 M total Na+ (optimum at 0.6 M). Phylogenetic analysis showed that strain HTRB-L1 is a novel species within the genus Sulfurospirillum (Epsilonproteobacteria) for which a name Sulfurospirillum alkalitolerans is proposed.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The full-scale Thiopaq bioreactors perform H2S removal from waste gases. The process is based on recently developed biotechnology of preferential oxidation of sulfide to insoluble sulfur by lithoautotrophic sulfide-oxidizing bacteria (SOB) under severe oxygen limitation (Janssen et al. 2009). H2S from the contaminated gas stream is first absorbed at a pH of around 8.2–9 by an alkaline solution, containing 0.5–1 M NaHCO3. The resulting alkaline sulfide solution is then fed into the bioreactor operating at a redox potential of −(300–400) mV, whereby SOB perform the partial oxidation of sulfide to insoluble sulfur. Despite that the lithoautotrophic SOB are the dominant populations in these bioreactors, a very low redox potential and the presence of high concentrations of sulfur compounds with intermediate valency, such as bio-sulfur (Janssen et al. 1999) and thiosulfate, provide a perfect conditions for the development of sulfidogenic anaerobes. Decomposition of the biomass of SOB grown on inorganic carbon may provide intermediate products that could be used by sulfidogens as electron donors.
Haloalkaline conditions prevailing in the Thiopaq bioreactors favor the growth of haloalkaliphilic sulfur-cycling bacteria. A recent analysis of a Thiopaq bioreactor in Eerbeek (The Netherlands) identified obligately haloalkaliphilic Thioalkalivibrio sulfidophilus (Gammaproteobacteria) as the dominant lithoautotrophic SOB species (Sorokin et al. 2012), and a moderately haloalkaliphilic bacterium Desulfurispirillum alkaliphilum (phylum Chrysiogenetes) as the dominant sulfur-respiring anaerobe (Sorokin et al. 2007). Both groups have also been detected in natural soda lakes (Sorokin et al. 2011a).
In this work, a microbiological analysis of a Thiopaq bioreactor in Lelystad (The Netherlands) resulted in the isolation of two obligately anaerobic haloalkalitolerant sulfidogenic bacteria which are described here as novel taxa in the Delta and in the Epsilonproteobacteria.
Methods
Cultivation
Enrichment and routine cultivation of haloalkaliphilic sulfur-reducing bacteria was performed at 30 °C on a mineral medium containing 0.5 M sodium bicarbonate, 0.1 M NaCl, and 0.5 g l−1 of K2HPO4. The pH of the medium after sterilization was around 8.5. The sterilized base medium was supplemented with electron donors (10–50 mM), 5–20 mM electron acceptors, 20 mg l−1 of yeast extract, 4 mM NH4Cl, 1 mM MgSO4, and 1 ml l−1 of acidic (Pfennig and Lippert 1966) and alkaline (Plugge 2005) trace metal solutions. Routine anaerobic cultivation was performed in 20 ml serum bottles with 10–15 ml medium made anoxic by 5 cycles of evacuation-flushing with argon gas. Solid alkaline media with a final salt concentrations of 0.5 M Na+ was prepared by 1:1 mixing of 4 % (w/v) agarose and 1 M Na+ mineral medium at 50 °C. The plates were incubated in closed jars under argon atmosphere with an oxygen-scavenging catalyzer (Oxoid). The pH dependence was examined at Na+ content of 0.6 M, using the following filter-sterilized mineral medium: for pH 6–8, 0.1 M HEPES and NaCl/Na2CO3; for pH 8–11, sodium carbonate-bicarbonate buffer containing 0.1 M NaCl. To study the influence of salt concentration on growth, sodium carbonate-bicarbonate/NaCl-based mineral media, containing 0.1 and 3.0 M of total Na+ at pH 9 were mixed in different proportions.
Analytical procedures
Sulfide, nitrite, ammonium and cell protein were analyzed according to Trüper and Schlegel (1964), Gries-Romijn van Eck (1966), Weatherburn (1967) and Lowry et al. (1951), respectively. Thiosulfate and sulfite were determined after removal of sulfide as ZnS by acidic iodimetric titration with formaldehyde block. Fatty acid composition of the membrane polar lipids was analyzed by GC–MS according to Zhilina et al. (1997). Phase contrast photographs were obtained with a Zeiss Axioplan Imaging 2 microscope (Göttingen, Germany).
Genetic and phylogenetic analysis
Isolation of genomic DNA and determination of the G + C content of the DNA were performed according to Marmur (1961) and Marmur and Doty (1962). For PCR, genomic DNA was extracted from the cells by alkaline SDS method using the UltraClean Soil DNA Extraction Kit (MolBio Laboratories, USA), following the manufacturer’s instructions. The PCR products were purified using the Qiagen Gel Extraction Kit (Qiagen, the Netherlands). The nearly complete 16S rRNA gene was obtained using general bacterial primers GM3f (5′-AGAGTTTGATCCTGGCTCAG-3′) and GM4r (5′–TACGGTTACCTT GTTACGACTT-3′). The sequences were first compared to all sequences stored in GenBank using the BLAST algorithm and were consequently aligned using CLUSTALW. A phylogenetic tree was reconstructed using TREECON W package (van de Peer and de Wachter 1994) and the neighbor-joining algorithm. DGGE analysis of enrichment cultures was performed according to Schäfer and Muyzer (2001).
Results and discussion
General bioreactor characteristics
The Thiopaq bioreactor in Lelystad differs, in some respects, from the well-studied model bioreactor in Eerbeek (Janssen et al. 2009; Sorokin et al. 2012). The pH and the total carbonate alkalinity were lower (8.3 versus 8.7–9 and 0.5 versus 0.9 M, respectively). In contrast, the redox potential was obviously higher than in the Eerbeek reactor, which was evident from high thiosulfate-oxidizing activity of the Lelystad SOB biomass, which is absent in the Eerbeek reactor. As a result, only a slightly alkalitolerant and fully aerobic Thiomicrospira sp. from the Tm. pelophila group has been identified as a dominant SOB in the Lelystad bioreactor (both by the DGGE analysis of whole reactor biomass and by the cultivation approach), in contrast to permanent domination of obligately haloalkaliphilic micro-aerophilic Thioalkalivibrio sulfidophilus in Eerbeek. Thiomicrospira pelophila type phylotype was also present in the Eebeek reactor but as a minor component. The density of the SOB population in the Lelystad reactor estimated in HS−/O2 gradient tubes was 1010 cell/ml.
Enrichment and isolation of the dominant sulfidogens
Primary anaerobic enrichment cultures from the Lelystad biomass with formate/sulfate, formate/thiosulfate and formate/sulfur at pH 8.5 and 0.6 M total Na+ were positive up to 10−7. Although this was 3 orders of magnitude lower than the density of active SOB population, its presence in the reactor is an indication of a complete sulfur cycle. The DGGE analysis of sulfate- and sulfur-reducing primary cultures showed a domination of two organisms, related to the genera Desulfonatronum (Deltaproteobacteria) and Sulfurospirillum (Epsilonproteobacteria), respectively (Fig. 1). From the highest positive dilution, 3 pure cultures were obtained by plating and isolation of single colonies: strain HSRB-L was obtained with sulfate, strain HTRB-L1—with thiosulfate and strain HTRB-L2—with sulfur as e-acceptor. The latter two were identical in their 16S-rRNA gene sequence, and therefore, only one of them was characterized further.
Characterization of strain HSRB-L
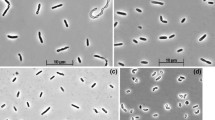
The bacterium has vibrio-shaped cells with a polar flagellation (Fig. 2a). Phylogenetic analysis (Fig. 3) placed strain HSRB-L into the genus Desulfonatronum, which so far includes five haloalkaliphilic SRB species from soda lakes (Zhilina et al. 2005; Pikuta et al. 1998, 2003; Sorokin et al. 2010). The sequence of the isolate was also 100 % similar to the sequence obtained from the dominant DGGE band. The closest relative is D. cooperativum (97 % sequence identity). The PLFA profile resembled those for the other Desulfonatronum species, but also had some clear differences, for example, the low content of 14:0 and 16:1ω7c (Supplementary Table).
Phylogenetic position of strain HSRB-L based on 16S rDNA sequence analysis within the order Desulfovibrionales in the Deltaproteobacteria. The tree was constructed using maximum likelihood method. The scale bar represents 5 nucleotide changes per 100 nucleotides. The percentage of bootstraps was derived from 1000 resampling using neighbor joining algorithm, only values >70 % are given
Strain HSRB-L is a typical sulfate reducer growing best lithoheterotrophically with formate/acetate or H2/acetate and sulfate accumulating up to 10 mM sulfide in 6 days. Sulfite and thiosulfate, but not sulfur, can also serve as e-acceptors. From other e-donors tested C2–C6 fatty acids, C2–C4 alcohols, pyruvate, malate, succinate, fumarate, only lactate and pyruvate can be used for sulfate-reducing growth. Pyruvate can be fermented, but the growth was much less active than in the presence of sulfate. The ability to grow by dismutation of thiosulfate and sulfite, typical for the genus, was not observed.
In contrast to the Desulfonatronum species obtained from soda lakes, the bioreactor isolate HSRB-L was not able to grow at pH 10 and had a typical alkalitolerant pH profile with a pH optimum of 8.7–9 (Fig. 4a). On the other hand, its salt tolerance was very similar to that of other species (Fig. 4b). These properties qualify the reactor isolate as a moderately haloalkalitolerant type. Table 1 summarizes the main properties of HSRB-L in comparison to the other Desulfonatronum species.
Characterization of strain HTRB-L1
When grown with formate and thiosulfate or sulfur, the bacterium had comma-shaped, actively motile cells (Fig. 2b); while in faster growing cultures on lactate, the cells were slightly bent rods (Fig. 2c). Phylogenetic analysis placed strain HTRB-L1 into the Epsilonproteobacteria with the nearest validly described species of the genus Sulfurospirillum as closest relatives (91 % 16S rRNA gene sequence identity). The 16S rRNA gene sequence of the isolate was 100 % similar to the sequence obtained from the dominant DGGE band. Several sequences deposited in the GenBank under the names “Sulfurospirillum sp.” were much closer to HTRB-L1, but the isolates were not characterized. Furthermore, the large phylogenetic distance indicates that both strain HTRB-L1 and its closely related uncharacterized “Sulfurospirillum” isolates belonged to a different genus-level group forming a clear separate cluster with many uncultured sequences within the family Campylobacteraceae (Fig. 5). On the other hand, the PLFA profile of strain HTRB-L1, in general, resembled the profiles of the described Sulfurospirillum species with a domination of the 16:0, 16:1ω7c and 18:1ω7c (Supplementary Table).
Phylogenetic position of strain HTRB-L1 based on 16S rDNA sequence analysis within the Epsilonproteobacteria. The tree was constructed using maximum likelihood method. The scale bar represents 5 nucleotide changes per 100 nucleotides. The percentage of bootstraps was derived from 1000 resampling using neighbor joining algorithm, only values >70 % are given
In its physiology, HTRB-L is a typical representative of anaerobic respirers from the Epsilonproteobacteria (Table 2). It grew with formate and H2 (with acetate ac C-source) most effectively with thiosulfate as e-acceptor, while reducing thiosulfate partially to sulfide (up to 15 mM sulfide in 4 days) and sulfite. Sulfite cannot be used as the e-acceptor. The ability to respire sulfite has been reported for several sulfurospirilla, including the type species S. deleyianum (Schumacher et al. 1992), and it became clear now that a periplasmic octaheme cytochrome c (MCC) is responsible for this function (Simon and Kroneck 2013) Sulfur also served as a good e-acceptor with formate and lactate as the e-donors, and those cultures accumulated up to 30 mM sulfide in 6 days. The fastest growth with lactate was observed with nitrate and fumarate as e-acceptors. Nitrate was reduced to ammonia via nitrite, as is typical for the epsilonproteobacterial anaerobes. From the other tested e-acceptors, arsenate supported growth with lactate. Pyruvate and fumarate can be fermented, but the growth was much less active than in the presence of sulfur or thiosulfate. While growing with lactate and nitrate, the sulfide added as a reductant was oxidized to elemental sulfur accumulating inside the cells similar to what was observed previously for Desulfurispirillum alkaliphilum (Sorokin et al. 2007). Washed cells of HTRB-L1 grown on formate and thiosulfate reduced elemental sulfur five times more actively than thiosulfate (0.28 μmol HS−/mg protein min), which might indicate that both acceptors are reduced via a Phs-like (thiosulfate-reductase) enzyme (Hinsley and Berks 2002).
With formate and thiosulfate, HTRB-L grew within a pH range from 7.1 to 9.7 with an optimum at pH 8.5, thus being a typical alkalitolerant bacterium. Sulfidogenic activity of washed cells had a much broader pH range and a significant alkaline shift of the pH optimum (Fig. 6a). In respect to salt tolerance, the organism is a moderate salt-tolerant organism (Fig. 6b).
Overall, our results demonstrated the presence of active populations of haloalkalitolerant respiratory sulfidogens in a full-scale Thiopaq bioreactor. The isolated strains are represented by a deltaproteobacterial SRB, and an epsilonproteobacterial sulfur/thiosulfate reducer, which, on the basis of their distinct phylogeny and phenotypic properties, are suggested here as two new species.
Description of Desulfonatronum alkalitolerans sp. nov
(Arabic n. alkali (al-qaliy), the ashes of saltwort; L. part. adj. tolerans, tolerating; N.L. part. adj. alkalitolerans, alkali-tolerating).
Cells are vibrio-shaped, 0.5–0.6 × 1.5–2.5 μm, motile by means of a polar flagellum. Strictly anaerobic utilizing H2 and formate (with acetate as C-source), lactate and pyruvate as electron donor and sulfate, sulfite and thiosulfate as electron acceptor. Pyruvate can be fermented. Alkalitolerant with a pH range for growth between 7.5 and 9.7 (opt. 8.5–9). Moderately salt-tolerant with range for growth at pH 9 from 0.1 to 1.75 M total Na+ (optimum at 0.3–0.6 M). Maximum temperature for growth at pH 9 is 42 °C. The dominant PLFA are i17:1ω8, 18:0, i15:0 and 16:0. The G + C content of the genomic DNA is 55.8 mol % (T m). The type strain HTRB-LT (DSM 24646T = UNIQEM U799T) is isolated from the Thiopaq bioreactor in Lelystad (The Netherlands). The GenBank 16S rRNA gene sequence accession number of the type strain is GQ863488.
Description of Sulfurospirillum alkalitolerans sp. nov
(Arabic n. alkali (al-qaliy), the ashes of saltwort; L. part. adj. tolerans, tolerating; N.L. part. adj. alkalitolerans, alkali-tolerating).
Cells are motile, Gram-negative, comma-to-rod shaped, 0.5–0.7 × 1.2–2.5 μm. Strictly anaerobic with respiratory metabolism. Use thiosulfate (incomplete reduction), sulfur, nitrate, nitrite, arsenate and fumarate as electron acceptors. Nitrate is reduced via nitrite to ammonium. Utilizes formate and H2 (with acetate as C-source), lactate, pyruvate, and fumarate as e-donor. Can oxidize sulfide to elemental sulfur intracellularly in the presence of nitrate as electron acceptor. Alkalitolerant, with a pH range for growth between 7.1 and 9.7 (opt. 8.5) and moderately salt-tolerant with a range for growth at pH 9 from 0.6 to 1.75 M total Na+ (optimum at 0.6 M). Maximum temperature for growth at pH 9 is 41 °C. The predominant fatty acids in the membrane lipids include 16:0, 16:1ω7c and 18:1ω7c. The G + C content of the genomic DNA is 47.6 mol % (T m). The type strain is HTRB-L1T (UNIQEM U795T). Isolated from the Thiopaq bioreactor in Lelystad (The Netherlands). The 16S rRNA gene sequence accession number of the type strain is GQ863490.
References
Gries-Romijn van Eck (1966) Physiological and chemical test for drinking water. NEN 1056, IY-2 Nederlandse Normalisatie Instituut Rijswijk
Hinsley AP, Berks BC (2002) Specificity of respiratory pathways involved in the reduction of sulfur compounds by Salmonella enterica. Microbiology 148:3631–3638
Janssen AJH, Lettinga G, de Keizer A (1999) Removal of hydrogen sulfide from wastewater and waste gases by biological conversion to elemental sulfur. Colloidal and interfacial aspects of biologically produced sulfur particles. Colloids Surf A Physicochem Eng Asp 151:389–397
Janssen AJH, Lens P, Stams AJM, Plugge CM, Sorokin DY, Muyzer G, Dijkman H, van Zessen E, Luimes P, Buisman CJN (2009) Application of bacteria involved in the biological sulfur cycle for paper mill effluent purification. Sci Tot Environ 407:1333–1343
Kodama Y, Ha LT, Watanabe K (2007) Sulfurospirillum cavolei sp. nov., a facultatively anaerobic sulfur-reducing bacterium isolated from an underground crude oil storage cavity. Int J Syst Evol Microbiol 57:827–831
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with Folin phenol reagent. J Biol Chem 193:265–275
Luijten MLGC, de Weert J, Smidt H, Boschker HTS, de Vos WM, Schraa G, Stams AJM (2003) Description of Sulfurospirillum halorespirans sp. nov., an anaerobic, tetrachloroethene- respiring bacterium, and transfer of Dehalospirillum multivorans to the genus Sulfurospirillum as Sulfurospirillum multivorans comb. nov. Int J Syst Evol Microbiol 53:787–793
Marmur J (1961) A procedure for isolation of DNA from microorganisms. J Mol Biol 3:208–214
Marmur J, Doty P (1962) Determination of the base composition of deoxyribonucleic acid from microorganisms. J Mol Biol 5:109–118
Pfennig N, Lippert KD (1966) Über das Vitamin B12—Bedürfnis phototropher Schwefelbakterien. Arch Mikrobiol 55:245–256
Pikuta EV, Zhilina TN, Zavarzin GA, Kostrikina NA, Osipov GA, Rainey FA (1998) Desulfonatronum lacustre gen. nov., sp. nov.: a new alkaliphilic sulfate-reducing bacterium utilizing ethanol. Microbiology (Moscow, English translation) 67:105–113
Pikuta EV, Hoover RB, Bej AK, Marsic D, Whitman WB, Cleland D, Krader P (2003) Desulfonatronum thiodismutans sp. nov., a novel alkaliphilic, sulfate-reducing bacterium capable of lithoautotrophic growth. Int J Syst Evol Microbiol 53:1327–1332
Plugge CM (2005) Anoxic media design, preparation, and considerations. Methods Enzymol 397:3–16
Schäfer H, Muyzer G (2001) Denaturing gradient gel electrophoresis in marine microbial ecology. In: Paul JH (ed) Methods in microbiology. Academic, New York, pp 425–468
Schumacher W, Kroneck PMH, Pfennig N (1992) Comparative systematic study on ‘Spirillum’ 5175, Campylobacter and Wolinella species. Description of ‘Spirillum’ 5175 as Sulfurospirillum deleyianum gen. nov., spec. nov. Arch Microbiol 158:287–293
Simon J, Kroneck PMH (2013) Microbial sulfite respiration. Adv Microb Physiol 62:45–117
Sorokin DY, Foti M, Tindall BJ, Muyzer G (2007) DeSulfurospirillum alkaliphilum gen. nov. sp. nov., a novel obligately anaerobic sulfur- and dissimilatory nitrate-reducing bacterium from a full-scale sulfide-removing bioreactor. Extremophiles 11:363–370
Sorokin DY, Kuenen JG, Muyzer G (2011a) The microbial sulfur cycle in soda lakes. Front Microbial Physiol 2, article 44
Sorokin DY, Tourova TP, Detkova EN, Kolganova TV, Galinski EA, Muyzer G (2011b) Culturable diversity of lithotrophic haloalkaliphilic sulfate-reducing bacteria in soda lakes and the description of Desulfonatronum thioautotrophicum sp. nov., Desulfonatronum thiosulfatophilum sp. nov., Desulfonatronovibrio thiodismutans sp. nov., and Desulfonatronovibrio magnus sp. nov. Extremophiles 15:391–401
Sorokin DY, Panteleeva AN, Muntyan MS, Muyzer G (2012) Thioalkalivibrio sulfidophilus sp. nov., a haloalkaliphilic sulfur-oxidizing gammaproteobacterium from alkaline habitats. Int J Syst Evol Microbiol 62:1884–1889
Stolz JF, Ellis DJ, Switzer Blum J, Ahmann D, Lovley DR, Oremland RS (1999) Sulfurospirillum barnesii sp. nov. and Sulfurospirillum arsenophilum sp. nov., new members of the Sulfurospirillum clade of the ε-Proteobacteria. Int J Syst Bacteriol 49:1177–1180
Trüper HG, Schlegel HG (1964) Sulfur metabolism in Thiorhodaceae. 1. Quantitative measurements on growing cells of Chromatium okenii. Antonie Van Leeuwenhoek 30:225–238
van de Peer Y, de Wachter R (1994) TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci 10:569–570
Weatherburn MV (1967) Phenol-hypochlorite reaction for determination of ammonia. Anal Chem 39:971–974
Zhilina TN, Zavarzin GA, Rainey FA, Pikuta EN, Osipov GA, Kostrikina NA (1997) Desulfonatronovibrio hydrogenovorans gen. nov., sp. nov., an alkaliphilic, sulfate-reducing bacterium. Int J Syst Bacteriol 47:144–149
Zhilina TN, Zavarzina DG, Kuever J, Lysenko AM, Zavarzin GA (2005) Desulfonatronum cooperativum sp. nov., a novel hydrogenotrophic, alkaliphilic, sulfate-reducing bacterium, from a syntrophic culture growing on acetate. Int J Syst Evol Microbiol 55:1001–1006
Acknowledgments
This work was supported by the RFBR Grant 13-04-00049 to DS, and by an Advanced ERC Grant to GM. We are grateful to Erik van Zessen for providing samples from the Thiopaq bioreactor.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Oren.
Nucleotide sequence accession number: the GenBank/EMBL accession numbers of the 16S rRNA gene sequences of strains HSRB-LT and HTRB-L1T are GQ863488 and GQ863490, respectively.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sorokin, D.Y., Tourova, T.P. & Muyzer, G. Isolation and characterization of two novel alkalitolerant sulfidogens from a Thiopaq bioreactor, Desulfonatronum alkalitolerans sp. nov., and Sulfurospirillum alkalitolerans sp. nov. Extremophiles 17, 535–543 (2013). https://doi.org/10.1007/s00792-013-0538-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-013-0538-4