Abstract
Archaea are known to play important roles in carbon cycling in marine sediments. The main compositions of archaeal community in five deep-sea sediment samples collected from west Pacific Warm Pool area (WP-0, WP-1, WP-2, WP-3, WP-4), and in five sediment layers (1 cm-, 3 cm-, 6 cm-, 10 cm-, 12 cm- layer) of the 12 cm sediment core of WP-0 were checked and compared by denaturing gradient gel electrophoresis and 16 S rRNA gene sequencing. It was revealed that all the deep-sea sediment samples checked contained members of non-thermophilic marine group I crenarchaeota as the predominant archaeal group. To further detect groups of archaea possibly relating with C1 metabolism, PCR amplification was carried out using primers targeting methane-oxidizing archaea. Although no methane-oxidizing archaea was detected, a group of novel archaea (named as WPA) was instead identified from all these five WP samples by clone analysis. They could be placed in the euryarchaeota kingdom, separated into two distinct groups, the main group was peripherally related with methanogens, the other group related with Thermoplasma. The vertical distributions of WPA, archaea and bacteria along the WP-0 sediment column were determined by quantitative-PCR. It was found that bacteria dominated at all depths, the numbers of bacteria were 10–104 times more than those of archaea. The proportion of archaea versus bacteria had a depth related increasing tendency, it was lowest at the first layer (0.01%), reached highest at the 12 cm- layer (10%). WPA only constituted a small proportion of the archaeal community (0.05% to 5%) of west Pacific Warm Pool sediment.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Deep-sea sediments that are primarily formed through the continual deposition of particles from the productive ocean surface cover approximately 70% of the earth’s surface. The organic matter settled on the sea floor is remineralized by the benthic microbial communities that colonize the sediments. The activities of ocean sediment microorganisms play important roles in the global cycling of carbon and nutrients (Vetriani et al. 1999; D‘Hondt et al. 2002). Although several recent studies have conducted to investigate the microbial communities involved in carbon and sulfur cycling in deep-sea sediments of the hydrothermal vents, methane hydrate regions, and so on (Marchesi et al. 2001; Dhillon et al. 2003; Schouten et al. 2003), the microbial populations, especially the archaeal populations in open ocean deep-sea sediments remain largely unknown. The archaea are divided into two kingdoms: Crenarchaeota and Euryarchaeota, and a third kingdom named as Korarchaeota has been proposed based on sequences retrieved from hot spring environments (Barns et al. 1996; Massana et al. 2000). Euryarchaeota is a diverse group, composed of all known methanogens, extreme halophiles, and some thermophiles; all cultivated Crenarchaeota are thermophiles (Massana et al. 1997). By using molecular markers, particularly 16S rRNA gene markers, archaea were found ubiquitous in marine environments (DeLong 1992; Massana et al. 2000; DeLong 2004). Euryarchaeota were found most abundant in surface waters (Massana et al. 1997; Vetriani et al. 1999), whereas Crenarchaeota prevalent in the deep (Massana et al. 1997; Vetriani et al. 1999; Massana et al. 2000; Takai et al. 2004). One crenarchaeotal lineage, designated as marine crenarchaeota group I, which is branched deeply from cultivated crenarcheota was found prevalent in cold and moderate-temperature environments (Massana et al. 1997; Takai et al. 2004). Although new lineages of archaea were increasingly reported (Grobkopf et al. 1998; Reysenbach et al. 2001), their possible functions need to be elucidated in the future. Recently, two groups of archaea named as ANME1 and ANME2 were found involving in the anaerobic oxidation of methane by using several culture-independent methods (Boetius et al. 2000; Orphan et al. 2001; Orphan et al. 2002). ANME1 and ANME2 are peripherally related to methanogens, and it is hypothesized that these relatives of methane-producing archaea conduct methane oxidizing by developing the capacity to reverse methanogenesis (Hallam et al. 2003; Hallam et al. 2004). Methane oxidizing archaea (MOA) are found in sedimentary sites such as gas hydrates (Nauhaus et al. 2002), cold methane seeps (Orphan et al. 2002), hydrothermal sites (Schouten et al. 2003) and in anoxic waters where methane is venting out into anoxic zones (Schouten et al. 2001), however it is not known if their distributions are only restricted to these high methane anoxic environments.
Ocean-atmosphere interactions in the tropical Pacific region have a strong influence on global heat and water vapor transport and thus constitute an important component of the climate system (Lea et al. 2000; Visser et al. 2003). The west Pacific Ocean has incredible rich of species diversity in the world (Briggs et al. 1999). Previously, the bacteria involving in C1 metabolism at one west Pacific Warm Pool site has been investigated and the archaea population was checked by 16S rRNA gene library analysis (Wang et al. 2004b). In this study, in order to get a more comprehensive picture of archaea in this area, the archaeal diversities were analyzed horizontally in five deep-sea sediments of west Pacific Warm Pool area, and vertically in a 12 cm sediment core by using denaturing gradient gel electrophoresis (DGGE) of amplified 16S rRNA gene fragments. Furthermore, PCR amplification was carried out using specific primers for MOA in the west Pacific Warm Pool sediments. The PCR products were cloned, the newly detected archaea were analyzed phylogenetically.
Materials and methods
Sample collection and characterization
Sediment samples from the tropical west Pacific Warm Pool site WP-0 and WP-1, WP-2, WP-3, WP-4 (Table 1) were taken during the cruise of DAYANG number1 by multi-core samplers in 2001 and 2002 respectively. The sediment cores were fractioned and transferred to sterile falcon tubes in clean bench and stored aseptically at −20°C till usage. pH and salinity of each sample were measured according to Munson et al (1997). The total organic carbon contents (TOC) of the samples were determined by the method of Gaudette et al (1974).
PCR-DGGE analysis
DNA templates for PCR were extracted from sediments according to the methods described before (Zhou et al. 1996). Archaeal 16S rRNA gene fragments were amplified using primer pairs ARC344f-ACGG GAGCAGCAGGCGCGA and ARC915r-GTGCTCCCCCGCCAATTCCT(Hendrik and Muyzer 2001). For DGGE analysis, a 40 bp GC-rich sequence (“GC-clamp”): 5’- CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACG GGGGG was attached to the 5’end of primer ARC344f to prevent the complete dissociation of the two DNA strands. A hot-start PCR was performed at 95°C for 10 min, and touchdown PCR was performed as follows: the annealing temperature was initially set at 65°C and then decreased by 0.5°C every cycle until reach to 55°C.Twenty additional cycles were carried out at 55°C. Denaturing was carried out at 94°C for 1 min. Primer annealing was performed using the scheme described above for 1 min, and primer extension was performed at 72°C for 3 min. The final extension step was 10 min at 72°C. DGGE was performed using the Bio-Rad Dcode universal mutation detection System (Bio-Rad, USA), according to the protocol of Mako et al (2002). PCR products were loaded onto a 6% (wt / vol) polyacrylamide gel in 1x TAE buffer (40 mM Tris, 20 mM acetic acid, and 1 mM EDTA at pH8.0). The 6% (wt / vol) polyacrylamide gels (acrylamide / bisacrylamide ratio, 37.5:1) were made with denaturing gradients ranging from 40% to 60% gradient of urea and formamide. Denaturant (100%) contained 40% (vol / vol) formamide and 7 M urea. The electrophoresis was run at 60°C, for 30 min at 20 V, and subsequently for 14 h at 100 V. After electrophoresis, the gels were stained for 20 min with ethidium bromide (0.5 μg / ml). DGGE gels were visualized with a UV transilluminator. The unique bands were reamplified with primer ARCf344 and ARCr915 and cloned by using the pGEM-T TA cloning kit (Promega) according to the manufacturer’s instructions.
PCR for WPA
PCR primers used to selectively amplify the 16S rRNA gene fragments of MOA were according to published reference (Thomsen et al. 2001) (ANMEF: GGCUCAGUAACACGUGGA, 907R: CCGTCAATTCCTTTRAGTTT). PCR reaction was set in 50μL reaction volume containing 50–100ng DNA template, 10×reaction buffer (with 1.5 mM MgCl2), 1 mM dNTP, 10 pmol each of primers and 5 units of Taq polymerase. Reactions were carried out in a T3 thermocycler (Biometra, Germany) with 35 cycles of 94°c 60 s, 55°C 90 s, 72°C 120 s and a final extension at 72°C for 10 min. Reaction products were checked by agarose gel electrophoresis.
16S rRNA gene sequencing and phylogenetic analysis
The amplified PCR products were cloned into the pGEM-T vector by using a 2× rapid ligation kit according to the instructions of the manufacturer (Promega). Ligation mixtures were used to transform competent cells of E. coli XL1 Blue according to the suggestions of the manufacturer (Promega). The inserted fragments of the clones were amplified, purified by ethanol precipitation. Then the fragments were digested for 8 h or more at 37°C with restriction endonuclease RsaI. The reaction results were visualized byelectrophoresis on a 5% agarose gel containing ethidiumbromide (0.5 mg/liter). The representative clones with different restriction fragment length polymorphism (RFLP)patterns of each library were selected to sequence (Sangon Inc., China). Each sequence was submitted to the CHECK-CHIMERA program of the RDP (Maidak et al. 1999) to detect the presence of possible chimeric artifacts. Phylogenetic trees were constructed from a matrix by least-squares distance matrix analysis (Olsen et al. 1988) and the neighbor-joining method (Saitou and Nei 1987) using the DNAMAN program (version 5.1, Lynnon Biosoft), and 1000 trial of bootstrap analysis was used to provide confident estimatesfor phylogenetic tree topologies.
Quantification of WPA, archaea and bacteria by QC-PCR
The quantitative competitive PCR (QC-PCR) method (Johnsen et al. 1999; Han and Semrau 2004; Wang et al. 2004) was used to determine the relative quantity of archaea, WPA and bacteria in samples. QC-PCR quantification uses a mixture of target DNA with varying amount of an internal competitor DNA as template for amplification by the same primer set in the same reaction. The quantity of target DNA is then measured according to the amount of the competitor DNA.
The internal competitor for WPA, archaea and bacteria was constructed to quantify WPA, archaea and bacteria respectively. A plasmid pWPA26 isolated from a clone of the WPA library was digested by NheI to get a 97 bp deletion in the region between the primer set ANMEF and 907R, self-ligated and digested by HindIII. The linear plasmid pWPA26Δ97 bp was used as the internal competitor DNA for WPA quantification. The competitor template DNA pA35Δ180 bp for quantification of archaea was constructed using a plasmid pMBWPA35 extracted from a clone of the archaeal library of WP-0 constructed with the universal archaeal primer ARC21f and ARC915r (Wang et al. 2004 b). pMBWPA35 was digested by PstI to get a 180bp deletion, it was then self-ligated and digested by HindIII to get the competitor pA35Δ180bp. The competitor pE106Δ300bp for quantification of bacteria was constructed from pE106, which was attained by cloning of a 450bp 16S rRNA gene fragment amplified from a bacteria strain WP8 of WP-0 site (Wang et al. 2004a) using primer EUB f933 and EUB r1387, into the pGEM-T vector. The plasmid pE106 was digested with Kpn2I to get a 300 bp deletion between the primer set, self-ligated and then digested with HindIII. The linear plasmid pE106Δ300bp was used as the internal competitor DNA for bacteria quantification. The quantities of DNA templates were determined by using spectrophotometer (Ultrospec 2100, Amersham Pharmacia).
Nucleotide sequence accession number
The sequence obtained in this study has been deposited in the EMBL / Genebank / DDBJ database under accession no: WPA: AJ783650, AJ783651, AJ783653- AJ783665; AW: AJ783666-AJ783674.
Results
Description of sediment samples
The sediment samples of WP-0 were mainly composed of foraminifera while the other four sediments from WP-1, WP-2, WP-3, and WP-4 were composed of dark brown siliceous clay. All samples had nearly equivalent pH and salinity but WP-0 had a relatively higher temperature than the other four sites (Table 1). All samples showed low total organic carbon content (TOC) ranging from 0.11% to 0.29% which was much lower than those of Pacific coast area (Liu et al. 2003). There was no correlation between site depth and organic carbon content which was also different from the coast area (Liu et al. 2003).
Population structure of archaea in west Pacific Warm Pool area (WP)
The archaeal communities in the deep-sea sediments of the five different sites of WP area (WP-0, WP-1, WP-2, WP-3, WP-4) and in the five layers (1 cm-, 3 cm-, 6 cm-, 10 cm-, 12 cm- layer) of the 12 cm sediment core of WP-0 were profiled by DGGE. DGGE fingerprints of the samples showed that most bands were common in all of the samples, demonstrating that the main archaeal populations in the five different WP sites and along the sediment core of WP-0 were similar (Fig. 1). Nine dominant bands, each representing a putative organism, were sequenced. Phylogenetic affiliation analysis showed that all clones from the five sites of west Pacific WP belonged to crenarchaeotal kingdom, most of the clones clustered into the non-thermophilic crenarcheotal marine group1 α subgroup (MGI-α), two clones AW1 and AW4 affiliated with members of marine benthic group A (Fig. 2).
The DGGE profile of the 16S rRNA gene fragment of archaea 16S rRNA gene fragments of archaea amplified from five geologically different west Pacific deep-sea sediments and five different layers of a sediment core were compared by DGGE as described in the materials and methods. lane1 WP-1; lane2 WP-2; lane-3 WP3; lane4 WP-4; lane5 WP-0–1cm; lane6 WP-0-3cm;lane7 WP-0-6 cm; lane8 WP-0-10 cm; lane9 WP-0-12 cm. Bands a, b, c, d, e, f, g, h, i were excised from the gel and cloned for sequencing as described above. They correspond to clone AW1, AW2, AW3, AW4, AW5, AW6, AW7, AW8, AW9 respectively.
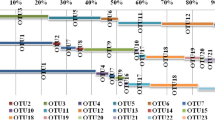
Phylogenetic tree constructed based on 16S rRNA gene sequcences of archaea Archaeal 16S rRNA gene clones retrieved from the Pacific Warm Pool sediments by DGGE method were named as AW. Nine AW clones together with relative clones in the databank were used for phylogenetic tree construction. The phylogenetic tree was constructed from a matrix by least-squares distance matrix analysis and the neighbor-joining method using the DNAMAN program. 1000 trial of bootstrap analysis was used to provide confident estimatesfor phylogenetic tree topologies. Only bootstrap values above 50 were shown. The scale bar represents 0.05 substitution per nucleic acid site. The accession number of AW1-AW9 was AJ783666-AJ783674. clones APA2-17 cm, ACA10-0 cm were retrieved from the Atlantic ocean sediment (4500 m); pCIRA-102, pCIRA-104 from hydrothermal plume; SB95-1, from Santa Barbara Channel, Pacific Ocean (522 m); AM-20A, from Arthur Harbor, Antarctica; Mariana archaeon no.11, from Mariana Trench (11000 m); PMC2A1, from hydrothermal vent of Myojin Knoll; pIVW1, from deep-sea hydrothermal vent of Iheya Basin (972 m), ANTACTIC12, from Antarctic marine; PVA_OTU_3, from hydrothermal vent environments; Ar18–4 and Cenarchaeum symbiosum, from marine sponges; 4B7, from marine sediment; MBWPA9, MBWPA17, MBWPA18, MBWPA24, MBWPA36, MBWPA51, MBWPA53, MBWPA67, MBWPA121 and MBWPA137, from the sediment of west Pacific Warm Pool WP-0.
Phylogenetic and signature analysis of newly detected archaea (WPA)
To further detect archaea relating with C1 metabolism, PCR amplifications were performed using primers targeting the MOA 16S rRNA gene fragments. Specific DNA bands of around 810 bp were amplified from all five WP sites (WP-0, WP-1, WP-2, WP-3, WP-4), and from all selected sediment layers of WP-0 by using MOA specific 16S rRNA gene primers (data not shown), indicating that these newly detected archaea were present in all the WP sites and from the surface to the bottom of the sediment core. PCR products from each sediment layer (1 cm-, 3 cm-, 6 cm-, 10 cm-, 12 cm- layer) of WP-0 were cloned and libraries were constructed. From each library, 50 clones were randomly selected and screened by RFLP analysis. No relation could be found between the clone RFLP types and the sediment layers (Table 2). According to the RFLP types, 15 clones were selected for sequencing. The sequences of the clones were checked and analyzed. It could be seen from Table 2 that the 16S rRNA gene sequences of all the clones retrieved had relatively low identities with those of known archaea (highest identities with those of known environmental clones were around 74 – 84 %). These newly detected archaea were named as WPA. The phylogenetic analysis classified all of the sequences into two distinct lines of descent in Euryarchaeotal kingdom (Fig. 3). One cluster WPA-I formed a Themoplasma-associated branch; however this cluster of archaea were only distantly related to the Thermoplasma-associated clone WCHD3-02 by overall levels of rRNA gene sequence similarity of 82%-83%. The intralineage levels of rRNA gene similarity among clones of WPA-I were between 90% and 95%. The other cluster WPA-II contained 12 clones forming a distant branch related with methanogens. This cluster could be further divided into six sub-clades, as shown in Fig. 3: clone WPA18–3, WPA4–12, WPA35–6,WPA4–3 each formed single group (WPAII-1, WPAII-2, WPII-4, WPAII-6); clone WPA38–6, WPA32–10, WPA4–10, WPA3–10, WPA21–3, WPA8–10 could be clustered as one group (WPAII-3); and clone WPA5–3, WPA15–2 formed one group (WPAII-5).
Phylogenetic tree showing the relationship of WPA and related clones and strains. Archaeal 16S rRNA gene clones retrieved by PCR using primers ANMEF and 907R from the Pacific Warm Pool sediment were named as WPA. 15 sequences representing 15 different RFLP types of WPA and 16S rRNA gene sequences of reference clones or strians in the databank were used for the dendrogram construction. The phylogenetic tree was constructed from a matrix by least-squares distance matrix analysis and the neighbor-joining method using the DNAMAN program, and 1000 trial of bootstrap analysis was used to provide confident estimatesfor phylogenetic tree topologies. Only bootstrap values above 50 were shown. The scale bar represents 0.05 substitution per nucleic acid site.
Quantification of WPA, archaea and bacteria along the sediment core
The abundances of WPA, archaea and bacteria along the 12 cm-sediment core were determined by QC-PCR. For the accurate quantification by QC-PCR, the target and competitor DNA must be amplified by the same efficiency. Therefore, the sensitivity and accuracy test of the three competitor standard for WPA, archaea and bacteria each was first carried out and data showed that as less as 10 molecules of standard DNA fragment could be tested, the target and competitor DNA could be amplified with equal efficiency (data not shown). This indicated that the competitors constructed were qualified and suitable to quantify WPA, archaea and bacteria in the environment. Then, DNAs extracted from the sediments were mixed with different amounts (101, 102, 103, 104, 105, 106, 107, 108, 109 molecules) of internal competitor (linear plasmid pE106Δ300bp for bacteria; pA35Δ180bp for archaea; pWPA26Δ97bp for WPA), the mixed DNAs were used as PCR templates for amplification using the same primer set. When the quantity of internal competitor was 10 times less than that of the target in the sediment, only the target fragment could be amplified; in contrast, when the quantity of internal competitor was 10 times higher than that of the target, only the competitor fragment could be amplified. When the quantities of the target and internal competitor were in the same range, both fragments could be amplified (photo not shown). The quantification data showed that the WP-0 sediments were dominated by bacteria at all depth intervals (Fig. 4). The number of bacteria was 10–104 times more than that of archaea. In the surface layer, the number of bacteria was highest, whereas the number of archaea was highest at the 3 cm- layer. The proportion of archaea versus bacteria had a depth related increasing tendency, it was lowest at the first layer (0.01%), reached highest at the 12 cm- layer (10%). WPA only constituted a small proportion of the archaeal community (0.05% to 5%) of west Pacific WP sediment.
Vertical distribution of WPA, archaea and bacteria along sediment core The numbers of WPA, archaea and bacteria in the five sediment layers (1 cm-, 3 cm-, 6 cm-, 10 cm-, 12 cm- layer) of the 12 cm sediment core of WP-0 were determined by QC-PCR as described in the text. WPA, archaea and bacteria represent the quantities of WPA, archaea and bacteria in samples respectively.
Discussion
Of the three major evolutionary domains of life on earth, the archaea are the least understood in terms of their diversity, physiology, genetics and ecology. Open ocean deep-sea sediments are generally organic poor, low methane, low oxygen, dark and cold (1–2°C) environments (Kato et al. 1999; D’Hondt et al. 2002). The microbial activities in these environments are extremely low. However, considering the large areas they cover, the microbes in these environments should not be ignored. Tropical west Pacific Warm Pool, with surface water temperature > 28°C, is a geological important area which is fundamental to the climate of the earth (Visser et al. 2003). The archaeal communities in five sites of west Pacific Warm Pool area and in different vertical sediment layers of one sediment core were investigated in our study. Our results clearly showed that MGI were the predominant members in the deep-sea sediments of west Pacific Warm Pool area. Members of MGI were found ubiquitous in marine environments, from the surface to the bottom of water, deep-sea sediments, subsurface sediments, hydrothermal systems, and so on (Vetriani et al. 1999; Inagaki et al. 2001; Takai et al. 2004). MGI is potentially the largest population of archaea on earth, however the physiological, metabolic modes and ecological significance of members of MGI are still unclear due to lack of cultivation, although recent studies have begun to reveal some of their properties (Pearson et al. 2001; Ouverney and Fuhrman 2000; Shouten et al. 2000). Using culture-independent molecular methods, distribution of MGI in the vicinity of deep-sea hydrothermal systems was investigated, it was hypothesized that deep-sea hydrothermal systems were potential sources and sinks of marine planktonic archaea (Moyer et al. 1998). Takai et al. (2004) further checked the distribution and abundance of MGI in two geologically and geographically distinct hydrothermal systems. It was found that the proportion of MGI was higher in the ambient water than in the normal deep sea water, and it was suggested that the ambient seawater habitats of the deep-sea hydrothermal sites may be a possible source environment of some groups of MGI. In this study, the MGI detected in the five distantly located deep-sea sediments of west Pacific WP area are nearly the same and all of them belong to MGI-α group (Massana et al. 2000; Takai et al. 2004). Among the seven MGI-α clones retrieved from the WP sediment samples, AW2 and AW9 showed highest 16S rRNA gene sequence homology identities with that of Mariana archaeon no.11, which was detected from Mariana Trench (11000 m) (92–94%); AW3 and AW8 had highest identities with SB95–1 (91–94%) detected from deep waters of Santa Barbara Channel, Pacific Ocean (522 m); AW5, AW6 and AW7 was most closely related with clone MBWPA36 (96%), MBWPA24 (93%) and MBWPA137 (97%) which were retrieved from WP-0 in our previous study (Wang et al. 2004b). Our data once again suggested that the MGI members of the deep ocean may have the same origin.
The west Pacific deep-sea sediment surface is slightly oxic, we have detected the occurrence of oxic methylotrophs in the sediment surface layer (Wang et al, 2004b). However, the properties of sediment core are very complex, as to the oxygen level, moving down the sediment, oxygen is typically depleted within millimeters below the surface by aerobic respirers and diffusional limitation of oxygen transport from the above, and the sediment structure is seldom uniform, it contains complex and different microenvironments even in the same sediment layer (Glud et al. 1996, Fenchel et al. 2002). Therefore, it seems more important to detect the anaerobic microbial community taking part in the C1 metabolism in the oxygen limited deep-sea sediment cores. It has been hypothesized that MOA are relatives of methanogens and methane oxidization was carried out by these methanogens reversely by producing hydrogen and carbon dioxide from methane. This hypothesis was further supported by genomic analysis of MOA from deep-sea sediments (Hallam et al. 2003; Hallam et al. 2004). However, the distribution of MOA or related methanogens has not been investigated systematically. We used a specific primer targeting anaerobic methane oxidizers (ANMEF) designed by Thomsen et al, together with a universal primer 907R to detect MOA in the west Pacific deep-sea sediments. It was announced by Thomsen et al that in the Ribosomal Database Project, only sequences of tentative methane oxidizer from the Eel River Basin match the ANMEF sequence. We made a Probe Match analysis again in the RDP, it was found that 126 sequences match the probe, not only include the putative methane oxidizers, but also many methanogens such as Methanobacterium thermoautotrophicum, Methanobacterium formicicum etc, and one sequence of clone VC2.1 Arc13 which was closely affiliated with Thermoplasma acidophile. Therefore, the primer ANMEF was not highly specific for MOA, it could be used not only to retrieve methane oxidizers, but also many methanogens. By using this primer, we got specific bands from all the five deep-sea sediment samples. However, none of the sequences was closely affiliated with any known archaeal methanotrophs or methanogens. These newly detected Euryarcheota could be divided into two groups: group I (WPA-I) was related with Thermoplasma; group II (WPA-II) was far related with Methanobacterium. Domain signatures and phylogenetic analysis showed that members of WPA may be new lineages within kingdom Euryarchaeota. Cluster WPA-II showed highest similarity with methanogens (16S rRNA gene sequence identities around 71%-77%), then with ANME (64%-74%). The function of the newly discovered archaea WPA could not be determined yet. From the phylogenetic position, it could be only suggested that they might have function in C1 metabolism in organic poor deep oceans, however as they were not very closely related with any known MOA or methanogens, no conclusion could be made yet. WPA was detected in all the five west Pacific deep-sea sediments that are geologically distinct located, suggesting that they may be ubiquitous group of archaea in deep-sea environments.
The abundance of bacteria, archaea and WPA in the sediment core of WP0 was assayed by QC-PCR. We must state here that the QC-PCR used in this study could only roughly quantify (or semi-quantify) the microbes in the environment. However, it is enough for our analysis. It was shown that the biomass of WPA group was only 0.05% to 5% of archaea and the archaeal constitution in the west Pacific Warm Pool sediment only ranged from 0.01% to 10% of the prokaryote. This low percentage of the novel group WPA in the environment is probably the reason for its escaping of detection by the PCR-DGGE method in our study because only numerically dominant populations will be detected by DGGE (Sievert et al. 1999).
References
Barns SM, Delwiche, CF, Palmer, JD, Pace NR (1996) Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci USA 93:9188–9193
Boetius A, Ravenschlag K, Schubert GJ, Rickert D, Widdel F, Gieseke A, Amann R, Jergensen BB, Wittle U, Pfannkuche O (2000) A maine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623–626
Briggs JC (1999) Coincident biogeographic patterns: Indo-West Pacific Ocean. Evolution 53:326–335
DeLong EF (1992) Archaea in coastal marine environments. Proc Natl Acad Sci USA 89:5685–5689
DeLong EF (2004) Archaeal means and extremes. Science 280:542–543
Dhillon A, Teske A, Dillon J, Stahl DA, Sogin ML (2003) Molecular characterization of sulfate-reducing bacteria in the Guaymas Basin. Appl Environ Microbial 69:2765–2772
D’Hondt S, Rutherford S, Spivack AJ (2002) Metabolic activity of subsurface life in deep-sea sediments. Science 295:2067–2070
Fenchel T (2002) Microbial behavior in a heterogeneous world. Science 296:1068–1071
Gaudette HE, Flight WR, Toner L, Folger DW (1974) An inexpensive titration method for determination of organic carbon in recent sediments. J Sediment Petrol 44:249–253
Glud RN, Ramsing NB, Gundersen JK, Klimant I (1996) Planar optrodes: a new tool for fine scale neasurements of two-dimensinal O2 distribution in benthic communities. Mar Ecol Prog Ser 140:217–226
Grobkopf R, Stephan S, Liesack K (1998) Novel Euryarchaeotal lineages detected on rice roots and in the anoxic bulk soil of flooded rice microcosms. Appl Environ Microbial 64:4983–4989
Hallam SJ, Girguis PR, Preston CM, Richardson PM, DeLong EF (2003) Identification of methyl coenzyme M reductase A (mcrA) genes associated with methane-oxidizing archaea. Appl Environ Microbial 69:5483–5491
Hallam SJ, Putnam N, Preston CM, Detter JC, Rokhsar D, Richardson PM, DeLong EF (2004) Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 305:1457–1462
Han JI, Semrau JD (2004) Quantification of gene extression in methanotrophs by competitive reverse transcription-polymerase chain reaction. Environ Microbiol 6:388–399
Hendrik S, Muyzer G (2001) Denaturing gradient gel electrophoresis in marine microbial ecology. In: Paul JH (eds) Methods in microbiology: Marine microbiology. Academic Press, San diego San Francisco New York Boston London Sydney Tokyo, pp 425–468
Inagaki F, Takai K, Komatsu T, Kanamatsu T, Fujioka K, Horikoshi K (2001) Archaeology of archaea: geomicrobiological record of Pleistocene thermal events concealed in a deep-sea subseafloor environment. Extremophiles 5:385–392
Johnsen K, Enger O, Jacobsen CS, Thirup L, Torsvik V (1999) Quantitative selective PCR of 16S ribosomal DNA correlates well with selective agar plating in describing population dynamics of indigenous pseudomonas spp In soil hot spots. Appl Environ Microbiol 65:1786–1789
Kato C (1999) Barophiles (piezophiles). In: Horikoshi K, Tsujii K (eds) Extremophiles in deep-sea environments. Springer-Verlag, Tokyo, pp 91–111
Lea DW, Pak DK, Spero HJ (2000) Climate impact of late quaternary equatorial pacific sea surface temperature variations. Science 289:1719–1724
Liu X, Bagwell CE, Wu L, Devol AH, Zhou J (2003) Molecular diversity of sulfate-reducing bacteria from two different continental margin habitats. Appl Environ Microbiol 69:6073–6081
Maidak BL, Cole JR, Parker CT, Garrity GM, Larsen N, Li B, Lilburn TG, Mccaughey MJ, Olsen GJ, Overbeck R, Pramanik S, Schmide TM, Tiedje JM, Woese CR (1999) A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res 27:171–173
Mako K, Eiichi M, Hisashi K, Nobuyasu Y, Katsuji T, Masao N (2002) Ribosomal DNA-based analysis of bacterial diversity in purified water used in pharmaceutical manufacturing processes by PCR and denaturing gradient gel electrophoresis. Appl Environ Microbiol 68:699–704
Marchesi JR, Weightman AJ, Cragg BA, Parkes RJ, Fry JC (2001) Methanogen and bacterial diversity and distribution in deep gas hydrate sediments from the Cascadia Margin as revealed by 16S rRNA molecular analysis. FEMS Microbiol Ecol 34:221–228
Massana R, Murray AE, Preston CM, Delone EF (1997) Vertical distribution and phylogenetic characterization of marine planktonic archaea in the Santa Barbara Channel. Appl Environ Microbial 63:50–56
Massana R, DeLong EF, Aliq cq (2000) A few cosmopolitan phylotypes dominate planktonic archaeal assemblages in widely different oceanic provinces. Appl Environ Microbial 66:1777–1787
Moyer CL, Tiedje JM, Dobbs FC, Karl DM (1998) Diversity of deep-sea hydrothermal vent archaea form Loihi Seamout, Hawaii. Deep-sea Rse 45:303–317
Munson MA, Nedwell DB, Embley TM (1997) Phylogenetic diversity of archaea in sediment samples from a coastal salt mash. Appl Environ Microbial 63:4729–4733
Nauhaus K, Boetius A, Kruger M, Widdel F (2002) In vitro demonstration of anaerobic oxidation of methane coupled to sulphate reduction in sediment from a marine gas hydrate area. Environ Microbiol 4:296–305
Olsen GJ (1988) Phylogenetic analysis using ribosomal RNA. Methods Enzymol 164:793–812
Orphan VJ, House CH, Hinrichs KU, Mckeegan KD, DeLong EF (2001) Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293:484–487
Orphan VJ, House CH, Hinrichs KU, Mckeegan KD, DeLong EF (2002) Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc Natl Acad Sci USA 99:7663–7668
Ouverney CC, Fuhrman JA (2000) Marine planktonic archaea take up amino acids. Appl Environ Microbial 66:4829–4833
Pearson A, McNichol AP, Benitez-Nelson BC, Hayes JM, Eglinton TI (2001) Origins of lipid biomarkers in Santa Monica Basin surface sediments: a case study using compound-specific 14 C analysis. Geochim Cosmochim Acta 65:3123–3137
Reysenbach AL, Longnecker K, Kirshein J (2001) Novel bacterial and archaeal lineages from an in situ growth chamber deployed at a mid-Atlantic ridge hydrothermal vent. Appl Environ Microbial 66:3798–3806
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425
Schouten S, Wakeham SG, Damste JSS (2001) Evidence for anaerobic methane oxidation by archaea in euxinic waters of the Black Sea. Org Geochem 32:1277–1281
Schouten S, Wakeham SG, Hopmans EC, Damste JSS (2003) Biogeochemical evidence that thermophilic archaea mediate the anaerobic oxidation of methane. Appl Environ Microbial 69:1680–1686
Shouten S, Hopmans EC, Pancost RD, Damste JSS (2001) Widerspread occurrence of structurally diverse tetraether membrane lipids: evidence for the ubiquitous presence of low-temperature relatives of hyperthermophiles. Proc Natl Acad Sci USA 97:14421–14423–6
Sievert SM, Brinkhoff T, Muyzer G, Ziebis W, Kuever J (1999) Spatial heterogeneity of bacterial populations along an environmental gradient at a shallow submarine hydrothermal vent near milos island (Greece). Appl Environ Microbial 65:3834–3842
Takai K, Oida H, Suzuki Y, Hirayama H, Nakagawa S, Nunoura T, Inagaki F, Nealson KH, Horikoshi K (2004) Spatial distribution of marine crenarchaeota group I in the vicinity of deep-sea hydrothermal systems. Appl Environ Microbiol 70:2404–2413
Thomsen TR, Finster K, Ramsing EB (2001) Biochemical and molecular signatures of anaerobic methane oxidation in a marine sediment. Appl Environ Microbial 67:1646–1656
Vetriani C, Jannasch HW, Macgregor BJ, Stahl DA, Resysenbach AL (1999) Population structure and phylogenetic characterization of marine benthic archaea in deep-sea sediments. Appl Environ Microbiol 65:4375–4384
Visser K, Thunell R, Stott L (2003) Magnitude and timing of temperature change in the indo-Pacific Warm Pool during deglaciation. Nature 421:152–155
Wang F, Wang P, Chen M, Xiao X (2004a) Isolation of extremophiles with the detection and retrieval of Shewanella strains in deep-sea sediments from the west Pacific. Extremophiles 8:165–168
Wang P, Wang F, Xu M, Xiao X (2004b) Molecular phylogeny of methylotrophs in a deep-sea sediment from a tropical west Pacific Warm Pool. FEMS Microbiol Ecol 47:77–84
Winker S, Woese CR (1991) A definition of the domains archaea, bacteria and eucarya in terms of small subunit ribosomal RNA characteristics System. Appl Microbilol 14:305–310
Zhou J, Bruns MA, Tiedje JM (1996) DNA recovery from soils of diverse composition. Appl Environ Microbiol 62:316–322
Acknowledgements
We would like to thank the crews on DaYang No.1 for assisting in collecting the samples. This work was supported in part by the Chinese high Tech (863) program (2004AA621010), Specific fund for preliminary fundamental research (202CCA05000) and COMRA Fund (DY105-04-02-7).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Horikoshi
Peng Wang and Xiang Xiao contributed equally to this paper
Rights and permissions
About this article
Cite this article
Wang, P., Xiao, X. & Wang, F. Phylogenetic analysis of Archaea in the deep-sea sediments of west Pacific Warm Pool. Extremophiles 9, 209–217 (2005). https://doi.org/10.1007/s00792-005-0436-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-005-0436-5