Abstract
Phage-host systems from extreme cold environments have rarely been surveyed. This study is concerned with the isolation and characterization of three different phage-host systems from Arctic sea ice and melt pond samples collected north-west of Svalbard (Arctic). On the basis of 16S rDNA sequences, the three bacterial phage hosts exhibited the greatest similarity to the species Shewanella frigidimarina (96.0%), Flavobacterium hibernum (94.0%), and Colwellia psychrerythraea (98.4%), respectively. The host bacteria are psychrophilic with good growth at 0°C, resulting in a rapid formation of visible colonies at this temperature. The phages showed an even more pronounced adaptation to cold temperatures than the bacteria, with growth maxima below 14°C and good plaque formation at 0°C. Transmission electron microscopy (TEM) examinations revealed that the bacteriophages belonged to the tailed, double-stranded DNA phage families Siphoviridae and Myoviridae. All three phages were host-specific.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the past few years, investigations on microbial communities of the permanently cold Southern and Arctic Ocean have increased. Sea ice has proved to be one of the most important and striking polar habitats, with extraordinary high microbial activities despite subzero temperatures (Grossi et al. 1984; Kottmeier et al. 1987; Helmke and Weyland 1995; Priddle et al. 1996; Thomas and Dieckmann 2002). Different studies in Arctic and Antarctic sea ice have indicated high primary and secondary production as well as high algal and bacterial abundance (Kottmeier and Sullivan 1989; Smith and Clement 1990; Grossmann and Dieckmann 1994). The bacterial sea ice communities were found to be remarkably diverse (Bowman et al. 1997; Staley and Gosink 1999; Brown and Bowman 2001) with an unusually high percentage of cultivatable bacteria (Helmke and Weyland 1995). Most of the bacteria were previously unrecognized species closely adapted to their site of occurrence.
Using direct counting with transmission electron microscopy (TEM), viruses were recognized as the most abundant biological entities in marine systems (Bergh et al. 1989) and temporal changes in viral counts indicated that these biological agents are dynamic components of marine microbial communities (Bratbak et al. 1996). Moreover, it could be shown that viral lysis has a significant impact on bacterial abundance as well as species composition and plays an important role in microbial nutrition webs (Proctor and Fuhrman 1990; Suttle et al. 1990; Moebus 1991; Wichels et al. 1998; Noble and Fuhrman 2000; Wommack and Colwell 2000). Although only a few studies have been conducted on viruses in polar waters (Smith et al. 1992; Bird et al. 1993; Steward et al. 1996; Marchant et al. 2000; Guixa-Boixereu et al. 2002), it is obvious that viruses are no less abundant in the Southern and Arctic Ocean and therefore are likely to be as important as elsewhere in the world's oceans. Two studies documented the presence of viruses in sea ice (Maranger et al. 1994; Vincent et al. 2000). Considering the high percentages of active and cultivatable bacterial cells in sea ice communities, it was not surprising to find that the virus-to-bacteria ratios (VBR) in sea ice are among the highest ever reported for natural samples and that viruses are dynamic components of sea-ice microbial communities (Maranger et al. 1994). However, so far, no cultivation of sea-ice phages has been reported.
The objective of the present study was the isolation and characterization of psychrophilic bacteriophage-host systems from Arctic sea-ice and melt-pond habitats. The host bacteria were characterized by 16S rDNA sequences, morphological appearance, as well as growth response to temperature, pH, and salinity. Morphological examinations with TEM, studies on growth temperature limits, as well as DNA analysis were used to describe the phages.
Materials and Methods
Isolation of bacterial hosts
Samples from bottom sections of sea ice and melt ponds on top of sea-ice floes were collected during the R. V. Polarstern cruise ARKXIII/2 (June/July 1997) in the northern Fram Strait and northeast of Svalbard, Arctic. Sea ice was collected and processed as previously described (Helmke and Weyland 1995). Water samples from melt ponds were taken with 100-ml syringes and transferred into small glass bottles. Before storage the sea-ice samples were melted in sterile sea water (1:1 v/v) to reduce mortality of bacteria due to osmotic shock during thawing. The melt pond and sea-ice samples were stored for about 6 months at 4°C in the dark. Potential bacterial host strains were isolated by spreading 200 µl of each sample on pre-cooled ZoBell agar plates. The ZoBell medium was prepared as described by Truong et al. (2001). Inoculated agar plates were incubated at 4°C in the dark. After 1–4 weeks, colonies of different appearance in color, size, and morphology were selected for isolation. Fifty pure bacterial strains were obtained, which were used as hosts in the bacteriophage screening.
Isolation of phages
To increase the number of isolated bacteriophages, enrichment cultures were set up by adding 1 ml of sample to 4 ml liquid ZoBell medium. The cultures were incubated over a period of 2–6 weeks at 4°C without shaking. Volumes of 20 µl of these enrichment cultures were spotted by a modified 20-fold multiapplicator onto soft agar (ZoBell medium containing 0.6% agar), which had been inoculated with the different host bacteria. During preparation of the soft agar, the utmost care was taken not to kill the psychrophilic bacteria with warm agar. The screening plates were incubated for 3–7 days at 4°C. Plaques that appeared during incubation were picked and re-isolated twice.
Phage lysates of high titer were produced as liquid lysates from shaking cultures or by elution of agar plates with 5 ml SM buffer (950 ml distilled water, 50 ml 1 M Tris-HCl (pH 7.5), 5.8 g NaCl, 2 g MgSO4 × 7 H2O, 0.1 g gelatin (Difco, Detroit, USA). For long-term storage at 4°C the lysates were filtered through filters of 200 nm pore size (Red Rim FP 030/3; Schleicher and Schuell, Dassel, Germany).
Scanning electron microscopy (SEM) of bacteria
Growth of bacteria in shaking cultures was stopped by the addition of NaN3 (final concentration 25 mM) and cell concentration was adjusted to 109 ml−1. The bacteria were bound for 10 min onto poly-l-lysine-coated glass slides, which were placed in Petri dishes with 100% air humidity. Unattached bacteria were removed from the glass slide by a brief transfer into cacodylate buffer (pH 7.4, 0.1 M cacodylate, 25 mM NaN3, and 32% of salt solution containing 821 mM NaCl, 52 mM MgCl2, 57 mM MgSO4, and 13 mM KCl). The cells were fixed with glutaraldehyde (3% glutaraldehyde in cacodylate buffer) for 2 h, rinsed in washing buffer (5×5 min), treated with 2% tannic acid (Mallinckrodt Baker, Phillipsburg, KY, USA) in washing buffer for 60 min and washed again with washing buffer (5×5 min). After postfixation with osmium tetroxide (1% in washing buffer, 60 min), washing with doubly-distilled water (5×5 min), and treating with aqueous thiocarbohydrazid (1% in water, 15 min), the washing with doubly-distilled water and the fixation with osmium tetroxide were repeated. The samples were again washed with doubly-distilled water (7×5 min), treated with aqueous uranyl actetate (1%, 60 min) and finally washed with bi-distilled water (3×5 min). All preparation steps were carried out at room temperature. The specimens were dehydrated with graded concentrations of ethanol, transferred from 100% ethanol through a graded series into isoamyl acetate and finally critical-point dried in a Polaron apparatus CPD7501 (VG Microtech, East Sussex, UK). The glass slides with the bacteria were mounted onto stubs. After addition of conductive silver paint they were sputtered with 10 nm gold using a Polaron SC7640 sputter coater (VG Microtech) applying 1 kV and 13 mA. The samples were examined in a Zeiss DSM 940A scanning electron microscope (Oberkochen, Germany) at 7 kV. Pictures were made with the camera equipment of Point Electronic (Halle, Germany). Bacterial morphologies were surveyed at magnifications of 5,000× to 10,000×. Besides SEM microscopy, light microscopy at 400× magnification was also used for morphological examination.
Taxonomic classification of bacterial strains
The bacterial isolates were taxonomically affiliated on the basis of 16S rDNA sequence analysis. Extraction of genomic DNA, amplification of 16S rDNA using PCR, and purification of PCR products was performed as described by Rainey et al. (1996). Purified 16S rDNA was sequenced as recommended by the supplier using ABI PRISM Dye Terminator cycle sequencing ready reaction kits (Applied Biosystems, Weiterstadt, Germany). DNA fragments of the sequence reactions were electrophoretically separated and analyzed in an Applied Biosystems 373A DNA sequencer. Sequences were analyzed and compared with data from representative bacteria of the Cytophaga-Flexibacter-Bacteroides (CFB) group (isolate 11B) and the gamma-Proteobacteria group (isolates 1A and 21C) according to Maidak et al. (1999). The 16S rDNA nucleotide sequences of the bacterial isolates reported in this study have been submitted to the Genbank/EMBL data bank with the accession numbers AF542201 for 1A, AF542202 for 11B, and AF542200 for 21C.
Physiological fingerprinting
The ability of the strains to utilize a panel of carbon sources was tested with Biolog GN MicroPlates (Biolog, Hayward, CA, USA). Testing was performed as recommended by the manufacturer but conditions were adapted to marine bacteria (Tan 1997). Suspensions of cells were prepared with artificial seawater (pH 7.6) from well-developed colonies grown on ZoBell agar plates for 7–10 days at 4°C. Microplates filled with cell suspensions of the isolates 1A, 11B, and 21C were incubated at 5°C for 60 h. Carbon utilization was scored positive if the specific well turned to purple (reduction of the tetrazolium violet redox dye).
Temperature, pH, and salt tolerance of bacteria
Temperature characteristics of the bacteria were determined with fresh precultures spotted on ZoBell agar and incubated in the temperature range 0°–37°C. Salt and pH tolerance of the bacterial isolates were tested in 1 ml cultures with liquid ZoBell medium varying in pH from 4.6 to 8.5 and in the salt concentration from 0.25× to 2× sea water salinity (Truong et al. 2001). Sea water contains approximately 23 g NaCl/l as well as other salts and components. In this study 23 g NaCl/l is set as 1× NaCl sea water salinity. The different media were inoculated with 100 µl preculture and incubated at 5°C with slight shaking (30 rpm) for 48 and 96 h, if not indicated otherwise. Growth was determined by means of OD at 600 nm.
Transmission electron microscopy (TEM) of phages
Phage suspensions from fresh lysates (phage particle concentration 1010 ml−1) were passed through filters of 200 nm pore size (Schleicher and Schuell, Dassel, Germany). Fresh glow-discharged formvar-coated grids that had been stabilized with a thin carbon film were wetted with doubly-distilled water. Phages were adsorbed for 5 min. The samples were washed twice with 50 µl doubly-distilled water and negatively stained with 1% aqueous uranyl acetate for 20 s. The excess stain was blotted off, and the grids were air-dried. Observations and micrographs were made with a Zeiss EM 906 (Oberkochen, Germany) at 80 kV. Photomicrographs of phages were taken at magnifications of 60,000× to 100,000×. Morphological characteristics of phages were compiled from multiple photomicrographs.
Phage development at different temperatures
Temperature response of the phages was examined by plaque formation at seven different temperatures in the range from 0° to 25°C, according to Greer (1983).
Genomic DNA size of phages
Restriction enzyme digestion was used to characterize the genomic size of the phages. The phage DNA was extracted from 50 or 100 ml of lysed cultures. Phages were concentrated with PEG precipitation (Ausubel et al. 2001). The phages 1a and 11b were purified using DEAE cellulose chromatography as described by Ausubel et al. (2001). Phage 21c was concentrated directly by a PEG precipitation step. The phage DNAs were isolated by phenol extraction. DNA was stored in TE buffer at 4°C until further use.
Aliquots of purified DNA were digested for about 1 h with the restriction endonucleases BamH1, Dra1, EcoR1, HindIII, Xba1 (Roche Diagnostics, Penzberg, Germany) according to the manufacturer's instructions. The resulting fragments as well as the uncut DNA were separated on 0.4% or 0.8% agarose gels. The restriction patterns were used to determine the genomic size of the phages. Molecular weight standards were EcoR1-digested SPP1 phage DNA (marker 1), HindIII-digested lambda DNA (marker 2) and molecular weight marker VI (marker 3) from Roche Diagnostics.
Results
Isolation of psychrophilic phage-host-systems
About 50 different bacterial isolates from sea ice and melt ponds were used for the isolation of phages. Five bacterial isolates were sensitive to phage infection. Three isolates were used for further examinations. Each bacterium was named after the sampling station. Bacteriophages were named by using the initials of the host bacterium in lower-case letters. The first phage-host-system 1a/1A was isolated from the same sea-ice sample collected at 81°41.1′N and 10°21.4′E. The phage-host-system 11b/11B originated from two different melt-pond samples, the bacterial isolate 11B came from 80°55.0′N and 09°49.4′E, and the phage isolate 11b came from 81°16′N and 13°00′E. Phage and host bacterium 21c/21C were also isolated from two different samples. The bacterium 21C came from sea ice collected at 81°04′N and 10°02′E, and the phage isolate 21c originated from sea ice taken at 81°07′N and 05°03′E.
Characterization of the phage-sensitive bacteria
All host bacteria were Gram-negative. Examination of the 16S rDNA sequence of the host bacteria 1A, 11B, and 21C indicated highest identities to the species Shewanella frigidimarina LMG 19867 (96.0%), Flavobacterium hibernum (94.0%), and Colwellia psychrerythraea (98.4%), respectively.
The 16S rDNA sequence of isolate 1A revealed further high homologies to the following species: Shewanella livingstonis LMG 19866T (95.0%), S. frigidimarina ACAM 584 (95.0%), S. baltica (94.0%), and S. putrefaciens (94.0%).
More distantly related bacteria of isolate 11B are Flavobacterium johnsoniae (93.1%), F. hydatis (92.6%), F. columnare (92.4%), F. aquatile (92.1%), and F. flevense (91.4%). Furthermore, isolate 11B showed 99.6% identity to an unvalidated isolate called Cytophaga sp. strain BAL13 (Pinhassi et al. 1997).
Other closely related species to isolate 21C were identified as Colwellia psychrerythraea ATCC 27364T (97.4%) (Bowman et al. 1997), C. demingiae (97.4%), C. hornerae (96.0%), C. maris (95.8%), C. rossensis (94.6%), and C. psychrotropica (94.4%).
The relatively low 16S rDNA homology of 1A (96.0%) and 11B (94.0%) to their closest validated relatives suggests that these strains represent new species. More detailed taxonomic studies, however, will be necessary to decide whether 21C (98.4% 16S rDNA similarity) is a new species or an ecosubspecies of Colwellia psychrerythraea.
In addition to the genotypic affiliation, the strains were characterized according to their physiological phenotype. Biolog GN plates were used for metabolic fingerprinting to analyze the capability of the strains for carbon source utilization. Corresponding with the genotypic results, our strains differed considerably from their closest relatives in their carbon source utilization pattern.
The isolate 1A developed (but only in 10 out of 95 Biolog GN microplate wells) a purple color, indicating that the following compounds can be oxidized: alpha-d-glucose, methyl pyruvate (weak), acetic acid (weak), formic acid, d,l-lactic acid, glycyl-l-aspartatic acid, l-serine, inosine (weak), uridine (weak), and thymidine (weak). A markedly higher number of compounds were metabolized by the strain 11B: α-cyclodextrin (very weak), dextrin, glycogen, tween 40, tween 80 (weak), cellobiose (weak), d-galactose, gentiobiose, α-d-glucose, maltose, d-mannose (weak), methyl pyruvate (weak), mono-methyl succinate (weak), acetic acid (weak), d-gluconic acid, p-hydroxy-phenylacetic acid (weak), d,l-lactic acid (weak), propionic acid (weak), succinic acid, alaninamide (weak), l-alanine, l-alanyl glycine, l-asparagine, l-aspartic acid, l-glutamic acid, glycyl-l-aspartic acid, glycyl-l-glutamic acid, l-ornithine, l-proline, l-serine, l-threonine (weak), uridine (weak), glycerol (weak), glucose-1-phosphate (weak), glucose-6-phosphate (weak). In contrast, the isolate 21C was unable to oxidize any of the 95 substrates on the Biolog GN microplate, indicating a very poor metabolizing capability for the Biolog GN components at least under the test conditions applied.
The morphology of the host bacteria (Fig. 1) was examined by light microscopy and scanning electron microscopy (SEM). The host bacteria 1A and 21C were highly motile, whereas the thin rods of isolate 11B were non-motile. Rod lengths ranged from approximately 1 to 4 µm. The host bacteria differed in the color, smell, and visible autolysis of colonies on agar plates. In young liquid cultures, cells of isolate 1A aggregated frequently. Strain 11B possessed a distinctive aromatic smell. Autolysis of old colonies on agar plates was observed with the host strains 1A and 21C. Strain 11B was highly sensitive to lysozyme, indicated by a significant decrease of OD at 600 nm after addition of lysozyme (Serva, Heidelberg, Germany). The isolates 1A and 21C were not susceptible to lysozyme.
Classification of the bacteriophages
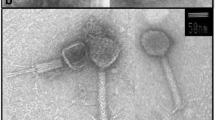
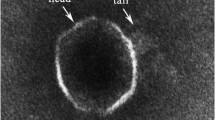
The bacteriophages were classified on the basis of their morphology examined with TEM at 60,000× or 100,000× magnifications (Fig. 2). Head and tail length were determined for the three phages. The data are compiled in Table 1. Head diameters ranged from 40 to 70 nm. Tail length ranged from 75 to 188 nm. The morphology of the phages 11b and 21c suggested an affiliation to the Siphovirida. Phage 1a, with a relatively large head and short contractive tail, is a typical representative of the Myovirus group.
The phage-typing assay confirmed that each phage could only infect its own host. No cross-infection of hosts was observed. The phages can therefore be regarded as host-specific. As expected, all phages failed to infect Escherichia coli K12.
All plaques showed a clear center surrounded by a turbid halo that progressed in the course of time. At the edges of plaques of 11b, tiny satellite plaques appeared.
Digestion with specific restriction enzymes revealed that the genomic nucleic acid of all three phages was DNA. All phages were double-stranded DNA viruses. The genomes of the phages showed distinct restriction patterns (Fig. 3). The higher intensity of selected bands is most probably due to the formation of more than one restriction fragment with the same molecular size and which therefore runs as an intensely staining band. All phage genomic DNAs could be digested with Dra1 and HindIII. The DNA of phage 21c could not be digested with Sal1. Genome sizes were calculated by the addition of estimated sizes of single fragment bands. For size estimation of the phage genome, the fragmentation patterns with Xho1 for phage 1a, EcoR1 for phage 11b, and Cla1 for phage 21c have been used. The biggest genome size, at about 70 kb, was found for isolate 1a. The smallest genome was phage 11b with about 30 kb. The genome size of isolate 21c is approximately 40–50 kb.
Phage genome analysis by electrophoretic separation of restriction fragments in a 0.8% agarose gel. The positions of molecular weight standards (kb) are indicated on both sides of the gel. Lane 1, marker 1; lane 2, 1a DNA uncut; lanes 3 through 6, 1a DNA cut with EcoR1, Dra1, HindIII, and Xba1, respectively; lane 7, 11b DNA uncut; lanes 8 through 11, 11b DNA cut with Dra1, EcoR1, HindIII, and Xba1, respectively; lane 12, 21c DNA uncut; lanes 13 through 15, 21c DNA cut with Cla1, Dra1, and HindIII, respectively; lane 16, marker 2; lane 1, marker 3. Molecular weight standards were EcoR1-digested SPP1 phage DNA (marker 1), HindIII-digested lambda DNA (marker 2) and molecular weight marker VI (marker 3) from Roche Diagnostics (Penzberg, Germany)
Bacterial growth characteristics
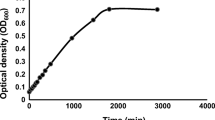
The host bacteria 1A, 11B, and 21C showed psychrophilic growth characteristics. The bacterial isolates were able to grow at 0°C (Table 2). All three strains developed visible colonies within 5–7 days at 0°C. The isolates 1A and 21C showed a significantly faster growth at 5°C than isolate 11B (Fig. 4). The specific growth rates (μ) of the bacterial isolates were about 0.09 h−1 for 1A and 21C and 0.06 h−1 for 11B. The maximal growth temperature of the strains 11B and 21C was at about 15°–20°C. The bacterial isolate 1A still grew weakly at 1°C above 20°C, which is the maximal temperature for psychrophiles according to the definition of Morita (1975). All three isolates were irreversibly inactivated after 2 days at 25°C. No viable bacteria were obtained from colonies that were incubated at 25°C for 2 days on agar dishes. Cell material exposed under these conditions was used to inoculate fresh medium at 5°C. This did not result in the development of a bacterial culture after 1 week.
The three bacterial host strains also showed differences in their pH adaptation. The strains 1A and 11B grew better at pH 4.6 than at pH 8.2. In contrast, the preferred pH of isolate 21C was 8.5. This strain showed poorer growth at a more acidic pH (range around pH 6.1).
Salinity tolerance was tested in the range of 0.25× to 2× sea-water salinity. The three strains could grow over the complete salt concentration range tested. The isolates 1A and 21C reached their highest final optical density at 0.5× sea-salt concentration. Isolate 11B showed the highest final optical density in the medium with a 0.25× sea-salt concentration. The lower salinity optimum of strain 11B corresponds well with the less saline conditions in the melt pond from which the strain originates. Furthermore, isolate 11B failed to grow in LB (5 g NaCl/l) medium.
Effect of temperature on phage development
The effect of temperature on phage growth was estimated by the visible development of phage plaques (Table 3). Phage 1a showed the biggest temperature range for plaque formation. Phage 1a formed plaques at 1A lawns in a temperature range of 0°–14°C. At 13°–14°C, the 1a plaques were smaller than plaques formed at 10°C. The most psychrophilic bacterial isolate, 21C, hosted the phage (21c) with the strongest dependency on low temperature conditions. Phage 21c did not form plaques on 21C lawns incubated at 10°C. The plaque size and number of phage 21c at 5°C was similar to those at 0°C.
Discussion
Phage-host systems have been described from diverse habitats including extreme environments such as the Dead Sea and ice-covered alpine as well as Antarctic lakes (Oren et al. 1997; Hofer and Sommaruga 2001; Kepner et al. 1998). It is supposed that bacteriophages are ubiquitous in permanently cold marine environments including sea ice. Maranger et al. (1994) revealed that the abundance of viruses in sea ice can be high. However, this group did not isolate viruses from the permanent cold environment sea ice. It has also been observed that the abundance of viruses changes during the season, indicating that viruses are dynamic components of the microbial sea-ice communities. Our knowledge about psychrophilic phages is very limited concerning their host specificity, their phenotypic and genotypic diversity, as well as their physiological adaptations and peculiarities. Our study describes the first isolation of cold-adapted phage-host systems from the permanently cold environments of Arctic sea ice and melt ponds. An isolation of psychrophilic bacteriophages from the marine environment (seawater and sediment samples) was reported by Olsen (1967). Greer (1983) described the isolation of cold-adapted phages from non-marine samples like refrigerated products, which lyse the psychrotrophic bacterium Brochothrix thermospacta.
Five different phage-host systems were found out of 50 different bacterial isolates, from which three phage-host systems were investigated in more detail in this study. We assume that the determination of further psychrophilic phage-host systems was hampered by the problematic establishment of the special marine environmental conditions under ordinary laboratory conditions. The high thermosensitivity of phages and bacteria from sea ice could be a reason, which significantly limited the identification of further phage-host systems. The two phages isolated in parallel to the three phages investigated in this study could not be cultured longer than approximately 4 weeks under the laboratory conditions used (data not shown).
Our investigation indicates that the identified indigenous sea-ice bacteria are associated with very specific bacteriophages. The phages identified in this study were host-specific, and no cross-infection of other bacterial isolates could be observed. In comparison with other cultured marine phages, the head diameter of the three isolated sea-ice phages was relatively small (Jiang et al. 1998), being 37–73 nm. Moreover, the sea-ice phage populations appear to be more diverse than those from sea water in the North Sea. The hosts of the three sea-ice phages identified in this study belong to three different genera in the gamma-Proteobacteria and CFB groups. 16S rDNA sequencing indicated that the psychrophilic phage-host bacteria may represent new species of the genera Shewanella, Flavobacterium, and Colwellia. In comparison, the bacterial hosts of the two North Sea studies with 22 and 31 marine phages, respectively, were members of the genera Pseudoaltermonas and Vibrio, respectively (Wichels et al. 1998, Moebus and Nattkemper 1983).
Phage development was clearly restricted to low temperatures. Furthermore, the growth experiments revealed a significant lower temperature maximum for phage development in comparison to the maximal growth temperature of the host bacterium. The highest thermosensitivity was shown by phage 21c, whose plaque formation could only be observed at 5°C but not at 10°C. The maximal temperature for optimal phage development was, in all three cases, approximately 10°C lower than the maximal growth temperature of the host bacterium. However, a gradually increasing plaque number at lower temperatures, as described by Greer (1983) for the psychrotolerant bacterium Brochothrix thermospacta, could not be observed with our phage-host systems (data not shown).
Although several studies have been undertaken to understand the diversity of microbial populations in sea ice, virtually no information exists on the isolation and cultivation of bacteriophages residing in sea-ice habitats. This study demonstrates that psychrophilic phage-host systems exist in sea ice and that they are highly dependent on low temperature conditions.
References
Ausubel FM, et al (2001) Current protocols in molecular biology. Wiley: New York
Bergh O, Boersheim KY, Bratbak G, Heldal M (1989) High abundance of viruses found in aquatic environments. Nature 6233:467–468
Bird DF, Maranger R, Karl DM (1993) Palmer LTER: aquatic virus abundances near the Antarctic Peninsula. Antarct J US 28:234–235
Bowman JP, McCammon SA, Brown MV, Nichols DS, McMeekin TA (1997) Diversity and association of psychrophilic bacteria in Antarctic sea ice. Appl Environ Microbiol 63:3068–3078
Bratbak G, Heldal M, Thingstad TF, Tuomi P (1996) Dynamics of virus abundance in coastal seawater. FEMS Microbiol Ecol 19:263–269
Brown MV, Bowman JP (2001) A molecular phylogenetic survey of sea-ice microbial communities (SIMCO). FEMS Microbiol Ecol 35:267–275
Greer G (1983) Psychrotrophic Brocothrix thermospacta bacteriophages isolated from beef. Appl Environ Microbiol 46:245–251
Grossi SM, Kottmeier ST, Sullivan CW (1984) Sea ice microbial communities. III. Seasonal abundance of microalgae and associated bacteria, McMurdo Sound, Antarctica. Microb Ecol 10:231–242
Grossmann S, Dieckmann G (1994) Bacterial standing stock, activity, and carbon production during formation and growth of sea ice in the Weddell Sea, Antarctica. Appl Environ Microbiol 60:2746–2753
Guixa-Boixereu N, Vaque D, Gaso JM, Sanchez-Camara J, Pedros-Alio C (2002) Viral distribution and activity in Antarctic waters. Deep-Sea Res 49:827–845
Helmke E, Weyland H (1995) Bacteria in sea ice and underlying water of the eastern Weddell Sea in midwinter. Mar Ecol Prog Ser 117:269–287
Hofer JS, Sommaruga R (2001) Seasonal dynamics of viruses in an alpine lake: importance of filamentous forms. Aquat Microb Ecol 26:1–11
Jiang SC, Kellogg CA, Paul JH (1998) Characterization of marine temperate phage-host systems isolated from Mamala Bay, Oahu, Hawaii. Appl Environ Microbiol 648:535–542
Kepner RL Jr, Wharton RA Jr, Suttle CA (1998) Viruses in Antarctic lakes. Limnol Oceanogr 43:1754–1761
Kottmeier ST; Sullivan CW (1987) Late winter primary production and bacterial production in sea ice and seawater west of the Antarctic Peninsula. Mar Ecol Prog Ser 36:287–298
Kottmeier ST, Grossi SM, Sullivan CW (1987) Sea ice microbial communities. VIII. Bacterial production in annual sea ice of McMurdo Sound, Antarctica. Mar Ecol Prog Ser 35:175–186
Maidak BL, et al (1999) A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res 27:171–173
Maranger R, Bird DF, Juniper SK (1994) Viral and bacterial dynamics in Arctic sea ice during the spring algal bloom near Resolute, NWT, Canada. Mar Ecol Prog Ser 111:121–127
Marchant H, Davidson A, Wright S, Glazebrook J (2000) The distribution and abundance of viruses in the Southern Ocean during spring. Antarct Sci 12:414–417
Moebus K (1991) Preliminary observations on the concentration of marine bacteriophages in the water around Helgoland. Helgol Wiss Meeresunters 45:411–422
Moebus K, Nattkemper H (1983) Taxonomic investigations of bacteriophage sensitive bacteria isolated from marine waters. Helgol Meeresunters 36:357–373
Morita RY (1975) Psychrophilic bacteria. Bact Rev 39:144–167
Noble RT, Fuhrman JA (2000) Rapid virus production and removal as measured with fluorescently labeled viruses as tracers. Appl Environ Microbiol 66:3790–3797
Olsen RH (1967) Isolation and growth of psychrophilic bacteriophage. Appl Microbiol 15:198
Oren A, Bratbak G, Heldal M (1997) Occurrence of virus-like particles in the Dead Sea. Extremophiles 1:143–149
Pinhassi J, Zweifel UL, Hagstrom A (1997) Dominant marine bacterioplankton species found among colony-forming bacteria. Appl Environ Microbiol 63:3359–3366
Priddle J, Leakey R, Archer S, Murphy E (1996) Eukaryotic microbiota in the surface waters and sea ice of the Southern Ocean: aspects of physiology, ecology and biodiversity in a 'two-phase' system. Biodivers Cons 5:1473–1504
Proctor LM, Fuhrman JA (1990) Viral mortality of marine cyanobacteria and bacteria. Nature 343:60–62
Rainey FA, Ward-Rainey N, Kroppenstedt RM, Stackebrandt E (1996) The genus Nocardiopsis represents a phylogenetically coherent taxon and a distinct actinomycete lineage: proposal of Nocardiopsaceae fam. nov. Int J Syst Bacteriol 46:1088–1092
Smith RE, Clement P (1990) Heterotrophic activity and bacterial productivity in assemblages of microbes from sea ice in the high Arctic. Polar Biol 10:351–357
Smith DC, Steward GF, Azam F, Hollibaugh JT (1992) Virus and bacteria abundance in the Drake Passage during January and August 1991. Antarct J US 27:125–127
Staley JT, Gosink JJ (1999) Poles apart: biodiversity and biogeography of sea ice bacteria. Annu Rev Microbiol 53:189–215
Steward FG, Smith DC, Azam F (1996) Abundance and production of bacteria and viruses in the Bering and Chukchi Seas. Mar Ecol Prog Ser 131:287–300
Suttle CA, Chan AM, Cottrell MT (1990) Infection of phytoplankton by viruses and reduction of primary productivity. Nature 347:467–469
Tan TL (1997) Biolog metabolic fingerprints for clustering marine oligotrophic bacteria from polar regions In: Insam H, Rangers A (eds) Microbial communities: function versus structural approaches. Springer, Berlin Heidelberg New York, pp 161–170
Thomas DN, Dieckmann GS (2002) Antarctic sea ice habitat for extremophiles. Science 5555:641–644
Truong LV, Tuyen H, Helmke E, Binh LT, Schweder T (2001) Cloning and characterization of two cold-adapted pectate lyases from the marine Antarctic bacterium Pseudoalteromonas haloplanktis strain ANT/505. Extremophiles 5:35–44
Vincent WF, Gibson JAE, Pienitz R, Villeneuve V (2000) Ice shelf microbial ecosystems in the high Arctic and implications for life on snowball earth. Naturwissenschaften 87:137–141
Wichels A, Biel SS, Gelderblom HR, Brinkhoff T, Muyzer G, Schütt C (1998) Bacteriophage diversity in the North Sea. Appl Environ Microbiol 64:4128–4133
Wommack KE, Colwell RR (2000) Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev 64:69–114
Acknowledgment
This work was supported by the Bundesministerium für Bildung und Forschung (03F0278B).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Horikoshi
Rights and permissions
About this article
Cite this article
Borriss, M., Helmke, E., Hanschke, R. et al. Isolation and characterization of marine psychrophilic phage-host systems from Arctic sea ice. Extremophiles 7, 377–384 (2003). https://doi.org/10.1007/s00792-003-0334-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-003-0334-7