Abstract
The corticotropin-releasing factor (CRF) and its CRF1 receptor (CRF1R) play a central role in the maintenance of homeostasis. Malfunctioning of the CRF/CRF1R unit is associated with several disorders, such as anxiety and depression. Non-peptide CRF1R-selective antagonists have been shown to exert anxiolytic and antidepressant effects on experimental animals. However, none of them is in clinical use today because of several side effects, thus demonstrating the need for the development of other more suitable CRF1R antagonists. In an effort to develop novel CRF1R antagonists we designed, synthesized and chemically characterized two tripeptide analogues of CRF, namely (R)-LMI and (S)-LMI, having their Leu either in R (or D) or in S (or L) configuration, respectively. Their design was based on the crystal structure of the N-extracellular domain (N-domain) of CRF1R/CRF complex, using a relevant array of computational methods. Experimental evaluation of the stability of synthetic peptides in human plasma has revealed that (R)-LMI is proteolytically more stable than (S)-LMI. Based on this finding, (R)-LMI was selected for pharmacological characterization. We have found that (R)-LMI is a CRF antagonist, inhibiting (1) the CRF-stimulated accumulation of cAMP in HEK 293 cells expressing the CRF1R, (2) the production of interleukins by adipocytes and (3) the proliferation rate of RAW 264.7 cells. (R)-LMI likely blocked agonist actions by interacting with the N-domain of CRF1R as suggested by data using a constitutively active chimera of CRF1R. We propose that (R)-LMI can be used as an optimal lead compound in the rational design of novel CRF antagonists.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The type 1 receptor (CRF1 receptor, or CRF1R) for corticotropin-releasing factor (CRF) mediates the effects of CRF in the regulation of the hypothalamic–pituitary–adrenal axis (Ramot et al. 2017; Chrousos 2009), as well as, the gastrointestinal (Martinez and Tache 2006), behavioral, immune (Slominski et al. 2013; Karalis et al. 1991; Dermitzaki et al. 2014; Tsatsanis et al. 2007), endocrine (Schmid et al. 2011; Huising et al. 2010) and central nervous (CNS) (Stengel et al. 2009; Deussing and Chen 2018; Dedic et al. 2018) systems, thus playing crucial role in the maintenance of homeostasis. An aberrant regulation of homeostatic mechanisms renders us vulnerable to various disorders, including anxiety and depression, with CRF1R being involved in these mechanisms. Indeed, the use of CRF1R-antisense oligonucleotides or knockout mice deficient of CRF1R suggested the involvement of CRF1R in the previously mentioned disorders (Ramot et al. 2017; Arborelius et al. 1999; Reul and Holsboer 2002; Muller et al. 2003; Timpl et al. 1998). It addition, non-peptide CRF1R-selective antagonists display antidepressant and anxiolytic properties (Zorrilla and Koob 2010; Fahmy et al. 2012). CRF1R-selective non-peptide antagonists have also been reported to reduce colonic transit time and defecation induced by various stressors or centrally injected CRF, and could be potentially used for the treatment of stress-induced metabolic syndrome manifestations and pituitary ACTH secreting tumors (Stengel et al. 2009; Martinez and Tache 2006; Grammatopoulos and Chrousos 2002).
CRF1R belongs to the family B of G protein-coupled receptors (GPCRs) (Grammatopoulos and Chrousos 2002; Liapakis et al. 2011). As all GPCRs, CRF1R is a plasma membrane protein, consisting of an amino-terminal extracellular region (N-domain), a carboxyl-terminal intracellular tail and seven, mostly hydrophobic, transmembrane domains (TMs), which are connected by alternating intracellular (ILs) and extracellular loops (ELs). The ELs and the upper parts of TMs form the J-domain of the receptor. CRF and related peptide agonists, such as sauvagine, interact with the N- and J-domains of CRF1R (Liapakis et al. 2011). In contrast, non-peptide, micromolecular, CRF1R-selective antagonists, such as antalarmin, bind deep in the binding site-crevice of CRF1R, which is formed by receptor TMs, and allosterically antagonize CRF-stimulated cAMP signaling (Hoare et al. 2003; Gkountelias et al. 2010). Although many allosteric non-peptide blockers exist, micromolecular orthosteric small molecule antagonists that bind to the extracellular parts of CRF1R have not yet been reported.
The interaction of CRF1R with CRF or its related peptide-agonists has been proposed to involve two steps (Hoare et al. 2004, 2008). In the first step, the C-terminal region of peptides interacts with the N-domain of the receptor. This “first-step” interaction orients the N-terminal region of peptides toward the J-domain of the receptor, resulting in the interaction (second-step) between each other. This “second-step” interaction is responsible for activation of CRF1R. N-terminally truncated CRF-related peptides, such as astressin or CRF(27–41), are antagonists, because they bind to the N-domain of the receptor in the “first-step” interaction, thus blocking agonist binding, but cannot proceed to the “second-step” as they lack the N-terminal region that binds to the J-domain and activates the receptor. Supporting evidence for the two-step model of ligand/receptor interaction is provided by a study, which removed the N-domain of CRF1R and tethered the first 16 N-terminal residues (signaling region) of CRF to the resting receptor (J-domain), thus creating the chimeric receptor, CRF(1–16)/R1ΔN (Nielsen et al. 2000). Such a tethering mimicked the “first-step” of CRF1R-peptide interaction, which placed the signaling region of CRF close to the J-domain of receptor, to interact with each other and activate the CRF1R. Thus, the CRF(1–16)/R1ΔN chimera is a constitutively active mutant, the activation of which was blocked by antalarmin but not by astressin (Nielsen et al. 2000).
A previous crystallization study of the N-domain of CRF1R/CRF complex has unveiled its three dimensional (3D) architecture, which includes a hydrophobic surface interacting with the peptide (Pioszak et al. 2008). In this study, CRF has been proposed to bind to CRF1R in such way so that its 26–41 C-terminal amino acids interact with the N-domain of the receptor and the 1–25 N-terminal residues of the peptide point towards the TMs of CRF1R. Among the C-terminal amino acids, Leu37, Met38 and Ile41 of CRF have been shown to play the major role in its interaction with the receptor (Pioszak et al. 2008). Structure–function relationship and NMR spectroscopy studies have also highlighted the functional importance of Leu37, Met38 and Ile41 of CRF and its related peptide analogues (Pioszak et al. 2008; Rijkers et al. 2004; Mesleh et al. 2007; Kornreich et al. 1992). Based on these studies, Leu37, Met38 and Ile41 most likely define a hot-spot binding epitope of CRF and its related peptides. On the basis of this epitope we rationally designed, synthesized and pharmacologically characterized for the first-time tripeptides that target the N-domain of CRF1R and antagonize CRF binding.
Materials and methods
Computational chemistry
The structures of the two diastereoisomers of tripeptide H3N+-Leu-Met-Ile-COO− (Leu adopts an R or S configuration, thus creating the (R)-LMI or (S)-LMI) were constructed on Maestro subroutine (Schrödinger Suite 2012 update 2). Protonation states of the residues were created using Epik software program (https://doi.org/10.1007/s10822-007-9133-z) at 7.4 pH and the peptides were energy-minimized using the Optimized Potential for Liquid Simulations (OPLS-2005) force field as it is implemented in MacroModel 9.0 software (Schrödinger Suite 2012 update 2) using water as a solvent. The compounds were subjected to 500 iterations by applying the Polak–Ribière conjugate gradient (PRCG) general method for energy minimization. Molecular Dynamics (MD) simulations were performed with the Dynamics panel of the subroutine Maestro software (Schrödinger Suite 2012 update 2). The number of structures to sample was set at 2000. The equations of motion were integrated with a time step equal to 1.5 fs while the average temperature was kept constant at 300 K, with equilibration and simulation times equal to 20 ns and 1000 ns, respectively. The structural clustering of the conformers was obtained using the conformer_cluster.py script available in the Schrödinger Suite. To generate the root-mean-square deviation (RMSD) of atomic positions matrix, all heavy atoms were considered. The average linkage method was used for clustering and the best number of clusters was 10.
The crystal structure of the extracellular domain of CRF1R (CSD reference code 3EHU (Pioszak et al. 2008)) was downloaded and sketched in the Maestro utility of the Schrödinger Suite 2012.2 (Schrödinger Suite 2012). (R)-LMI and (S)-LMI peptides were designed and optimized by the same utility. The optimized potentials for liquid simulations 3 (OPLS3) force field (Shivakumar et al. 2010) was used for the minimization of the structures. The disulfide bonds of the protein were created according to the crystal structure of the extracellular domain of CRF1R. GlideXP software (Friesner et al. 2004) was used for docking calculations. The docking site of CRF in CRF1R was considered as the center of the docking grid box surrounding residues Tyr99, Pro69 and Cys68/Cys102. The ten tripeptide conformers of the lowest energy underwent docking simulations. The Induced Fit Docking (IFD) protocol (Sherman et al. 2006) based on GlideXP and the software “Refinement module in Prime” aided the prediction of ligand binding modes and concomitant structural changes in the receptor. The following steps were used (as indicated from the induced fit docking (IFD) protocol): (i) Optional constrained minimization of the receptor (protein preparation, refinement only) with an RMSD cutoff of 0.18 Å. (ii) Initial GlideXP docking of each ligand using a softened potential (van der Waals radii scaling). A maximum of 20 poses per ligand were retained. (iii) Prime side-chain prediction for each protein/ligand complex, on residues within a 6 Å distance of any ligand pose. (iv) Glide redocking of each protein/ligand complex structure within 30 kcal/mol of the lowest energy structure.
MD simulations of receptor complexes were carried out with the PMEMD module of AMBER 12 simulation package (Case et al. 2005). The initial geometry of peptides was optimized with the HF/6-31G* basis set (Gaussian 09) (Frisch et al. 2009). The force field ff99SB was utilized to represent the behavior of complexes. The systems were solvated using the transferable intermolecular potential with 3 points (TIP3P) water model in a truncated octahedron (Jorgensen et al. 1983), with each protein atom being at least 10 Å away from the edges of the periodic box. The minimization of systems was carried out for 5000 steps, using a nonbonded cutoff of 10 Å. The solvated complexes were next heated under constant volume for 100 ps. The temperature was gradually increased from 0 to 315 K, and was controlled using a Langevin thermostat (Izaguirre et al. 2001); the collision frequency was set at 2 ps−1. During heating, restraints of 10 kcal mol−1 Å−2 were applied to receptor and peptides ((R)-LMI or (S)-LMI). Subsequently, the systems were equilibrated under constant pressure in two steps of 100 ps each. In the first step, constraints of 10 kcal mol−1 Å−2 were applied to the complexes, while in the second step restraints were removed. Finally, two independent MD calculations of 200 ns for (R)-LMI–CRF1R and (S)-LMI–CRF1R complexes were run at 315 K under constant pressure. The bonds involving hydrogen atoms were constrained at their equilibrium distance using the SHAKE algorithm and a 2 fs time step was used (Ryckaert et al. 1977). The ptraj module of AMBER software was used for the analysis of trajectories(Roe and Cheatham 2013).
The MM-PBSA (Molecular Mechanics Poisson-Boltzmann Surface Area) method was used for the calculation of the binding energy between receptor and peptides (Kollman et al. 2000; Gohlke et al. 2003; Wang and Kollman 2001). The calculation of enthalpy contribution (ΔH) was performed by processing 1000 evenly spaced snapshots of each simulation, while a sub-group of 100 snapshots, belonging to the last 20 ns of calculations, were processed for entropy estimation ( – ΤΔS). The entropy term was calculated by the NMODE module of AMBER. For each snapshot, the free energy of the system, the ligand and the receptor was calculated. The sum of the resulting enthalpy and entropy terms for the system yielded the total binding energy (ΔGbind).
Synthesis of peptides
Peptides 1 [(S)-LMI] and 2 [(R)-LMI] were synthesized following the fluorenylmethyloxycarbonyl chloride/tert-butyl (Fmoc/tBu) solid phase methodology utilizing the 2-chlorotrityl chloride (CLTR-Cl) resin (1.00 g) (Kordopati et al. 2015; Laimou et al. 2012; Tselios et al. 2014; Friligou et al. 2011, 2013; Ieronymaki et al. 2015). The first Nα-Fmoc-protected amino acid, Fmoc-Ile-OH (1 equiv), was coupled (esterified) to the resin in the presence of Diisopropylethylamine (DIEA, 4.5 equiv) in Dichloromethane (CH2Cl2, 14 mL) for 1 h at room temperature (RT). Subsequently, Methanol (CH3OH, 2 mL) and DIEA (1 mL) were added and the mixture was stirred for another 30 min at RT. The Fmoc-Ile-resin was successively filtered and washed with a mixture of CH2Cl2/CH3OH/DIEA (80:15:5, 3 × 20 mL 5 min), Dimethylformamide (DMF, 5 × 15 mL) and isopropanole (iPrOH, 3 × 15 mL). The remaining peptide chain was coupled with the appropriate amino acids (2.5 equiv) in the presence of Diisopropylcarbodiimide (DIC, 2.75 equiv) and 1-Hydroxybenzotriazole (HOBt, 3.75 equiv) in Dimethyacetamide (DMA) for 4–6 h followed by Fmoc deprotection with 3 × piperidine (20% in DMF) for 5, 15 and 10 min, respectively. The following Fmoc protected amino acids were used for the synthesis: Fmoc-Met-OH, Fmoc-Leu-OH and Fmoc-DLeu-OH. Completeness of each coupling and deprotection cycle was verified by the Kaiser test and the retention time and ninhydrin test of TLC [Acetonitrile (CH3CN)/H2O 2:1]. The synthesized protected peptide on the resin was dried under vacuum and then was cleaved with CH2Cl2/Trifluroethanole (TFE)—7:3 for 3 h at RT. The mixture was filtered, the solvent was removed on a rotary evaporator and the obtained oily product was precipitated by the addition of cold diethyl ether (Et2O) as an amorphous white solid.
The crude products were further purified by semi preparative reversed phase HPLC (RP-HPLC) (column: Nucleosil C-18, 7 μm, 250 × 10 mm, eluents: A: 0.08% TFA/H2O, B: 0.08% Trifluroacetic acid (TFA)/CH3CN, gradual gradient: from 10 to 70% B in 45 min, flow rate: 3 mL/min, detection: 230 nm, 254 nm, identification with ESI–MS) (Kordopati et al. 2015; Laimou et al. 2012; Tselios et al. 2014; Friligou et al. 2011, 2013 ; Ieronymaki et al. 2015). The purity of the final products were assessed by analytical RP-HPLC (column: Lichrosorb C18, 5 μm, 250 × 4.6 mm, eluents: A: 0.08% TFA/H2O, B: 0.08% TFA/CH3CN, gradual gradient: from 10 to 100% B in 30 min, flow rate: 1 mL/min, detection: 214 nm or 254 nm) and the purity was ~ 98%. The final products were identified by electrospray ionization mass spectrometry (ESI–MS).
The synthesis of NH3+-Aib-LMI-COO− peptide, which was used as internal standard (IS) for the liquid chromatography–tandem mass spectrometry (LC–MS/MS) assays was achieved using the general procedure described for the synthesis of studied peptides 1 and 2.
NMR Spectroscopy
Standard 1D (one-dimensional) and 2D (two-dimensional) homo- and hetero-nuclear NMR (Nuclear Magnetic Resonance) experiments were performed for the NMR characterization of the peptides in solution. The NMR samples were obtained by diluting the tripeptides in 90%/10% H2O/DMSO at pH 7, at a final concentration of 2–3 mM. 1D 1H NMR, 2D TOCSY (total correlation spectroscopy) and NOESY (Nuclear Overhauser Effect Spectroscopy) as well as 1H-15 N and 1H-13C natural abundance HSQC (Heteronuclear Single Quantum Coherence), were performed with 64 and 32 number of scans (NS), respectively, at 298 K with a Bruker Avance III HD 700 MHz spectrometer equipped with a 5 mm TCI cyroprobe. All the 1D 1H NMR spectra acquired using a spectral width (SW) of 13.49 ppm with suppression of the residual H2O signal using excitation sculpting water suppression pulse sequences and consisted of 2048 data points (TD) with 16 NS. The 2D HSQC spectra were acquired using a SW of 13.99 ppm for the F2 (1H) dimension and 40.00 ppm for the F1 (15 N) at the 1H-15 N natural abundance HSQC. The 1H-13C HSQC spectrum acquired with 9.99 ppm and 199.98 ppm for the F2 (1H) and F1 (13C) dimension, respectively. The TD for the F2 dimension was 2 k data points in both 1H-13C HSQC spectra and for the F1 128 and 256 for the 15 N and 13C respectively. 2D 1H-1H TOCSY experiments were carried out by using MLEV-17 spin-lock sequence with a mixing time of 80 ms, 32 NS and 12.02 ppm SW (spectral width) for both dimensions. The data points acquired in both samples were 2 k for the F2 dimension and 256 for the F1. Finally, for both tripeptides, a set of 2D NOESY spectra were carried out using three different mixing times, 150, 250 and 400 ms. For all these spectra, the SW was the same with that used for TOCSY, while each increment was acquired with 16 NS. The increments in F1 dimension was 512 and for the F2 also a total number of 2 k was used. Raw NMR data processed with the standard Bruker software (Topspin 3.2).
Mass spectrometry (MS)
The mass characterization of the two tripeptides along with IS were conducted on an EVOQ Elite ER (Bruker, Germany) triple quadrupole MS, operated in positive electrospray ionization mode (ESI) by direct infusion of the compounds at concentration of 500 ng mL−1 (data not shown).
Peptide stability assays
A. Stock, calibration and working solutions
Stock solutions of 1 mg mL−1 for the two tripeptides and IS were prepared by weighting and dissolving the appropriate amounts in DMSO. These solutions were further diluted with acetonitrile to prepare the working solutions of the two tripeptides (10–300 μM) and IS (140 μM). Calibration standards (0.05–1.5 μM) for the two tripeptides were prepared by spiking appropriate amounts of working solutions into the blank human plasma (plasma that does not contain the compounds under study or IS). The final working solutions (200 μM) of two tripeptides for the plasma stability assay were prepared by further dilution with acetonitrile but from different stock solutions. All solutions were stored at – 20 °C.
B. In vitro stability in human plasma
The in vitro human plasma stability of two tripeptides was conducted by LC–MS/MS in a thermoshaker set up at 37 °C in triplicates. In 80 μL of blank human plasma from healthy donors (a kind offer from Blood Donation Center of the University Hospital of Ioannina), 10 μL of the respective working solutions (200 μM) of two tripeptides were added separately, and incubated for 0, 1, 2, 4, 6, 12 and 24 h at 37 °C. Samples were incubated with 400 μL ice-cold acetonitrile for protein precipitation and subsequently 10 μL of IS working solution (140 μM) were added into the mixtures. Samples were vortexed-mixed and centrifuged at 10.000 rpm for 10 min. In a new vial containing 300 μL water (LC–MS grade), 100 μL of the supernatant were added, vortex-mixed and filtered with 0.2 μm Minisart RC 4 syringe filters (Sartorius). Samples were tranfered in LC–MS vials and 5 μL were injected into the LC–MS system. The concentration of compounds was calculated by standard curves that were prepared by plotting tripeptides versus IS areas against nominal concentration of the calibration standards. The plot of the percentage remaining of the parent compound against time was designed for the two tripeptides.
HEK 293 cell culture, transfection, and harvesting
Human embryonic kidney 293 (HEK 293) cells transiently or stably expressing the wild type (WT) CRF1R or transiently expressing the chimera CRF(1–16)/R1ΔN (Nielsen et al. 2000) were grown in Dulbecco’s modified Eagle’s medium/F-12 (1:1), (DMEM/F-12), containing 3.15 g/liter glucose and 10% bovine calf serum (BCS) at 37 °C and 5% CO2. For transient expression, 60-mm dishes of HEK 293 cells at 80–90% confluence were transfected with 2.5 μg of plasmid containing the cDNA encoding the WT (pcin4 plasmid) or CRF(1–16)/R1ΔN (pCI plasmid) using 9 μL of LipofectAMINE™ (Life Technologies, Inc.) and 1 ml of Opti-MEM™ (Life Technologies, Inc.). Twenty-four hours after transfection, the medium was replaced by DMEM/F-12 containing glucose and BCS and the cells were incubated for 24 h at 37 °C and 5% CO2. Stable transfected cells were seeded in 60 dishes and incubated in DMEM/F-12 containing glucose and BCS at 37 °C and 5% CO2 for 24 h. At the end of the incubation, both stably and transiently receptor-expressing cells were seeded in 96-well plates (pretreated with 0.1 mg/ml poly-L-lysine) and incubated in DMEM/F-12 containing glucose and BCS at 37 °C and 5% CO2 for 16 h before cAMP accumulation assays.
cAMP accumulation assays
HEK 293 cells, stably or transiently expressing WT or transiently expressing CRF(1–16)/R1ΔN were incubated in 96-well plates in 100 μL assay buffer (25 mM HEPES, pH 7.4, 2 mM choline, 288 mM sucrose, 0.9 mM CaCl2, 0.5 mM MgCl2, and 1 mM 3-isobutyl-1 methylxanthine). After 1-h incubation at 37 °C, more assay buffer with or without ligands was added to a total volume of 200 μl, and the incubation was continued for 30 min at 37 °C. Specifically, HEK 293 transiently expressing WT or CRF(1–16)/R1ΔN were incubated without or with 10 nM CRF, 1 μM antalarmin, or 1 μM (R)-LMI, whereas HEK 293 stably expressing WT were incubated with 10 nM CRF in the presence or absence of increasing concentrations of (R)-LMI. The latter experiments were performed to determine the half-maximal inhibitory concentration of (R)-LMI. At the end of the incubation, the assay buffer was removed. The cells were placed on ice and lysed with 3% trichloroacetic acid. Lysates were incubated on ice for 30–60 min and stored at – 20 °C. After 1–5 days, frozen lysates were thawed and centrifuged at 1800 g for 10 min at 4 °C and the supernatants were neutralized with 2 N NaOH. Quantification of cAMP in the neutralized supernatants was performed using a competitive binding assay as previously described (Spyridaki et al. 2014). In brief, supernatants were transferred to polypropylene mini-tubes (20 μl/tube) containing buffer A (100 mM Tris–HCl, pH 7.4, 100 mM NaCl, and 5 mM EDTA) with 1 to 1.5 nM [2,8-3H]cAMP (Perkin Elmer). Subsequently, cAMP-binding protein (~ 100 mg of crude bovine adrenal cortex extract in 500 mL of buffer A) was added to each tube. The cAMP-binding protein was prepared from bovine cortexes obtained from the local slaughterhouse according to the method of Liapakis et al. (Liapakis et al. 2000). After incubation on ice for 3 h, the mixtures were filtered through Whatman 934AH glass fiber filters, using a Brandel cell harvester. The filters were washed three times with 0.5 mL of ice-cold buffer C (120 mM NaCl and 10 mM Tris HCl, pH 7.4 at 4 °C), placed in 4 mL scintillation fluid and assessed for radioactivity in a beta counter (50% efficiency). The amount of cAMP in each sample (one tenth of a well) was determined by comparison with a standard curve of known concentrations of unlabeled cAMP (1–100 pmol/tube). The half-maximal inhibitory concentration (or antagonistic potency, -LogIC50) values were obtained by fitting the data to a one-site competition model using nonlinear regression analysis (Prism 4.0).
3T3L1 cells differentiation
3T3L1 pre-adipocytes (obtained from the American Type Culture Collection) were plated using a basal medium (DMEM, 4 mM L-Glutamine, 25 mM D-Glucose, 1 mM Sodium Pyruvate, 3.7 g/L NaHCO3, 50 units/ mL penicillin, and 50 µg/ mL streptomycin) supplemented with 10% NBS in a humidified atmosphere at 37 °C. The next day the media were changed to the differentiation media that composed of the basal medium supplemented with 10% FBS, 10 µg/ mL insulin, 1 µM DEX and 0.5 mM 3-Isobutyl-1-Methyl-xanthine (IBMX) (Sigma). Two days later, the media were changed to the basal medium supplemented with 10% FBS and 10 µg/ mL insulin. The media were replaced every 2 days for a period of 12 days.
CXCL1 measurement from 3T3L1 cells
Pre-adipocytes were cultured to 12-well plates at a density of 20,000 cells per well and forced to differentiate as described above. The fully differentiated adipocytes were exposed to CRF in the presence or absence of (R)-LMI and cell culture supernatants were collected and stored at − 80 °C until used for the determination of CXCL1 concentration by ELISA (DY453) from R&D. For normalization of the measurements, cells were harvested and sonicated for quantification of total cellular proteins as previously described.
Proliferation assays
The proliferation rate of RAW 264.7 cells was determined in the presence or absence of 1 µM (R)-LMI, or 1 µM astressin. Specifically, cells were plated in 96-well plates (Costar) at a concentration of 3000 cells/well, using DMEM-low glucose medium. After 1–3 days incubation of cells, MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) was added at a final concentration of 0.5 mg/ mL and the cells were incubated for 4 h at 37 °C. Mitochondria of live cells metabolize MTT to blue crystals. Crystals were dissolved by incubating cells with DMSO at room temperature for 20 min and determining the optical density in a Dynatech MicroElisa reader (Chantilly, VA) at the wavelength of 595 nm.
Results
Rational peptide design and synthesis
Eight possible structures of the tripeptide Leu-Met-Ile were in silico evaluated in molecular docking experiments at the extracellular part of CRF1R (Table 1), with the side chain of isoleucine adopting an S stereochemistry. The tripeptide with the most favorable binding score in these experiments was found to adopt the RSS stereochemistry. Using the RSS stereochemistry of the tripeptide, derivative compounds were produced by the software MUSE (Tripos 2011). 20,000 compounds were initially designed and then scored via TriposScore (Tripos 2011). Amongst them, only 2000 survived the filtration process. The selection was based on the spatial arrangement of the tripeptide and pharmacophore similarity. None of those provided better score than the RSS H3N+-Leu-Met-Ile-COO− tripeptide.
To probe the in silico findings, two diastereoisomers of the H3N+-Leu-Met-Ile-COO− tripeptide were synthesized, with Leu in its S or R configuration, thus creating the peptides, (S)-LMI and the (R)-LMI, respectively. The (S)-LMI has been synthesized as a reference compound, representing the natural tripeptide, using the Fmoc/tBu methodology and 2-chlorotrityl chloride (CLTR-Cl) resin.
The structural identification of the two diastereoisomers was achieved by using a combination of different NMR homo- and hetero-nuclear spectra, including two dimensional Nuclear Overhauser Effect Spectroscopy (2D NOESY). 2D NOESY was used for determining their low-energy average conformations. As it can be observed from the NMR experiments the spectra of the two diastereoisomers exhibit significant similarities in terms of chemical shifts, except the splittings for γCH2 Met and β CH2 of Leu (Fig. 1). NH chemical shifts are shifted downfield for the H3N+-R-Leu-Met-Ile-COO− (or (R)-LMI). 2D NOESY spectra revealed that the two molecules did not show medium- or long-distance Nuclear Overhauser Effects (NOEs) indicating absence of vicinal proximity between the side chains (spectra not shown), as expected for such short and mobile peptides.
Molecular modeling
Ten structural clusters were generated for each tripeptide in water using RMSD (root-mean-square deviation) as a criterion, and the lowest energy conformers from each cluster were used for docking experiments (utilizing the GlideXP software and the Induced Fit Docking (IFD) protocol). The RMSD values (Fig. 2) of the lowest energy docking poses in comparison to the lowest energy conformers derived from the Molecular Dynamics (MD) clusters ranged between 0.5 and 4.0 Å showing that docking results were in agreement with the experimental NOEs. The ten lowest energy MD conformers for each peptide are shown in Fig. 3 and a representative docking pose of the lowest energy conformers is shown in Fig. 4.
Molecular dynamics of peptide binding to the CRF1R. RMSD and RMSF values to depict the flexibility and stability of the receptor and peptides during the binding process. a All atom RMSD values versus trajectory time for CRF1R while docked with either (R)-LMI (CRF R-LMI) or (S)-LMI (CRF S-LMI) and peptides independently (R-LMI and S-LMI) b Root mean square fluctuations (RMSF) of the heavy atoms of CRF1R while docked with either (R)-LMI (CRF R-LMI) or (S)-LMI (CRF S-LMI). c Snapshot of the CRF1R/(R)-LMI system after 200 ns of simulation. d Root mean square fluctuations of tripeptides by residue name
Specifically, the RMSD plots from the MD experiments for the two peptide-receptor complexes in water (under NOE constraints) indicate that after the first 40 ns of the simulations, the MD runs eventually equilibrated and the receptor appeared significantly stable in both cases (Fig. 2a). Also, the independent structures of (R)-LMI and (S)-LMI ultimately are stabilized despite that (S)-LMI displayed a noticeable conformational change from 40 to 90 ns (Fig. 2a).
During the MD simulations of the CRF1R-peptide complexes, analysis of the hydrogen bonds (HBs) throughout the trajectories, revealed a lack of stable HBs between the peptides and the receptor (less than 10% occupancy), suggesting that peptide binding is mostly driven by non-polar interactions. Comparison of the binding energies of (R)-LMI and (S)-LMI receptor complexes (free energy calculations) using the Molecular Mechanics Poisson-Boltzmann Surface Area (MM-PBSA) method (Table 2) suggest that the binding of (R)-LMI peptide in the extracellular N-domain of CRF1R is more energetically favorable, than that of (S)-LMI. We also explored the flexibility of the docked peptides by calculating the root mean square fluctuations (RMSF) of the heavy atoms (C, N, O) (Fig. 2d). While methionine and isoleucine appear equally flexible in both peptides, leucine shows minimum RMSF in the R form, which seems practically stable. Snapshots of the systems toward the end of the simulation show that Leu is engulfed by the adjacent amino acids of CRF1R.
Evaluation of the stability of the synthetic peptide in human plasma
The quantification of the two tripeptides as also the evaluation of their stability in human plasma in a time dependent manner was accomplished by establishing an LC–MS/MS protocol. An internal standard (IS) was first constructed meeting the physicochemical characteristics of the two synthetic peptides. Specifically, we synthesized as IS the NH3+-Aib-(R)-Leu-Met-Ile-COO− peptide. Several trials were conducted to identify the optimal chromatographic conditions (mobile phase, column type, flow rate, etc.) for the highest possible resolution and the shortest run time. Next, tandem mass spectrometry was used for the detection of the tripeptides in positive electrospray ionization mass spectrometry (ESI–MS) since the precursor → product transition was optimum in this mode. The stability profile of the two tripeptides in human plasma after 24 h incubation at 37 °C is presented in Fig. 5. The degradation rate for (S)-LMI was significantly high since after 2 h incubation in human plasma, only approximately 14% from the initial concentration was still present. In contrast, (R)-LMI was profoundly more stable in human plasma, with 78% of the initial concentration still present after 24 h incubation.
Evaluation of the antagonistic properties of (R)-LMI
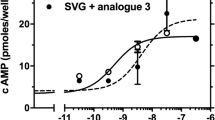
To evaluate the antagonistic properties of the (R)-LMI, we determined its ability to inhibit (1) the CRF-stimulated accumulation of cAMP in HEK 293 cells expressing the CRF1R, (2) the production of interleukins by adipocytes and (3) the proliferation rate of RAW 264.7 cells. We selected the (R)-LMI to determine its antagonistic properties, based on that it is proteolytically more stable than the (S)-LMI. In cAMP experiments, we determined the half-maximal inhibitory concentration (or antagonistic potency, -LogIC50) of (R)-LMI, which is the concentration of tripeptide required to reduce the CRF-stimulated cAMP accumulation by half. As shown in Fig. 6 the (R)-LMI inhibited CRF-stimulated cAMP accumulation in a dose–response manner, with an antagonistic potency (IC50) of = 1.17 μM (or – LogIC50 = 5.93 ± 0.21, n = 4).
Inhibition of CRF-induced cAMP accumulation by (R)-LMI. Stimulation of cAMP accumulation in HEK 293 cells stably expressing the CRF1R by 10 nM CRF in the absence or presence of increasing concentrations of (R)-LMI. The means and SE (duplicate determination) are shown from a representative experiment performed 4 times with similar results (n = 4)
To further determine the antagonistic properties of (R)-LMI we tested whether this peptide blocked the effects of CRF on the production of the interleukin CXCL1 from differentiated white adipocytes. 3T3L1 mouse pre-adipocytes were induced to differentiate to white adipocytes according to the method of Dermitzaki et al. (Dermitzaki et al. 2014). In these experiments, we used 10 nM CRF as previously described (Dermitzaki et al. 2014). Exposure of differentiated white adipocytes to 10 nM CRF for 24 h significantly reduced the production of CXCL1 compared to untreated (control) cells (Fig. 7). The CXCL1 levels after treatment of CRF-stimulated adipocytes with 5 μM (or 5 × 10−6 M) (R)-LMI were not statistically significant compared to controls. This suggests that (R)-LMI antagonized the effects of CRF on the production of CXCL1 from adipocytes (Fig. 7).
(R)-LMI antagonizes the effects of CRF on the production of CXCL1 from mouse mature adipocytes. Differentiated to white 3T3L1 mature adipocytes were exposed to 10 nM CRF in the presence or absence of 0.5 μΜ (5Χ10−7Μ) or 5 μΜ (5Χ10−6Μ) (R)-LMI for 24 h and the release of CXCL1 was determined using ELISA. Data are expressed as percentage change compared with control values (cells exposed only to vehicle). Bars indicate the mean ± SE values (n = 8) from 3 independent experiments. Bars with asterisk indicate that inhibition of CXCL1 release was significantly lower than control cells (*p < 0.05 depicts the statistical significant difference from control)
We also performed proliferation assays using RAW 264.7 monocyte/macrophage cells, which endogenously express CRF (Agelaki et al. 2002; Smith et al. 2006), to verify the antagonistic properties of (R)-LMI and compare the results of tripeptide with those obtained using astressin. As shown in Fig. 8, (R)-LMI at concentration of 1 μM significantly decreased the proliferation rate of RAW 264.7 cells the third day of culture, compared to vehicle-treated cells, at levels similar to those achieved with astressin at the same concentration.
(R)-LMI decreases the basal proliferation rate of RAW 264.7 cells. RAW 264.7 cells were exposed to 1000 nM (R)-LMI (R-LMI) or astressin and their proliferation was evaluated at 24, 48, and 72 h using the MTT method. Each point indicates the mean ± SE values from 4 wells of an independent experiment (n = 3 experiments). (*p < 0.05 depicts the statistical significant difference from control)
Experimental evaluation of the interaction of (R)-LMI with the N-domain of CRF1R
The design of (R)-LMI was based on a pharmacophore created from the interactions between the amino acids of CRF and residues of the N-domain of CRF1R. To experimentally evaluate the interaction between (R)-LMI and the N-domain of CRF1R, we determined the effects of the tripeptide on the functional properties of the CRF(1–16)/R1ΔN, which lacks the N-domain of CRF1R. In this chimeric receptor the first 16 N-terminal residues (signaling region) of CRF were tethered to the J-domain of CRF1R in place of its N-domain (Nielsen et al. 2000). Such a modification created a constitutively active receptor that was not able to be further stimulated by exogenously administered CRF. Stimulation of cAMP accumulation in the absence of agonists was threefold higher than that of WT CRF1R (control) and it was not further increased by 10 nM CRF (Fig. 9). As CRF, 1 μM (R)-LMI did not significantly alter the CRF(1–16)/R1ΔN-mediated constitutive stimulation of cAMP accumulation (Fig. 9). In marked contrast, the CRF(1–16)/R1ΔN-mediated constitutive stimulation of cAMP accumulation was significantly reduced by 1 μM antalarmin, which interacts with the transmembrane domains of the receptor.
(R)-LMI is unable to inhibit the elevated basal levels of cAMP in cells expressing the constitutive active CRF(1–16)/R1ΔN. Basal (solid bar), or ligand-modified intracellular levels of cAMP in HEK 293 cells transiently expressing the chimera of CRF1R, CRF(1–16)/R1ΔN, was compared relative to basal levels (value set at 100%) of wild type (WT) CRF1R (control, open bar). The ligands used in these experiments, were CRF (dotted bar), (R)-LMI (hatched bar on left), or antalarmin (ANTL), (hatched bar on right). The CRF(1–16)/R1ΔN has been created by replacing its N-domain of CRF1R by the first 16 N-terminal residues of CRF. Bars, indicate the mean ± SE values from 3 independent experiments. Bar with pound indicates that basal levels of cAMP were significantly higher than those of control, whereas bar with asterisk indicates that stimulation of cAMP by CRF(1–16)/R1ΔN in the presence of antalarmin was significantly lower than basal stimulation by CRF(1–16)/R1ΔN (*p and #p < 0.05 depict the statistical significant differences from chimera and control, respectively)
Discussion
In this study we designed, synthesized and pharmacologically characterized for first time two tripeptide CRF analogues targeting the N-domain of CRF1R. To achieve this aim, we have utilized an array of in silico tools to scrutinize a large number of molecules as potential binders to the extracellular N-domain of CRF1R. The molecular basis of our screening studies was based on the available X-ray 3D architecture of the N-domain of CRF1R in complex with the CRF (Pioszak et al. 2008). Among the amino acids of CRF, those in its carboxyl-terminal region (26–41) have been shown to interact with the N-domain of the receptor, with Leu37, Met38 and Ile41 of peptide being very important. Modification of these residues of CRF and its analogue CRF-(27–41) dramatically reduced peptide binding, supporting their important functional role (Pioszak et al. 2008; Rijkers et al. 2004). Similar results were obtained by Mesleh et al. who, using transferred NMR spectroscopy, characterized the binding of a 12-residue peptide antagonist (cCRF30–41) while bound to the isolated N-domain of the CRF1R. This study proposed that Met38 and Ile41 may play a crucial role in the binding of antagonists (Mesleh et al. 2007). The importance of Leu37, Met38, Ile41 of CRF in ligand-receptor interactions has also been suggested by an NMR study of the N-domain of CRF1R in complex with the CRF-related ligand, alpha helical cyclic CRF (Grace et al. 2010) and in a structure–function study, which showed that alanine substitution of these residues reduced or abolished the biological potency of CRF (Kornreich et al. 1992).
Guided from these findings, we have selected as the most potent binder the tripeptide H3N+-Leu-Met-Ile-COO−, the Leu, Met and Ile of which could mimic the architectural environment of the C-terminal Leu37, Met38 and Ile41 of CRF. Our in silico studies demonstrated that the configuration of the chiral center of Leu, D (or R) or L (or S), could alter the interaction potential of the tripeptide with the receptor. Thus, we have synthesized two tripeptides, the (S)-LMI (Leu in S configuration) and (R)-LMI (Leu in R configuration).
The structural identification and conformational properties of the two diastereoisomers, were carried out by NMR spectroscopy and molecular dynamics experiments. The NMR results indicated that the two molecules did not show medium or long-distance NOEs indicating absence of a structure in these short peptides in solution. The open form of peptide permits its hydrophobic side chains to suitably interact with hydrophobic amino acids of the receptor.
To determine the binding of (S)-LMI and (R)-LMI to CRF1R, we calculated their binding energy values in MM-PBSA free energy calculations (Table 2). We found that (R)-LMI bound to CRF1R with higher affinity than (S)-LMI. Specifically, MM-PBSA calculations illustrated that van der Waals interactions for (R)-LMI are more favorable ( – 35.62 kcal/mol) in comparison to (S)-LMI ( – 27.30 kcal/mol). The binding of tripeptides to CRF1R has been shown by our molecular modeling studies to be mediated through interactions involving Cys102, Tyr73 Tyr99, and Arg96 of the N-domain of CRF1R (Fig. 4). The impact of a simple alteration in the configuration (S–R) of a single chiral center of an amino acid (Leu) in the hot-spot of a peptide-protein (CRF-CRF1R) interaction interface on the binding potential is an interesting case to be reported. This could be due to the differential positioning of the tripeptides in the N-domain of CRF1R, associated with more enhanced van der Waals interactions of (R)-LMI versus (S)-LMI as estimated by MM-PBSA calculations (Table 2 and Fig. 4).
Substitution of L-Leucine of (S)-LMI by D-Leucine, thus creating the (R)-LMI, increased not only the binding of the peptide, but also its human plasma stability profile, abrogating its rapid enzymatic degradation, as revealed by the LC–MS/MS experiments. In addition, it is possible for this peptide to be stable from proteasomal degradation since a previous study suggested that the presence of D-amino acids at the N-terminus of a polypeptide abrogates its proteasomal degradation (Rabideau and Pentelute 2015). Based on these findings, (R)-LMI was selected for further pharmacological characterization.
(R)-LMI binding to the CRF1R most likely inhibits the first step interaction of C-terminal region of peptide-agonists, such as CRF and sauvagine, with the N-domain of receptor. This interaction has been proposed to orient the CRF peptides and all peptide-agonists for family B GPCRs such as their N-terminal residues (signaling region) to interact with the J-domain of receptors and activate them (Liapakis et al. 2017). Peptides, such as the CRF-(27–41) or the larger astressin, lacking the N-terminal signaling region are antagonists because they bind to the N-domain of the receptor in the first step of receptor–ligand interaction, thus blocking agonist binding, but cannot proceed to the second step as they are not able to interact with the J-domain and activate the receptor (Liapakis et al. 2017). Similarly, the small tripeptide, (R)-LMI, is expected to be a CRF antagonist by binding to the N-domain of receptor and blocking agonist binding, without however activating the receptor since it lacks the N-terminal signaling region of CRF. Indeed, (R)-LMI inhibited CRF-stimulated accumulation of cAMP in HEK 293 cells stably expressing the full length CRF1R in a dose–response manner, suggesting that it is a CRF antagonist.
Moreover, (R)-LMI was able to inhibit the CRF-stimulated production of interleukins by adipocytes. Specifically, (R)-LMI decreased the effect of CRF on CXCL1 production from mouse 3T3L1 in vitro differentiated white adipocytes, which has been shown to express CRF1R (Dermitzaki et al. 2014). CXCL1 is a chemokine produced from mouse cells in pro-inflammatory situations to support the establishment of inflammation. Similarly, the CRF1R-selective antagonist, antalarmin has been recently shown to block the effects of CRF on adipocytes function (Dermitzaki et al. 2014). To further verify the antagonistic properties of (R)-LMI, we determined its ability to block the effects of endogenous CRF on cell proliferation in RAW 264.7 monocyte/macrophage cells. (R)-LMI significantly decreased the basal proliferation rate of RAW 264.7 cells at similar levels with astressin. RAW 264.7 cells have been shown to express endogenous CRF and to respond to CRF1R antagonists suggesting that they express functional CRF1R receptors (Agelaki et al. 2002; Smith et al. 2006).
Supportive evidence for the interaction of (R)-LMI with the N-domain of CRF1R is provided by results from experiments using the CRF(1–16)/R1ΔN chimera. This chimera is a constitutive active receptor which has been created by excising the N-domain of CRF1R and tethering the first 16 N-terminal residues (signaling region) of CRF to the remaining portion (J-domain) of receptor (Nielsen et al. 2000). Such tethering mimics the first step of receptor–ligand interaction and brings the signaling region of CRF in close proximity to the J-domain of CRF1R, to interact with each other and activate the receptor. This chimera cannot be stimulated by CRF, since it lacks the N-domain of wild type CRF1R, in agreement with a previous study (Nielsen et al. 2000). Similarly, the constitutive activity of this chimera was not blocked by astressin which binds to the N-domain of wild type CRF1R (Nielsen et al. 2000). In contrast the constitutive activity of this construct was blocked by the non-peptide CRF antagonist, antalarmin, in agreement with a previous study (Nielsen et al. 2000). Antalarmin has been shown to bind to the TMs of CRF1R and block receptor activation (Spyridaki et al. 2014). Similar to astressin and in contrast to antalarmin, the (R)-LMI was not able to block the constitutive activity of the CRF(1–16)/R1ΔN chimera, suggesting its interaction with the N-domain of CRF1R.
Conclusively, we designed, synthesized and pharmacologically characterized the proteolytically stable tripeptide (R)-LMI, which blocks the ability of CRF to activate the receptor. The effects of (R)-LMI appear to be mediated through its interaction with the N-domain of CRF1R, in contrast to allosteric non-peptide antagonists, which block CRF effects by interacting with TM residues that are located deep in the binding site-crevice of CRF1R. These findings hold significant potential for clinical applications setting the basis for the development of novel non-peptide orthosteric antagonists of CRF, which could be potentially used for the treatment of several CRF-related diseases.
References
Agelaki S, Tsatsanis C, Gravanis A, Margioris AN (2002) Corticotropin-releasing hormone augments proinflammatory cytokine production from macrophages in vitro and in lipopolysaccharide-induced endotoxin shock in mice. Infect Immun 70(11):6068–6074
Arborelius L, Owens MJ, Plotsky PM, Nemeroff CB (1999) The role of corticotropin-releasing factor in depression and anxiety disorders. J Endocrinol 160(1):1–12
Case DA, Cheatham TE 3rd, Darden T, Gohlke H, Luo R, Merz KM Jr, Onufriev A, Simmerling C, Wang B, Woods RJ (2005) The Amber biomolecular simulation programs. J Comput Chem 26(16):1668–1688. https://doi.org/10.1002/jcc.20290
Chrousos GP (2009) Stress and disorders of the stress system. Nat Rev Endocrinol 5(7):374–381. https://doi.org/10.1038/nrendo.2009.106nrendo.2009.106
Dedic N, Chen A, Deussing JM (2018) The CRF family of neuropeptides and their receptors - mediators of the central stress response. Curr Mol Pharmacol 11(1):4–31. https://doi.org/10.2174/1874467210666170302104053
Dermitzaki E, Liapakis G, Androulidaki A, Venihaki M, Melissas J, Tsatsanis C, Margioris AN (2014) Corticotrophin-releasing factor (CRF) and the urocortins are potent regulators of the inflammatory phenotype of human and mouse white adipocytes and the differentiation of mouse 3T3L1 pre-adipocytes. PLoS ONE 9(5):e97060. https://doi.org/10.1371/journal.pone.0097060PONE-D-13-31684
Deussing JM, Chen A (2018) The corticotropin-releasing factor family: physiology of the stress response. Physiol Rev 98(4):2225–2286. https://doi.org/10.1152/physrev.00042.2017
Fahmy H, Spyridaki K, Kuppast B, Liapakis G (2012) The "homeostasis hormone" and its CRF(1) receptor. From structure function. Hormones 11(3):254–271
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47(7):1739–1749. https://doi.org/10.1021/jm0306430
Friligou I, Papadimitriou E, Gatos D, Matsoukas J, Tselios T (2011) Microwave-assisted solid-phase peptide synthesis of the 60–110 domain of human pleiotrophin on 2-chlorotrityl resin. Amino Acids 40(5):1431–1440. https://doi.org/10.1007/s00726-010-0753-6
Friligou I, Rizzolo F, Nuti F, Tselios T, Evangelidou M, Emmanouil M, Karamita M, Matsoukas J, Chelli M, Rovero P, Papini AM (2013) Divergent and convergent synthesis of polymannosylated dibranched antigenic peptide of the immunodominant epitope MBP(83–99). Bioorg Med Chem 21(21):6718–6725. https://doi.org/10.1016/j.bmc.2013.08.008
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JA Jr, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam JM, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas Ö, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ (2009) Gaussian Inc. Wallingford CT, Wallingford, CT
Gkountelias K, Papadokostaki M, Javitch JA, Liapakis G (2010) Exploring the binding site crevice of a family B G protein-coupled receptor, the type 1 corticotropin releasing factor receptor. Mol Pharmacol 78(4):785–793
Gohlke H, Kiel C, Case DA (2003) Insights into protein-protein binding by binding free energy calculation and free energy decomposition for the Ras-Raf and Ras-RalGDS complexes. J Mol Biol 330(4):891–913
Grace CR, Perrin MH, Gulyas J, Rivier JE, Vale WW, Riek R (2010) NMR structure of the first extracellular domain of corticotropin-releasing factor receptor 1 (ECD1-CRF-R1) complexed with a high affinity agonist. J Biol Chem 285(49):38580–38589. https://doi.org/10.1074/jbc.M110.121897M110.121897
Grammatopoulos DK, Chrousos GP (2002) Functional characteristics of CRH receptors and potential clinical applications of CRH-receptor antagonists. Trends Endocrinol Metab 13(10):436–444
Hoare SR, Fleck BA, Gross RS, Crowe PD, Williams JP, Grigoriadis DE (2008) Allosteric ligands for the corticotropin releasing factor type 1 receptor modulate conformational states involved in receptor activation. Mol Pharmacol 73(5):1371–1380
Hoare SR, Sullivan SK, Ling N, Crowe PD, Grigoriadis DE (2003) Mechanism of corticotropin-releasing factor type I receptor regulation by nonpeptide antagonists. Mol Pharmacol 63(3):751–765
Hoare SR, Sullivan SK, Schwarz DA, Ling N, Vale WW, Crowe PD, Grigoriadis DE (2004) Ligand affinity for amino-terminal and juxtamembrane domains of the corticotropin releasing factor type I receptor: regulation by G-protein and nonpeptide antagonists. Biochemistry 43(13):3996–4011
Huising MO, van der Meulen T, Vaughan JM, Matsumoto M, Donaldson CJ, Park H, Billestrup N, Vale WW (2010) CRFR1 is expressed on pancreatic beta cells, promotes beta cell proliferation, and potentiates insulin secretion in a glucose-dependent manner. Proc Natl Acad Sci U S A 107(2):912–917. https://doi.org/10.1073/pnas.0913610107
Ieronymaki M, Androutsou ME, Pantelia A, Friligou I, Crisp M, High K, Penkman K, Gatos D, Tselios T (2015) Use of the 2-chlorotrityl chloride resin for microwave-assisted solid phase peptide synthesis. Biopolymers 104(5):506–514. https://doi.org/10.1002/bip.22710
Izaguirre JA, Catarello DP, Wozniak JM, Skeel RD (2001) Langevin stabilization of molecular dynamics. J Chem Phys 114:2090–2098
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935
Karalis K, Sano H, Redwine J, Listwak S, Wilder RL, Chrousos GP (1991) Autocrine or paracrine inflammatory actions of corticotropin-releasing hormone in vivo. Science 254(5030):421–423
Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE 3rd (2000) Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res 33(12):889–897
Kordopati GG, Tselios TV, Kellici T, Merzel F, Mavromoustakos T, Grdadolnik SG, Tsivgoulis GM (2015) A novel synthetic luteinizing hormone-releasing hormone (LHRH) analogue coupled with modiied beta-cyclodextrin: insight into its intramolecular interactions. Biochim Biophys Acta 1850(1):159–168
Kornreich WD, Galyean R, Hernandez JF, Craig AG, Donaldson CJ, Yamamoto G, Rivier C, Vale W, Rivier J (1992) Alanine series of ovine corticotropin releasing factor (oCRF): a structure-activity relationship study. J Med Chem 35(10):1870–1876
Laimou D, Katsila T, Matsoukas J, Schally A, Gkountelias K, Liapakis G, Tamvakopoulos C, Tselios T (2012) Rationally designed cyclic analogues of luteinizing hormone-releasing hormone: enhanced enzymatic stability and biological properties. Eur J Med Chem 58:237–247. https://doi.org/10.1016/j.ejmech.2012.09.043,S0223-5234(12)00595-8
Liapakis G, Ballesteros JA, Papachristou S, Chan WC, Chen X, Javitch JA (2000) The forgotten serine A critical role for Ser-2035.42 in ligand binding to and activation of the beta 2-adrenergic receptor. J Biol Chem 275(48):37779–37788
Liapakis G, Matsoukas MT, Karageorgos V, Venihaki M, Mavromoustakos T (2017) Family B G protein-coupled receptors and their ligands: From structure to function. Curr Med Chem 24(31):3323–3355. https://doi.org/10.2174/0929867324666170303162416
Liapakis G, Venihaki M, Margioris A, Grigoriadis D, Gkountelias K (2011) Members of CRF family and their receptors: from past to future. Curr Med Chem 18(17):2583–2600
Martinez V, Tache Y (2006) CRF1 receptors as a therapeutic target for irritable bowel syndrome. Curr Pharm Des 12(31):4071–4088
Mesleh MF, Shirley WA, Heise CE, Ling N, Maki RA, Laura RP (2007) NMR structural characterization of a minimal peptide antagonist bound to the extracellular domain of the corticotropin-releasing factor1 receptor. J Biol Chem 282(9):6338–6346
Muller MB, Zimmermann S, Sillaber I, Hagemeyer TP, Deussing JM, Timpl P, Kormann MS, Droste SK, Kuhn R, Reul JM, Holsboer F, Wurst W (2003) Limbic corticotropin-releasing hormone receptor 1 mediates anxiety-related behavior and hormonal adaptation to stress. Nat Neurosci 6(10):1100–1107
Nielsen SM, Nielsen LZ, Hjorth SA, Perrin MH, Vale WW (2000) Constitutive activation of tethered-peptide/corticotropin-releasing factor receptor chimeras. Proc Natl Acad Sci U S A 97(18):10277–10281
Pioszak AA, Parker NR, Suino-Powell K, Xu HE (2008) Molecular recognition of corticotropin-releasing factor by its G-protein-coupled receptor CRFR1. J Biol Chem 283(47):32900–32912
Rabideau AE, Pentelute BL (2015) A d-amino acid at the N-Terminus of a protein abrogates its degradation by the N-end rule pathway. ACS Central Sci 1(8):423–430. https://doi.org/10.1021/acscentsci.5b00308
Ramot A, Jiang Z, Tian JB, Nahum T, Kuperman Y, Justice N, Chen A (2017) Hypothalamic CRFR1 is essential for HPA axis regulation following chronic stress. Nat Neurosci 20(3):385–388. https://doi.org/10.1038/nn.4491
Reul JM, Holsboer F (2002) Corticotropin-releasing factor receptors 1 and 2 in anxiety and depression. Curr Opin Pharmacol 2(1):23–33
Rijkers DT, Kruijtzer JA, van Oostenbrugge M, Ronken E, den Hartog JA, Liskamp RM (2004) Structure-activity studies on the corticotropin releasing factor antagonist astressin, leading to a minimal sequence necessary for antagonistic activity. ChemBioChem 5(3):340–348
Roe DR, Cheatham TE (2013) PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput 9(7):3084–3095. https://doi.org/10.1021/ct400341p
Ryckaert J-P, Ciccotti G, Berendsen HJC (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23(3):327–341. https://doi.org/10.1016/0021-9991(77)90098-5
Schmid J, Ludwig B, Schally AV, Steffen A, Ziegler CG, Block NL, Koutmani Y, Brendel MD, Karalis KP, Simeonovic CJ, Licinio J, Ehrhart-Bornstein M, Bornstein SR (2011) Modulation of pancreatic islets-stress axis by hypothalamic releasing hormones and 11beta-hydroxysteroid dehydrogenase. Proc Natl Acad Sci U S A 108(33):13722–13727. https://doi.org/10.1073/pnas.1110965108
Schrödinger Release 2012-2 (2012) LLC, New York, NY
Sherman W, Day T, Jacobson MP, Friesner RA, Farid R (2006) Novel procedure for modeling ligand/receptor induced fit effects. J Med Chem 49(2):534–553. https://doi.org/10.1021/jm050540c
Shivakumar D, Williams J, Wu Y, Damm W, Shelley J, Sherman W (2010) Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J Chem Theory Comput 6(5):1509–1519. https://doi.org/10.1021/ct900587b
Slominski AT, Zmijewski MA, Zbytek B, Tobin DJ, Theoharides TC, Rivier J (2013) Key role of CRF in the skin stress response system. Endocr Rev 34(6):827–884. https://doi.org/10.1210/er.2012-1092er.2012-1092
Smith EM, Gregg M, Hashemi F, Schott L, Hughes TK (2006) Corticotropin releasing factor (CRF) activation of NF-kappaB-directed transcription in leukocytes. Cell Mol Neurobiol 26(4–6):1021–1036. https://doi.org/10.1007/s10571-006-9040-1
Spyridaki K, Matsoukas MT, Cordomi A, Gkountelias K, Papadokostaki M, Mavromoustakos T, Logothetis DE, Margioris AN, Pardo L, Liapakis G (2014) Structural-functional analysis of the third transmembrane domain of the corticotropin-releasing factor type 1 receptor: role in activation and allosteric antagonism. J Biol Chem 289(27):18966–18977. https://doi.org/10.1074/jbc.M113.544460jbc.M113.544460
Stengel A, Goebel M, Million M, Stenzel-Poore MP, Kobelt P, Monnikes H, Tache Y, Wang L (2009) Corticotropin-releasing factor-overexpressing mice exhibit reduced neuronal activation in the arcuate nucleus and food intake in response to fasting. Endocrinology 150(1):153–160. https://doi.org/10.1210/en.2008-0723
Timpl P, Spanagel R, Sillaber I, Kresse A, Reul JM, Stalla GK, Blanquet V, Steckler T, Holsboer F, Wurst W (1998) Impaired stress response and reduced anxiety in mice lacking a functional corticotropin-releasing hormone receptor 1. Nat Genet 19(2):162–166
Tripos (2011) A New, Powerful software tool for molecular invention. 2.1 edn., St Louis, Missouri, USA
Tsatsanis C, Androulidaki A, Dermitzaki E, Gravanis A, Margioris AN (2007) Corticotropin releasing factor receptor 1 (CRF1) and CRF2 agonists exert an anti-inflammatory effect during the early phase of inflammation suppressing LPS-induced TNF-alpha release from macrophages via induction of COX-2 and PGE2. J Cell Physiol 210(3):774–783
Tselios T, Aggelidakis M, Tapeinou A, Tseveleki V, Kanistras I, Gatos D, Matsoukas J (2014) Rational design and synthesis of altered peptide ligands based on human myelin oligodendrocyte glycoprotein 35–55 epitope: inhibition of chronic experimental autoimmune encephalomyelitis in mice. Molecules 19(11):17968–17984. https://doi.org/10.3390/molecules191117968
Wang W, Kollman PA (2001) Computational study of protein specificity: the molecular basis of HIV-1 protease drug resistance. Proc Natl Acad Sci U S A 98(26):14937–14942. https://doi.org/10.1073/pnas.251265598
Zorrilla EP, Koob GF (2010) Progress in corticotropin-releasing factor-1 antagonist development. Drug Discov Today 15(9–10):371–383. https://doi.org/10.1016/j.drudis.2010.02.011S1359-6446(10)00074-7
Acknowledgements
This work was supported by ELKE grant K.A. 4373 (George Liapakis) and ELKE grant K.A. 3330 (Andrew Margioris). EU FP7 REGPOT CT-2011-285950 – “SEE-DRUG” project is acknowledged for financial support (Konstantinos D. Marousis, Georgios A. Spyroulias) as well as for the purchase of UPAT’s 700 MHz NMR equipment. We are grateful to Dr. Marilyn Perrin (for donating the pCI plasmid containing the cDNA encoding the chimera of CRF1R, CRF(1–16)/R1ΔN), George Chrousos (for donating antalarmin) and the blood Donation Center of the University Hospital of Ioannina (for donating human plasma from healthy donors).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Research involving human participants and/or animals informed consent
This research study does not involve human participants and/or animals and informed consent is not needed.
Additional information
Handling editor: T. Langer.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liapakis, G., Karageorgos, V., Andreadelis, I. et al. Discovery of a stable tripeptide targeting the N-domain of CRF1 receptor. Amino Acids 52, 1337–1351 (2020). https://doi.org/10.1007/s00726-020-02895-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-020-02895-4