Abstract
The SpAHA1 gene, encoding a plasma membrane (PM) H+-ATPase (AHA) in Sesuvium portulacastrum, was transformed into Arabidopsis plants, and its expression increased salinity tolerance of transgenic Arabidopsis plants: seed germination ratio, root growth, and biomass of transgenic plants were greater compared to wild-type plants under NaCl treatment condition. Upon salinity stress, both Na+ and H+ effluxes in the roots of SpAHA1 expressing plants were faster than those of untransformed plants. Transformed plants with SpAHA1 had lower Na+ and higher K+ contents relative to wild-type plants when treated with NaCl, resulting in greater K+/Na+ ratio in transgenic plants than in wild-type plants under salt stress. Extent of oxidative stress increased in both transgenic and wild-type plants exposed to salinity stress, but overexpression of SpAHA1 could alleviate the accumulation of hydrogen peroxide (H2O2) induced by NaCl treatment in transgenic plants relative to wild-type plants; the content of malondialdehyde (MDA) was lower in transgenic plants than that in wild-type plants under salinity stress. These results suggest that the higher H+-pumping activity generated by SpAHA1 improved the growth of transgenic plants via regulating ion and reactive oxygen species (ROS) homeostasis in plant cells under salinity stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soil salinity is a major environmental stress affecting crop growth worldwide. It is estimated that an area of 950 million ha of arable land throughout the world, including 250 million ha of irrigated land, is affected by salinity (Ruan et al. 2010; Bose et al. 2015). So, soil salinity presents an increasing threat to plant agriculture. Excessive sodium (Na+) in the soil can enter plant cells, where it is toxic to important physiological and biochemical processes; thus, the growth of plants under salt stress is involved in their ability to remove Na+ ions from cells (Zhou et al. 2015). Plasma membrane (PM) H+-ATPases (AHAs) are the primary active transporters that pump protons out of cell, providing proton motive force that drives Na+ extrusion mediated by PM Na+/H+ antiporter. So, the PM H+-ATPase is very important for energizing sodium extrusion and involved in salt tolerance in plants.
The PM H+-ATPase is encoded by a large gene family (Gaxiola et al. 2007; Fuglsang et al. 2010). Bioinformatics analyses of Arabidopsis and rice genome sequences showed 11 and 10 PM H+-ATPases in them, respectively (Axelsen and Palmgren 2001; Arango et al. 2003). The PM H+-ATPase generates an electrochemical potential gradient across the membrane through hydrolyzing ATP, which provides energy for metabolic processors in plants. In plant cells, nitrate transporter (NRT) is a NO3−/H+ symporter, which localizes at the plasma membrane, mediates NO3− uptake using the proton motive force generated by PM H+-ATPase (Santi et al. 2003); stomatal aperture involves regulation of osmotic pressure within guard cells, which is powered by H+-ATPase (Gaxiola et al. 2007); the PM H+-ATPase is involved in auxin-mediated cell elongation (Rober-Kleber et al. 2003); so, the PM H+-ATPase is necessary for nutrient uptake, photosynthesis, and cell growth. Besides regulation of many physiological processes, the PM H+-ATPases have a critical role in plant adaptation to salt stress conditions. The expression of three Arabidopsis AHA genes (AtAHA1, AtAHA2, and AtAHA3) was induced by NaCl treatment (Bose et al. 2015). The transcript level of PM H+-ATPase gene in salt-tolerant poplar was higher versus a salt-sensitive poplar (Ding et al. 2010). PM H+-ATPase mRNA abundance was greater in halophytes relative to glycophytes (Niu et al. 1993; Sahu and Shaw 2009). In contrast, salt stress induced AHA expression in all tested plants, but transcript levels of AHA genes were much lower in halophytes (quinoa) compared with glycophyte (Arabidopsis) (Bose et al. 2015). Besides upregulation of PM H+-ATPase gene, salinity induced its protein biosynthesis in some plants (Chen et al. 2010; Lopez-Pérez et al. 2009; Palmgren 2001). Salt treatment activated PM H+-ATPase pump activity in Thellungiella halophila (Vera-Estrella et al. 2005). Salt-tolerant rice had higher H+-ATPase activity than sensitive one (Pons et al. 2011). PM H+-ATPase activity was higher in halophytes than that in glycophyte under salinity stress, too (Bose et al. 2015). A PM H+-ATPase AHA4 mutant Arabidopsis showed a dramatic growth reduction when exposed to salt stress (Vitart et al. 2001). Transgenic tobacco plants expressing a constitutively activated plasma membrane H+-ATPase, lacking the autoinhibitory domain, displayed increased salt tolerance compared with untransformed plants (Gevaudant et al. 2007). These suggest that PM H+-ATPase activity is involved in plant adaptation to salinity; so theoretically, expression of wild-type (WT) PM H+-ATPase should confer salt tolerance to transgenic plants, but the reports regarding improving salt tolerance of transgenic plants via overexpressing PM H+-ATPase are rare.
Sesuvium portulacastrum is a halophyte with optimal growth at 200–300-mM NaCl (Yi et al. 2014). Upon grown in saline environment, S. portulacastrum cells accumulated lots of Na+, and there is neither salt glands nor bladders in all tissues of the halophyte (Slama et al. 2008; Lokhande et al. 2010; Rabhi et al. 2010), suggesting that S. portulacastrum may have special ability to extrude Na+ out of cells. SpAHA1 is a PM-bound protein found in S. portulacastrum; not only its expression could be induced by salinity but also SpAHA1 generated an increased proton gradient that stimulated the Na+/H+ exchange activity of SpSOS1 in transgenic yeast cells, such that the yeast cells coexpressing SpSOS1 and SpAHA1 grew better than the cells transformed with only SpSOS1 or SpAHA1 under NaCl treatment (Zhou et al. 2015). These suggest that SpAHA1 may be implicated in the plant salt tolerance. To test the hypothesis, the SpAHA1 gene was transformed into Arabidopsis plants. SpAHA1-expressing Arabidopsis plants displayed better growth relative to WT plants under salinity condition. So, our results provided direct genetic evidence about the salt-tolerant role of PM H+-ATPase in transgenic plants.
Materials and methods
Plasmid construction
The cDNA sequence of PM H+-ATPase gene SpAHA1 (accession no. JX628604) from S. portulacastrum plants was previously reported (Zhou et al. 2015). To construct its plant expression vector, the full length of SpAHA1 was obtained by PCR using the plasmid p414-SpAHA1 as a template and a pair of primers (SpAHA1-F and SpAHA1-R shown in Supplementary Table 1). The resulting fragment was digested with Spe I and Eco72 I and then inserted between the two restriction enzyme sites of the pCAMBIA1304 binary vector (CAMBIA Co., Australia) containing kanamycin and hygromycin resistance selectable markers to generate pCAMBIA1304-SpAHA1.
Arabidopsis transformation and gene expression assays
The recombinant plasmid pCAMBIA1304-SpAHA1 was introduced into Agrobacterium tumefaciens strain GV3101 (Konez and Schell 1986) using chemical transformation and then transformed into Arabidopsis thaliana (Col-0) via flower bud (Clough and Bent, 1998). Seeds from T0 generation were initially screened on MS medium containing 50-μg/mL hygromycin B. DNA was extracted from putative transgenic lines and used as the template to verify SpAHA1 transgenic plants via PCR amplification with SpAHA1 gene-specific primers (SpAHA1-TF and SpAHA1-TR shown in Supplementary Table 1) and the hygB gene primers (HygF and HygR shown in Supplementary Table 1). Total RNA was extracted from transgenic and WT plants using the TRIzol method (Invitrogen, Carlsbad, USA). SpAHA1 expression in transgenic plants was analyzed via reverse transcriptional (RT) PCR with an internal control of the housekeeping gene Actin as described previously (Albrecht et al. 2003). The PCR products with described size were examined by agarose gel electrophoresis, respectively. Quantitative real-time PCR (qRT-PCR) was performed for expression assay of Arabidopsis AtSOS1 gene. RNA was extracted from WT or SpAHA1-transgenic Arabidopsis plants treated with or without 200-mM NaCl. The PCR conditions were as follows: 95 °C for 30 s, followed by 40 cycles at 95 °C for 5 s, and 60 °C for 30 s. A dissociation curve from 60 to 95 °C was generated to verify primer specificity, and data analysis was performed using the SDS plate utility software, version 2.4. The value of relative expression of the AtSOS1 gene in untransformed plants without salt treatment was arbitrarily set to 1, and values of the relative expression of the gene in salt-treated WT plants and SpAHA1-transgenic plants treated with or without NaCl were expressed based on the value measured in the WT plants grown under control condition. The primers of SpAHA1 (SpAHA1-RT-F and SpAHA1-RT-R), AtSOS1 (AtSOS1-qRT-F and AtSOS1-qRT-R), and Actin (Actin-RT-F and Actin-RT-R; Actin-qRT-F and Actin-qRT-R) were shown in Supplementary Table 1.
Salt-tolerant determination of SpAHA1-expressing plants
To test the response of SpAHA1-transgenic Arabidopsis plants to salinity stress, seeds from T3 homozygous transgenic and WT plants were germinated on MS plates supplemented with 0-, 50-, 75-, or 100-mM NaCl in growth chamber at 22 °C with 16-h light/8-h dark. After 7 days, the plates were photographed, and germination ratios of these seeds were recorded. As for the experiments about the response of seedlings to salinity stress, 5-day-old seedlings grown on MS medium were transferred onto MS plates with 0-, 50-, and 75-mM NaCl, respectively, and allowed to grow for 7 days; after that the root length, lateral root number, and fresh weight of the seedlings were measured. Again, the 5-day-old seedlings were transferred into pots containing organic soil and sand (3:1, v/v) and grown in the greenhouse at 22 °C with 16-h light period for 4 weeks and then treated with 200-mM NaCl. After 10 days, the plants were photographed, and plant growth was assessed as fresh weight.
Determination of H+ and Na+ flux in roots
Seven-day-old seedlings grown on MS plates were transferred to MS medium with 0- or 100-mM NaCl. Three days after NaCl treatment, the roots from the seedlings were balanced in measurement buffer (0.1-mM KCl, 0.1-mM CaCl2, and 0.5-mM MES, pH 6.0) to balance for 5 min, and then, net H+ or Na+ flux in the roots was determined using Non-invasive Micro-test Technology (NMT100 Series, Younger USA LLC., Amherst, MA01002, USA) and iFluxes/imFluxes 1.0 (YoungerUSA, LLC, Amherst, MA 01002, USA) Software in the Younger USA (Xuyue Beijing) NMT Service Center as described previously (Sun et al. 2009; Zhou et al. 2016).
Assays of Na+ and K+ contents in Arabidopsis plants
The SpAHA1-expressing and WT seedlings were treated with or without 200-mM NaCl for 2 weeks, and then the Na+ and K+ contents in the plants were determined by atomic absorption spectrometry as previously described (Zhou et al. 2016).
Assays of hydrogen peroxide and MDA content
One-week-old seedlings of SpAHA1-expressing and WT Arabidopsis were exposed to 75-mM NaCl for 7 days, and then hydrogen peroxide (H2O2) in plants was analyzed by 3,3′-diaminobenzidine (DAB) staining method described by Daudi and O’Brien (2012); the MDA content was determined in the leaves according to the thiobarbituric acid (TBA) method (Dhindsa and Matowe 1981).
Statistical analysis
The two-tailed Student’s test was used to analyze the data expressed as the means ± SE, and differences with a p value < 0.05 were considered significant.
Results
Identification of SpAHA1 transgenic plants
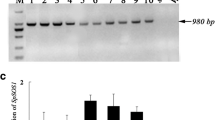
The gene SpAHA1, encoding a PM-bound H+-ATPase (Zhou et al. 2015), was transformed into Arabidopsis plants via mediation of a binary expression vector pCAMBIA1304 with the CaMV 35S promoter. Genomic DNA PCR analyses with SpAHA1 gene specific primers and the primers of hygB gene showed that 11 transgenic lines were gotten (Supplementary Fig. 1a). RT-PCR analysis was performed to study the expression level of SpAHA1 in transgenic plants. Except for the lower expression level in line 5 and line 8, SpAHA1 could significantly be expressed in other transgenic lines (Supplementary Fig. 1b). So, the T3 generation of homozygous transgenic plants of SpAHA1-line 1, SpAHA1-line 2, or SpAHA1-line 3 was used to analyze SpAHA1 function.
The effects of salinity stress on seed germination and the growth of seedlings
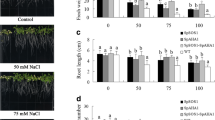
Seed germination and seedling growth are most sensitive stages during whole life period; so, the plant responses to salinity stress during the two early stages can clearly indicate plant salt tolerance. Seeds from three transgenic lines (line1, line2, and line3) and WT Arabidopsis plants could nearly germinate under normal MS medium plates, but NaCl treatment severely inhibited the germination of WT seeds, the germination ratio decreased with increasing NaCl concentrations, even all WT seeds exposed to 100-mM NaCl could not germinate (Fig. 1); the transgenic seedlings grew slowly after germination under salinity treatment relative to those grown on MS plates without NaCl, but their seeds nearly could germinate under 50- and 75-mM NaCl except for decreased germination ratio under 100-mM NaCl.
Effect of NaCl treatment on seed germination. The germination was initiated with seeds sown on MS plates with different NaCl concentrations as described. After 7 days, the plates were photographed (a), and germination ratio of these seeds was recorded (b). Data represent the mean ± SE of 60 replicates, and different letters above the columns indicate significant differences at p < 0.05 level among the different data. OE-1/2/3, SpAHA1-overexpressing line 1/2/3; WT, wild-type plants
In order to investigate the influence of salinity on seedlings growth, 5-day-old seedlings of SpAHA1-line 2 and SpAHA1-line 3 grown in normal medium were transferred to normal medium or MS medium containing 50- or 75-mM NaCl. Seven days after transferring, the plant growth parameters were analyzed. As seen from Fig. 2a–d, no growth difference was observed between WT and transgenic plants under normal condition. However, when grown on NaCl plates, SpAHA1-expressing seedlings grew better relative to WT plants. For example, the root length, fresh weight, and the number of lateral roots of SpAHA1-line 2 were 27, 40, and 38% greater than untransformed plants under 75-mM NaCl condition, respectively (Fig. 2). These indicated that SpAHA1 expression lightened the effect of salinity stress on plant growth.
Change in the growth of the seedlings treated with NaCl. Five-day-old seedlings grown MS plates were transferred to MS plates with 0-, 50-, and 75-mM NaCl, respectively. a After 7 days, the seedlings on the plates were photographed. Growth performance was expressed as fresh weight (b), root length (c), or lateral root number (d). Data represent the mean ± SE of 12 replicates, and different letters above the columns indicate significant differences at p < 0.05 level among the different data. OE-2/3, SpAHA1-overexpressing line 2/3; WT, wild-type plants
SpAHA1 increased salt tolerance of mature plants in soil
To address more accurately the effect of salinity stress on plant growth, 5-day-old uniform seedlings grown in normal medium were transferred into soil. After 4 weeks, the plants were treated with 200-mM NaCl for 10 days, and then the whole plants were harvested and weighted. The growths of WT and transgenic plants (line 2 and line 3) were inhibited while exposed to NaCl treatment (Fig. 3a, b), but the growth reduction of SpAHA1-expressing plants is 52 and 46% smaller relative to WT plants under NaCl treatment (Fig. 3c), indicating that SpAHA1 conferred salt tolerance to transgenic plants.
Change in the growth of Arabidopsis plants in soil supplied with NaCl. Five-day-old seedlings of wild-type and transgenic plants were transferred from MS plates to soil (four plants/pot) and allowed to develop for 4 weeks, and then the plants were treated with 0- or 200-mM NaCl. Ten days after NaCl treatment, the plants in pots were photographed (a); fresh weight was measured (b). To more accurately assess the salt-tolerance capacity of transgenic plants, c the relative change (the percentage reduction) of fresh weight under salt stress compared with the non-stressed control was determined. Data represent the mean ± SE of 12 replicates, and different letters above the columns indicate significant differences at p < 0.05 level among the different data. OE-2/3, SpAHA1-overexpressing line 2/3; WT, wild-type plants
Faster H+ efflux and Na+ extrusion in roots and lower Na+ content in transformed plants exposed to salt stress
The PM H+-ATPase generates proton gradient across the membrane, which energizes for many physiological processes, such as nutrient uptake, stomatal opening, phloem loading, and cell growth (Palmgren 2001). The proton motive force created by H+-ATPase is also essential for Na+ extrusion from root cells mediated by a PM Na+/H+ antiporter SOS1, which is the only Na+ efflux protein (Gaxiola et al. 2007). SpAHA1 could function as a PM H+-ATPase and provide energy for Na+/H+ exchange activity of SpSOS1 in co-transgenic yeast cells under salinity condition (Zhou et al. 2015). In the present investigation, the roots from the tested plants were equilibrated in measurement buffer of pH 6 for 5 min, and then proton flux signal was recorded. Acidification of the measurement buffer led to significant proton uptake of roots of plants grown in normal medium (Fig. 4). In contrast, salinity treatment led to H+ efflux in the roots (Fig. 4), which is consistent with the previous report that NaCl induced significantly H+ efflux in the roots of Arabidopsis plants (Bose et al. 2015), suggesting that PM H+-ATPase activity in roots from seedlings exposed to salinity stress was higher compared with unstressed plants. The increased H+-pumping activity in Arabidopsis treated with NaCl may be involved in salinity-induced AHA expression in Arabidopsis plants (Bose et al. 2015). Furthermore, H+ efflux in the roots of SpAHA1 transgenic plants (SpAHA1-line2) was faster than that in WT plants while treated with NaCl (Fig. 4b). H+ extrusion across plasma membrane is dependent on PM H+-ATPase (Fuglsang et al. 2007), that is, if the activity of PM H+-ATPase increases, the rate of H+ efflux at plasma membrane increases. These suggested that proton pump ability of PM H+-ATPase of SpAHA1-expressing seedlings was relatively higher compared to WT, which was involved in constitutive expression of SpAHA1 regulated by 35S promoter from the plasmid pCAMBIA1304 (Supplementary Fig. 1b). So, the extra H+-pumping activity mediated by SpAHA1 could provide more motive force for Na+ extrusion generated by PM Na+/H+ antiporter; correspondingly, Na+ was faster transported out of cells in SpAHA1 transgenic (SpAHA1-line2) roots than WT plants under salinity stress (Fig. 5a, b). Additionally, PM H+-ATPase higher activity in saline conditions reduced Na+ influx through repolarizing the NaCl-induced depolarization of the PM (Sun et al. 2009). These lead to less Na+ accumulation in SpAHA1-expressing plants exposed to salinity stress relative to WT plants (Fig. 6a); so, SpAHA1 could alleviate toxicity of salinity stress via decreasing Na+ concentration in cytosol.
H+ fluxes in roots of wild-type Arabidopsis plants and transgenic lines. Seven-day-old seedlings were transferred to MS medium with or without 100-mM NaCl for 3 days, and then H+ flux in roots was measured using the NMT technique as described in the “Materials and methods” section. a The change in the NMT signal is expressed as arbitrary units. b H+ flux activity is expressed as the efflux amount per second per square centimeter (pmol cm−2 s−1), and data represent the mean ± SE of six replicates, and different letters above the columns indicate significant differences at p < 0.05 level among the different data. SpAHA1, SpAHA1-overexpressing line 2; WT, wild-type plants
Na+ fluxes in roots of wild-type Arabidopsis plants and transgenic lines. Seven-day-old seedlings were transferred to MS plates containing 100-mM NaCl for 3 days, and then Na+ flux in roots was measured using the NMT technique as described in the “Materials and methods” section, respectively. a The change in the NMT signal is expressed as arbitrary units. b Na+ flux activity is expressed as the efflux amount per second per square centimeter (pmol cm−2 s−1), data represent the mean ± SE of six replicates, and different letters above the columns indicate significant differences at p < 0.05 level among the different data. SpAHA1, SpAHA1-overexpressing line 2; WT, wild-type plants
Na+ and K+ contents in wild-type Arabidopsis plants and transgenic lines. WT and transgenic plants grew in soil with 0- or 200-mM NaCl for 2 weeks, and Na+ (a) and K+ (b) in the above plants were determined as described in the “Materials and methods.” Data represent the mean ± SE of three replicates; different letters above the columns indicate significant differences at p < 0.05 level among the different data. SpAHA1, SpAHA1-overexpressing line 2; WT, wild-type plants
Increased K+ content in SpAHA1-expressing plants
High concentrations of Na+ in plant cells exposed to salinity stress caused plasma membrane depolarization, which increases K+ efflux through depolarization-activated outward rectifying K+ channels (Adams and Shin 2014), leading to K+ nutrient deficiency in plants under salt stress. Therefore, maintaining cellular K+ levels above a certain threshold in saline environments is critical for plant growth and salt tolerance (Zhu et al. 1998). The proton gradient created by H+-ATPase is largely responsible for a negative potential across the plasma membrane, which is essential for PM repolarization under salinity stress (Mansour 2014). Upon exposure to salinity stress, the roots of SpAHA1-expressing Arabidopsis had faster H+ efflux compared to WT plants (Fig. 4a, b), indicating that higher H+-pumping activity generated by SpAHA1 was helpful to PM repolarization; NaCl treatment decreased K+ content in both WT and transgenic plants, but SpAHA1-expressing plants (SpAHA1-line2) had increased K+ content relative to WT plant while treated with NaCl (Fig. 6b); the plants expressing SpAHA1 grew better than WT plants in saline environment. These suggest that SpAHA1 expression could relieve the inhibitory effect of salinity on plant growth via maintaining K+ homeostasis in plant cells.
SpAHA1 expression led to less reactive oxygen species (ROS) accumulation in transgenic plants treated with NaCl
Salt stress interferes with the homoeostasis of ROS and leads to ROS accumulation (Borsani et al. 2001). Hydrogen peroxide (H2O2) is one of several ROS and can be detected out by staining with 3,3′-diaminobenzidine (DAB) in plant tissues (Daudi and O’Brien 2012). In the present study, DAB-staining WT and transgenic plants exposed to salinity stress had darker color than non-stressed plants (Fig. 7a), indicating that H2O2 content was higher in NaCl-treated plants than in untreated plants. However, as seen from Fig. 7a, the brown color of DAB-staining plants of transgenic plants was lighter relative to WT plants under saline condition, suggesting that SpAHA1 might alleviate H2O2 accumulation in transgenic plants induced by salinity. Excessive ROS can damage membrane integrity via oxidizing membrane lipids, such that useful metabolites leak from plant cells (Velarde-Buendía et al. 2012). Malondialdehyde (MDA) is a critical oxide of membrane lipids; so, it can represent the membrane lipid damage to some extent. Here, the difference of MDA contents between WT and transgenic plants was not clear under non-stress condition, whereas upon exposure to salinity, WT plants accumulated more MDA compared with transgenic plants (Fig. 7b), suggesting that oxidative damage to membranes in SpAHA1-transgenic plants was less. These results indicate that SpAHA1 could increase plant salt tolerance via alleviating oxidative damage generated by salinity stress.
Assays of hydrogen peroxide (H2O2) and MDA content. Wild-type and transgenic seedlings were treated with 75-mM NaCl, and the plants were harvested. a Plant H2O2 was analyzed as described in the “Materials and methods.” One representative of three plants. After wild-type and transgenic Arabidopsis plants were treated with 75-mM NaCl for 10 days, b MDA contents in leaves were determined as described in the “Materials and methods.” Data represent the mean ± SE of three replicates; different letters above the columns indicate significant differences at p < 0.05 level among the different data. OE-2/3, SpAHA1-overexpressing line 2/3; WT, wild-type plants
Discussion
Sodium is a trace element in plants. However, excess sodium in the plant cells is toxic to some important physiological and biochemical processes; so, Na+ extrusion from cells is involved in plant salt tolerance. The proton motive force created by H+-ATPase under salt stress energizes Na+ extrusion mediated by a PM Na+/H+ antiporter SOS1 (Zhou et al. 2015). Therefore, the PM H+-ATPase is a kind of key plant halotolerance factor. T-DNA insertion mutant Arabidopsis of a PM H+-ATPase AHA4 displayed decreased growth relative to WT plant under salinity stress (Vitart et al. 2001). Hyperactive mutant of a tobacco plasma membrane H+-ATPase △PMA4 increased salt tolerance of transgenic tobacco compared to untransformed plants (Gevaudant et al. 2007), but it was not reported whether the overexpression of the WT H+-ATPase conferred strong salt tolerance to transgenic plants there. In the present study, in order to provide a direct genetic evidence for the involvement of full-length PM H+-ATPase gene in salt tolerance in transgenic plants, a PM H+-ATPase gene SpAHA1 was cloned from a halophyte S. portulacastrum and transformed into Arabidopsis plants. The growth of both WT and transformed Arabidopsis plants was inhibited under salinity stress, but SpAHA1-expressing plants exhibited increased salt tolerance relative to WT plants and displayed more than 80% higher germination rate and 40% greater biomass than WT plants under 75-mM NaCl, respectively (Figs. 1 and 2); the roots of transgenic plants with SpAHA1 had a faster net H+ efflux rate than WT plant under salt stress(Fig. 4); in these transgenic and non-transgenic plants treated with NaCl, the Na+ efflux rate in transgenic roots was faster relative to untransformed roots, such that SpAHA1-expressing plants accumulated less Na+ compared to WT plants (Fig. 5).
Our recent investigation showed that SpAHA1 functioned as a H+-ATPase at the plasma membrane and produced an increased electrochemical gradient of protons to promote the Na+/H+ exchange activity of SpSOS1 in yeast cells coexpressing both genes SpAHA1 and SpSOS1 (Zhou et al. 2015). NaCl treatment induced the expression of AtSOS1 in Arabidopsis plants, which is consistent with the previous report that Arabidopsis SOS1 was up-regulated at the transcription level in response to NaCl (Shi et al. 2000). Furthermore, mRNA abundance of AtSOS1 was higher in SpAHA1-transgenic plants than WT plants under salinity stress (Supplementary Fig. 2). These suggest that enhanced plasma membrane H+ pumping generated by SpAHA1 in transgenic plants provided extra energy to support Na+ export mediated by the native Na+/H+ antiporter at the plasma membrane; excess Na+ in the cytosol was transported out of cells, such that the toxicity of intracellular Na+ became lower; correspondingly, the plants expressing SpAHA1 grew better than untransformed plants in the saline environment (Figs. 1 and 2), indicating that the SpAHA1 may play a decisive role in plant salt tolerance and can be utilized as candidate gene for engineering salt tolerance in crops.
ROS are toxic when they accumulate in the cells. So, intracellular ROS levels under normal conditions are tightly regulated by numerous peroxidative and antioxidative reactions within the cell. Salinity can disequilibrate between ROS production and scavenging, resulting in the accumulation of ROS, which in turn negatively affects cellular structures and metabolism (Chaves and Oliveira 2004; Chaves et al. 2009). In order to protect a cell from oxidative damage generated by salinity, excess ROS should be scavenged by antioxidant mechanisms. Haem oxygenase (HO) is an important component of the antioxidant defense systems in plants; overexpression of AtHO gene enhanced salinity tolerance in Arabidopsis via increasing the activity and expression of the PM H+-ATPase (Bose et al. 2013). Here, H2O2 contents increased in both untransformed and transgenic plants when treated with NaCl, but comparative analyses showed that SpAHA1 expression in Arabidopsis plants led to lower H2O2 level relative to WT plants under salinity stress (Fig. 7a). Excessive ROS can damage membrane integrity, such that useful metabolites leak out of plant cells. It is reported that NaCl-induced K+ efflux is a result of the PM disintegrity and not due to ion channel mediation in rice (Coskun et al. 2013), indicative of the prime role of the PM stability maintenance in salt tolerance of plants. In the present study, the MDA content of non-transgenic Arabidopsis plants was higher than SpAHA1-expressing plants when exposed to salinity stress (Fig. 7b). These indicate that expression of SpAHA1 alleviated membrane damage via regulating ROS homeostasis in plant cells under salt stress and improved the growth of transgenic plants treated with NaCl.
Potassium (K+) is an essential macronutrient that plays an important role in plant growth and development (Barragan et al. 2012). Due to similarity in physicochemical properties between Na+ and K+, Na+ competes with K+ for major binding sites in key physiological and biochemical processes in the cytoplasm (Marschner 1995), especially, Na+ inhibits the activity of many enzymes that require K+ for functioning (Duggleby and Dennis 1973); so, excessive Na+ can interfere with the essential roles of K+ in the cytosol (Marschner 1995). It has been suggested that plant survival under salt stress requires a high cytosolic K+/Na+ ratio in the cytoplasm. So, restricting Na+ influx into cells may improve plant growth under salinity stress (Lutts et al. 1996; Tester and Davenport 2003). Under salt stress conditions, PM potential is strongly depolarized, which favors passive Na+ influx to cells and K+ efflux out of cells. Electrochemical potential gradient across PM generated by H+-ATPases can repolarize the NaCl-induced depolarization of PM (Mansour 2014). So, the maintenance of the PM potential through H+-ATPases activity can help to reduce Na+ influx via depolarization-activated NSCCs and K+ efflux via KORs and NSCCs (Sun et al. 2009). Net H+ efflux from root cells was faster in SpAHA1 transgenic plants relative to WT plants (Fig. 4), suggesting that SpAHA1 generated extra H+-ATPase activity and provided higher electrochemical potential and H+ gradient across PM, which may be helpful to reduce Na+ influx and K+ efflux, correspondingly, leading more K+ retention and lower Na+ concentration in cytosol of transgenic Arabidopsis plants than WT plants (Fig. 6), and restored higher K+/Na+ level in SpAHA1 transgenic plants than in WT plants, which is a strong indication of salt tolerance. These results suggest that SpAHA1 could increase salt tolerance through controlling K+/Na+ homeostasis in transgenic plants.
Taken together, the genetic evidence suggests that SpAHA1 function to decrease the accumulation of Na+ via producing an increased electrochemical gradient of protons to promote PM Na+/H+ exchange activity and plays a critical role in plant salt tolerance.
References
Adams E, Shin R (2014) Transport, signaling, and homeostasis of potassium and sodium in plants. J Integr Plant Biol 56:231–249. https://doi.org/10.1111/jipb.12159
Albrecht V, Weinl S, Blazevic D, D’Angelo C, Batistic O, Kolukisaoglu U, Bock R, Schulz B, Harter K, Kudla J (2003) The calcium sensor CBL1 integrates plant responses to abiotic stresses. Plant J 36:457–470. https://doi.org/10.1046/j.1365-313X.2003.01892.x
Arango M, Gevaudant F, Oufattole M, Boutry M (2003) The plasma membrane proton pump ATPase: the significance of gene subfamilies. Planta 216:355–365. https://doi.org/10.1007/s00425-002-0856-8
Axelsen KB, Palmgren MG (2001) Inventory of the superfamily of P-type ion pumps in Arabidopsis. Plant Physiol 126:696–706. https://doi.org/10.1104/pp.126.2.696
Barragan V, Leidi EO, Andrés Z, Rubio L, De Luca A, Fernandez JA, Cubero B, Pardo JM (2012) Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 24:1127–1142. https://doi.org/10.1105/tpc.111.095273
Borsani O, Valpuesta V, Botella MA (2001) Evidence for a role of salicylic acid in the oxidative damage generated by NaCl and osmotic stress in Arabidopsis seedlings. Plant Physiol 126:1024–1030. https://doi.org/10.1104/pp.126.3.1024
Bose J, Xie Y, Shen W, Shabala S (2013) Haem oxygenase modifies salinity tolerance in Arabidopsis by controlling K+ retention via regulation of the plasma membrane H+-ATPase and by altering SOS1 transcript levels in roots. J Exp Bot 64:471–481. https://doi.org/10.1093/jxb/ers343
Bose J, Rodrigo-Moreno A, Lai D, Xie Y, Shen W, Shabala S (2015) Rapid regulation of the plasma membrane H+-ATPase activity is essential to salinity tolerance in two halophyte species, Atriplex lentiformis and Chenopodium quinoa. Ann Bot 115:481–494. https://doi.org/10.1093/aob/mcu219
Chaves MM, Oliveira MM (2004) Mechanisms underlying plant resilience to water deficits: prospects for water-saving agriculture. J Exp Bot 55:2365–2384. https://doi.org/10.1093/jxb/erh269
Chaves MM, Flexas J, Pinheiro C (2009) Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann Bot 103:551–560. https://doi.org/10.1093/aob/mcn125
Chen M, Song J, Wang B (2010) NaCl increases the activity of the plasma membrane H+-ATPase in C3 halophyte Suaeda salsa callus. Acta Physiol Plant 36:27–36. https://doi.org/10.1093/jxb/erh269
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743. https://doi.org/10.1046/j.1365-313x.1998.00343.x
Coskun D, Britto D, Jean Y, Kabir I, Tolay I, Torun A, Kronzucker H (2013) K+ efflux and retention in response to NaCl stress do not predict salt tolerance in contrasting genotypes of rice (Oryza sativa L.). PLoS One 8:57767. https://doi.org/10.1371/journal.pone.0057767
Daudi A, O’Brien JA (2012) Detection of hydrogen peroxide by DAB staining in Arabidopsis leaves. Bio Protocol 2(18):e263. https://doi.org/10.21769/BioProtoc.263
Dhindsa R, Matowe W (1981) Drought tolerance in two mosses: correlated with enzymatic defence against lipid peroxidation. J Exp Bot 32:79–91. https://doi.org/10.1093/jxb/32.1.79
Ding M, Hou P, Shen X, Wang M, Deng S, Sun J, Xiao F, Wang R, Zhou X, Lu C, Zhang D, Zheng X, Hu Z, Chen S (2010) Salt-induced expression of genes related to Na+/K+ and ROS homeostasis in leaves of salt-resistant and salt sensitive poplar species. Plant Mol Biol 73:251–269. https://doi.org/10.1007/s11103-010-9612-9
Duggleby RG, Dennis DT (1973) Pyruvate kinase, a possible regulatory enzyme in higher plants. Plant Physiol 52:312–317. https://doi.org/10.1104/pp.52.4.312
Fuglsang AT, Guo Y, Cuin TA, Qiu Q, Song C, Kristiansen KA, Bych K, Schulz A, Shabala S, Schumaker KS, Palmgren MG, Zhu JK (2007) Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. Plant Cell 19:1617–1634. https://doi.org/10.1105/tpc.105.035626
Fuglsang AT, Paez-Valencia J, Gaxiola RA (2010) Plant proton pumps: regulatory circuits involving H+-ATPase and H+-PPase. In Book: Transporters and Pumps in Plant Signaling 39–64
Gaxiola RA, Palmgren MG, Schumacher K (2007) Plant proton pumps. FEBS Lett 581:2204–2214. https://doi.org/10.1016/j.febslet.2007.03.050
Gevaudant F, Duby G, Evon Stedingk E, Zhao R, Morsomme P, Boutry M (2007) Expression of a constitutively activated plasma membrane H+-ATPase alters plant development and increases salt tolerance. Plant Physiol 144:1763–1776. https://doi.org/10.1104/pp.107.103762
Konez C, Schell J (1986) The promoter of TL-DNA gene 5′ controls the tissue specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204:383–396. https://doi.org/10.1007/BF00331014
Lokhande VH, Nikam TD, Suprasanna P (2010) Biochemical, physiological and growth changes in response to salinity in callus cultures of Sesuvium portulacastrum L. Plant Cell Tiss Org 102:17–25. https://doi.org/10.1007/s11240-010-9699-3
Lopez-Pérez L, Martinez-Ballesta M, Maurel C, Carvajal M (2009) Changes in plasma membrane lipids, aquaporins and proton pump of broccoli roots, as an adaptation mechanism to salinity. Phytochemistry 70:492–500. https://doi.org/10.1016/j.phytochem.2009.01.014
Lutts S, Kinet J, Bouharmont J (1996) NaCl-induced senescence in leaves of rice (Oryza sativa L.) cultivars differing in salinity resistance. Ann Bot 78:389–398. https://doi.org/10.1006/anbo.1996.0134
Mansour MMF (2014) The plasma membrane transport systems and adaptation to salinity. J Plant Physiol 171:1787–1800. https://doi.org/10.1016/j.jplph.2014.08.016
Marschner H (1995) Mineral nutrition of higher plants. Ann Bot 78:527–528. https://doi.org/10.1006/anbo.1996.0155
Niu X, Narasimhan M, Salzman R (1993) NaCl regulation of plasma membrane H+-ATPase gene expression in glycophyte and halophyte. Plant Physiol 103:712–718. https://doi.org/10.1104/pp.103.3.713
Palmgren MG (2001) Plant plasma membrane H1-ATPases: powerhouses for nutrient uptake. Annu Rev Plant Physiol Plant Mol Biol 52:817–845. https://doi.org/10.1146/annurev.arplant.52.1.817
Pons R, Cornejo MJ, Sanz A (2011) Differential salinity-induced variations in the activity of H+-pumps and Na+/H+ antiporters that are involved in cytoplasm ion homeostasis as a function of genotype and tolerance level in rice cell lines. Plant Physiol Biochem 49:1399–1409. https://doi.org/10.1016/j.plaphy.2011.09.011
Rabhi M, Ferchichi S, Jouini J, Hamrouni MH, Koyro HW, Ranieri A, Abdelly C, Smaoui A (2010) Phytodesalination of a salt-affected soil with the halophyte Sesuvium portulacastrum L. to arrange in advance the requirements for the successful growth of a glycophytic crop. Bioresour Technol 101:6822–6828. https://doi.org/10.1016/j.biortech.2010.03.097
Rober-Kleber N, Albrechtova JT, Fleig S, Huck N, Michalke W, Wagner E, Speth V, Neuhaus G, Fischer-Iglesias C (2003) Plasma membrane H+-ATPase is involved in auxin-mediated cell elongation during wheat embryo development. Plant Physiol 131:1302–1312. https://doi.org/10.1104/pp.013466
Ruan CJ, da Silva JAT, Mopper S, Qin P, Lutts S (2010) Halophyte improvement for a salinized world. Crit Rev Plant Sci 29:329–359. https://doi.org/10.1080/07352689.2010.524517
Sahu B, Shaw B (2009) Salt-inducible isoform of plasma membrane H+-ATPase gene in rice remains constitutively expressed in natural halophyte, Suaeda maritima. J Plant Physiol 166:1077–1089. https://doi.org/10.1016/j.jplph.2008.12.001
Santi S, Locci G, Monte R, Pinton R, Varanini Z (2003) Inducation of nitrate uptake in maize roots: expression of a putative high-affinity nitrate transporter and plasma membrane H+-ATPase isoforms. J Exp Bot 54:1851–1864. https://doi.org/10.1093/jxb/erg208
Shi H, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci U S A 97:6896–6901. https://doi.org/10.1073/pnas.120170197
Slama I, Ghnaya T, Savoure A, Abdelly C (2008) Combined effects of long-term salinity and soil drying on growth, water relations, nutrient status and proline accumulation of Sesuvium portulacastrum. C R Biol 331:442–451. https://doi.org/10.1016/j.crvi.2008.03.006
Sun J, Dai S, Wang R, Chen S, Li N, Zhou X, Lu C, Shen X, Zheng X, Hu Z, Zhang Z, Song J, Xu Y (2009) Calcium mediates root K+/Na+ homeostasis in poplar species differing in salt tolerance. Tree Physiol 29:1175–1186. https://doi.org/10.1093/treephys/tpp048
Tester M, Davenport RJ (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot 91:503–527. https://doi.org/10.1093/aob/mcg058
Velarde-Buendía AM, Shabala S, Cvikrova M, Dobrovinskaya O (2012) Salt-sensitive and salt-tolerant barley varieties differ in the extent of potentiation of the ROS-induced K+ efflux by polyamines. Plant Physiol Biochem 61:18–23. https://doi.org/10.1016/j.plaphy.2012.09.002
Vera-Estrella R, Barkla BJ, Garcia-Ramirez L, Pantoja O (2005) Salt stress in Thellungiella halophila activates Na+ transport mechanisms required for salinity tolerance. Plant Physiol 139:1507–1517. https://doi.org/10.1104/pp.105.067850
Vitart V, Baxter I, Doerner P, Harper JF (2001) Evidence for a role in growth and salt resistance of a plasma membrane H+-ATPase in the root endodermis. Plant J 27:191–201. https://doi.org/10.1046/j.1365-313x.2001.01081.x
Yi XP, Sun Y, Yang Q, Guo AP, Chang LL, Wang D, Tong Z, Jin X, Wang LM, Yu JL, Jin WH, Xie YM, Wang XC (2014) Quantitative proteomics of Sesuvium portulacastrum leaves revealed that ion transportation by V-ATPase and sugar accumulation in chloroplast played crucial roles in halophyte salt tolerance. J Proteome 99:84–100. https://doi.org/10.1016/j.jprot.2014.01.017
Zhou Y, Yin X, Duan R, Hao G, Guo J, Jiang X (2015) SpAHA1 and SpSOS1 coordinate in transgenic yeast to improve salt tolerance. PLoS One 10:e0137447. https://doi.org/10.1371/journal.pone.0137447
Zhou Y, Lai Z, Yin X, Yu S, Xu Y, Wang X, Cong X, Luo Y, Xu H, Jiang X (2016) Hyperactive mutant of a wheat plasma membrane Na+/H+ antiporter improves the growth and salt tolerance of transgenic tobacco. Plant Sci 253:176–186. https://doi.org/10.1016/j.plantsci.2016.09.016
Zhu JK, Liu J, Xiong L (1998) Genetic analysis of salt tolerance in Arabidopsis: evidence for a critical role of potassium nutrition. Plant Cell 10:1181–1191. https://doi.org/10.1105/tpc.10.7.1181
Funding
This work was supported by the Natural Science Foundation of China (31660253, 31260218), the Scientific and Technological Foundation of Hainan Province (HNGDhs201502), Hainan Provincial Natural Science Foundation of China (318QN189), the National Key Technology Support Program (2015BAD01B02), and Startup funding from Hainan University (KYQD(ZR)1845).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling Editor: Peter Nick
Electronic supplementary material
Supplementary Table 1
(XLS 18 kb)
Supplementary Fig. 1
Identification of transgenic plants. a DNA was isolated from the leaves of SpAHA1-transgenic or WT plants. PCR was performed with primers for SpAHA1 or hygB genes with the related DNA as the template. PCR products were resolved by agarose gel electrophoresis. b Expression pattern of SpAHA1 in transgenic Arabidopsis lines. Total RNA extracted from the leaves of T3 generation of transgenic plants was used for RT-PCR analyses. The Arabidopsis Actin gene was amplified as an internal control. M: DL2000 marker; 1–11, different transgenic lines (lines 1–11); 12, positive control (pCAMBIA1304-SpAHA1); 13, negative control (wild-type plants) (GIF 56 kb)
Supplementary Fig. 2
Expression assay of Arabidopsis SOS1 (AtSOS1) in wild-type and transgenic plants. Total RNA was isolated from the leaves of SpAHA1-transgenic and wild-type plants treated with or without NaCl. Quantitative real-time PCR was performed with primers for AtSOS1. The Arabidopsis Actin gene was used as the reference gene. Data represent the mean ± SE of three replicates, different letters above the columns indicate significant differences at p < 0.05 level among the different data. OE-2/3, SpAHA1-overexpressing line 2/3; WT, wild-type plants (GIF 12 kb)
Rights and permissions
About this article
Cite this article
Fan, Y., Wan, S., Jiang, Y. et al. Over-expression of a plasma membrane H+-ATPase SpAHA1 conferred salt tolerance to transgenic Arabidopsis. Protoplasma 255, 1827–1837 (2018). https://doi.org/10.1007/s00709-018-1275-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-018-1275-4