Abstract
Cysteine (Cys) regulates plant growth, development, and various abiotic stress responses. However, the knowledge gained from genetic and molecular studies on the role of Cys metabolism in salinity response remains limited. Here, we introduce a sulfate metabolism-related component, Arabidopsis Halotolerance 2-like (AHL), which is involved in salt stress adaptation. AHL expression was experimentally induced under salt stress conditions using AHL promoter-β-glucuronidase transgenic plants. Phenotypic analysis revealed that the survival rate of AHL-overexpressing (AHL-OE) lines was greater than that of wild-type (WT) plants under high-salinity conditions, while ahl mutant and AHL-RNAi seedlings were more sensitive than WT seedlings. Accumulation of Cys and proline was increased in AHL-OE transgenic lines under salinity stress conditions, but malondialdehyde and hydrogen peroxide levels were lower in AHL-OE than in WT, ahl, and AHL-RNAi plants. In addition, the transcription levels of genes associated with Cys biosynthesis and sulfate metabolism were higher in AHL-OE than ahl and AHL-RNAi seedlings after salt stress treatment. Moreover, exogenous application of Cys could rescue the salt stress-induced sensitive phenotypes of ahl and AHL-RNAi lines, and return them to the WT phenotype. Taken together, our findings indicate that AHL positively regulates salinity response in Arabidopsis seedlings by modulating a Cys-dependent pathway.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Salinity-induced alterations occur during plant growth and development due to the cumulative effects of excess saline on a number of physiological and biochemical properties (Iqbal et al. 2015). Some examples of the effects of salinity on plant cells include mineral ion homeostasis, osmolyte accumulation, water balance, antioxidant metabolism, nitrogen fixation, and photosynthesis (Iqbal et al. 2015). Salt stress triggers overproduction of reactive oxygen species (ROS) and affects the redox balance of plant cells, leading to damage to proteins, lipids, nucleic acids, and cell membrane integrity (Mittler 2002; Ahmad et al. 2008). Excess production of ROS causes oxidative stress, leading to cellular damage and, ultimately, cell death. To prevent or alleviate ROS-induced damage and allow the beneficial functions of ROS to continue, plants have evolved an intriguing antioxidant defense system. These defense systems aim to keep levels of reactive or active oxygen species under control. Antioxidant defense systems comprise both enzymatic as well as non-enzymatic components (Ahanger and Agarwal 2017). Plants tend to accumulate free amino acids, free proline, non-structured carbohydrates, and quaternary ammonium compounds (betaines) in response to salt stress conditions (Ahanger and Agarwal 2017).
Sulfate metabolism is a vital nutritional metabolic process that is required for plant growth, development, and environmental response (Kopriva et al. 2012; Shin et al. 2019). In nature, sulfate is the most abundant form of sulfur (S) that can be taken up by plants. Sulfate metabolism is carried out in various subcellular compartments, such as the cytosol, chloroplast or plastids, depending on the demands of the specific plant cell (Kopriva et al. 2012). Cysteine (Cys) is the first organic compound in primary sulfate metabolism (Takahashi et al. 2011). Cys is an amino acid that participates in the process of protein synthesis. Cys is also a precursor for a large number of essential biomolecules, such as vitamins, cofactors, or Fe-S clusters (Romero et al. 2014). Moreover, Cys is involved in the formation of many important bio-components, including glutathione (GSH), methionine, polyamine, glucosinolates, and camalexin (Romero et al. 2014). Sulfur-containing metabolites are necessary for various aspects of plant growth and development, as well as responses to unfavorable environments. Some examples of such metabolites include the Cys derivatives biotin, ethylene, polyamine, and GSH, which can directly or indirectly regulate the salinity stress response (Khan et al. 2014). GSH is involved in antioxidant-regulated cellular redox homeostasis through its role in ROS product catalysis (Khan et al. 2014). Arabidopsis HAL2-like (AHL), encoding the At1g54390 gene (Shin et al. 2019), was identified as a 3′-phosphoadenosine 5′-phosphate (PAP) phosphatase that belongs to the secondary branch of sulfate metabolism. It shares more than 40% amino acid sequence identity with the Arabidopsis 3′ (2′), 5′-bisphosphate nucleotidase (SAL) family (Chen et al. 2011; Shin et al. 2019). AHL has been determined to share a much higher transcript level with SAL1 than with other SAL family members (Chen et al. 2011; Shin et al. 2019). However, AHL may not directly participate in constitutive PAP accumulation under both normal and stress conditions (Chen et al. 2011; Shin et al. 2019). Suppression of PAP content in AHL-RNAi plants may be caused by SAL1 enzyme activity (Shin et al. 2019). Instead, AHL activity is closely involved in AMP reproduction (Shin et al. 2019; Nguyen et al. 2019). Additionally, AHL was found to participate in Cys generation through primary sulfate metabolism. In fact, the accumulation of AMP and Cys in AHL-overexpressing transgenic plants granted Arabidopsis a high degree of tolerance to osmotic stress and high glucose response (Shin et al. 2019; Nguyen et al. 2019). However, whether AHL modulates Cys production under conditions of salinity stress needs to be clarified.
Here, we wonder whether Arabidopsis HAL2-like (AHL) might be able to the regulation of Cys biosynthesis pathway for mediation of the salt stress response. To determine this possibility, the current study evaluated the role of AHL in salt stress by investigating sulfate metabolism of WT, ahl mutant, and AHL transgenic plants using molecular biological and physiological assays. We provided evidence indicating that gain of AHL function leads to an increase in salinity tolerance phenotypes. The Cys content was higher in AHL-overexpressing lines than in wild-type (WT) plants during salt stress treatment, but the levels of ahl mutant and AHL-RNAi lines were lower in the mutant than WT under salt stress. In addition, exogenous application of Cys ameliorates the salt stress-induced sensitive phenotype of the ahl and AHL-RNAi lines.
Materials and Methods
Plant Materials, Growth Conditions, and Salt Stress Treatment
Here, we investigated the seeds of Arabidopsis thaliana (Col-0), ahl mutant, and AHL transgenic plants including AHL pro-β-glucuronidase (GUS2-1, GUS3-3), AHL-RNAi (ri2-3, ri5-2), and AHL-overexpressing (OE2-2, OE5-1) lines previously designated by Nguyen et al. (2019) and Shin et al. (2019). To analyze AHL expression levels in ahl and AHL transgenic plants, reverse transcription (RT)-PCR and quantitative real-time PCR (qPCR) assays were performed. These assays reveled that two individual AHL-overexpressing (OE2-2 and OE5-1) lines displayed high levels of AHL transgene expression compared to WT, while two individual AHL-RNAi (ri2-3 and ri5-2) lines exhibited low levels of AHL expression (Supplementary Fig. S1). In addition, AHL transcription was completely abolished in ahl mutant (Supplementary Fig. S1). They were selected for phenotype characterization. Plants were grown in a standard growth chamber (16 h light/8 h dark, 22 °C condition, 60% relative humidity, and 110 μmol m−2 s−1 intense light). For salt stress treatment, 10-day-old plants were submerged in 150 mM NaCl solution and collected at 0, 3, 6, 12, and 24 h.
Analysis of β-Glucuronidase (GUS) Staining
GUS staining in AHL pro-GUS transgenic plants was conducted as described previously (Shin et al. 2019). Whole seedlings were immersed in 100 mM sodium phosphate solution (pH 7.0) containing 1 mM 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (X-Gluc), and then incubated for 12 h at 37 °C. After removal of chlorophyll with 70% ethanol, seedlings were photographed using a microscope.
Analysis of Salt Stress-Induced Sensitivity
For salt stress-induced sensitivity experiments, seeds were sown on one-half strength MS (for Murashige and Skoog) plates with or without 150 mM NaCl and grown in a growth chamber. Seed germination and survival rates were measured. To investigate the salt stress response to Cys, seeds were sown on 150 mM NaCl-containing one-half strength MS plates containing 40 μM Cys and grown in a growth chamber. Survival rates were measured. Data were obtained from three biological replicates, with 50 seeds per genotype per replicate.
Amino Acid and Malondialdehyde (MDA) Measurements
Proline (Pro) concentration was measured as described by Bates et al. (1973). Pro was obtained from approximately 200 mg of seedlings homogenized in 2 mL of 3% sulfosalicylic acid and reacted with 200 μL of ninhydrin reagent solution. The reaction mixture was combined with 400 μL of toluene and vortexed. The absorbance of the toluene layer was read at 520 nm with a UV/VIS spectrophotometer (JASCO, Tokyo, Japan). The Pro concentration was calculated from a standard curve.
The Cys content of the seedling samples was estimated following the method of Priya (2016). Cys was obtained from 500 mg seedling samples by homogenization in 5% cold perchloric acid solution and centrifugation at 10,000 rpm for 20 min at 4 °C. The supernatant was mixed with ninhydrin reagent and glacial acetic acid in equal volumes (1:1, mL). The reaction mixture was then heated at 95 °C for 10 min and rapidly cooled on ice. Absorbance was measured at 560 nm. The Cys concentration was calculated from a standard curve.
The MDA content of the seedling samples was measured as described by Kim et al. (2017). MDA was obtained from approximately 200 mg of seedlings by grinding in 10% trichloroacetic acid (TCA) buffer, and centrifuged at 10,000 rpm for 15 min. The supernatant (0.5 mL) was mixed with 1 mL of 0.6% thiobarbituric acid and 10% TCA. The reaction mixture was then heated at 95 °C for 15 min and rapidly cooled on ice. The absorbance values of the colored supernatants were measured at a wavelength of 532 nm and were corrected for non-specific absorbance at 450 and 600 nm.
3′-Diaminobenzidine (DAB) Staining and Hydrogen Peroxide (H2O2) Measurement
H2O2 accumulation was detected by DAB staining buffer as described by Daudi and O’Brien (2012). Ten-day-old seedlings were submerged in 150 mM NaCl solution for 12 h, then stained with DAB solution for 12 h. After removal of chlorophyll by acetic acid, ethanol, and glycerol (v:v:v, 1:3:1), the seedlings were photographed.
To determine the H2O2 concentration of the seedlings, a colorimetric technique using potassium iodide buffer was used (Junglee et al. 2014). Five hundred milligrams of 150 mM NaCl-treated frozen samples were homogenized at 4 °C in 2 mL of extraction buffer containing 0.1% TCA, 1 M potassium iodide, and 10 mM potassium phosphate buffer (pH 8.0). The homogenate was centrifuged at 12,000 rpm for 20 min at 4 °C. Absorbance of the supernatant layer was read at 350 nm in a UV/VIS spectrophotometer (JASCO, Tokyo, Japan). A calibration standard curve obtained with H2O2 solution prepared in 0.1% TCA was used for quantification.
Total RNA Extraction and Quantitative Real-Time PCR (qPCR) Analysis
Total RNA was isolated from the seedling samples using the Plant RNeasy extraction kit (Qiagen, Valencia, CA). Genomic DNA was removed from the total RNA preparation by treatment with RNase-free DNase I enzyme according to the manufacturer’s instructions (Qiagen). The concentration of total RNA was quantified using spectrophotometric measurements, and 3 μg of total RNA was resolved on a 1.0% formaldehyde agarose gel to determine RNA concentration and visualize its integrity. qPCR was carried out using the CFX Connect quantitative PCR apparatus (Bio-Rad, Hercules, CA, USA). The IQ SYBR Green Supermix kit (Bio-Rad) was used for qPCR according to the manufacturer’s instructions. Actin 1 (ACT1) was used as an internal control. Quantitative analysis was carried out using the delta-delta-CT method of Livak and Schmittgen (2001). Data were obtained from three biological replicates with ≥ 12 pooled seedlings per genotype per replicate. The reaction primers are listed in Supplementary Table S1.
Statistical Analysis
Statistical analyses were performed using SPSS 23.0 software (IBM Co, Armonk, NY), and included one-way analysis of variance (ANOVA) and Duncan’s multiple-range test. Different letters indicate statistically significant differences at P < 0.05 by Duncan’s multiple-range test.
Results
AHL is Upregulated by NaCl Treatment
AHL encodes PAP phosphatase, which participates in secondary sulfate metabolism (Chen et al. 2011). Recently, we discovered that AHL regulates the responses of young Arabidopsis seedlings to abiotic stresses such as abscisic acid (ABA), drought, osmotic stress, oxidative stress, and high Glc treatment (Shin et al. 2019; Nguyen et al. 2019). Previously, Fatma et al. (2014) reported that excess sulfate supplementation improves photosynthetic capacity and growth in mustard plants under salt stress conditions via a reduction in glutathione content.
To investigate the function of AHL in response to salt stress, we initially measured the expression pattern of AHL in 10-day-old Arabidopsis seedlings subjected to 150 mM NaCl treatment. Quantitative real-time PCR (qPCR) showed that AHL transcript levels were significantly increased in samples within 6–12 h of salt stress treatment, but decreased thereafter (Fig. 1A). RD29A (Responsive to Dehydration 29A) was used as a positive control for salt stress treatment (Fig. 1A). The results indicate that AHL is up-regulated by salt treatment.
AHL is induced by salt stress. A Ten-day-old seedlings were harvested for total RNA extraction at the indicated time points under 150 mM NaCl treatment, and AHL expression levels were quantified by qPCR. RD29A served as a control for salt stress treatment. The bars represent the mean values (± SD) of three biological replicates. Different letters above the bars indicate a statistically significant difference at P < 0.05 by Duncan’s multiple-range test. B Histochemical GUS activity for AHL in response to salt stress. Ten-day-old whole seedlings of the AHLpro-GUS transgenic (2-1 and 3-3) lines were subjected to 150 mM NaCl. After 12 h of NaCl treatment, the seedlings were stained for GUS activity, with H2O-treated seedlings used as a control
To gain further insight into the expression of AHL in Arabidopsis seedling organs, we analyzed the GUS staining of 1925-bp AHL promoter (pro)-GUS transgenic lines (2-1 and 3-3) following water (H2O) or salt (150 mM NaCl) treatment. As shown in Fig. 1B, GUS staining was observed in the veins of leaves, the primary root and lateral roots under H2O treatment conditions in two individual AHL pro-GUS transgenic lines. When 150 mM NaCl was applied to AHL pro-GUS transgenic seedlings for 12 h, GUS activity increased to a greater extent than under salt-untreated conditions (Fig. 1B). Furthermore, GUS staining was detected in the hypocotyl (arrow) after salt treatment, but not under normal conditions. Thus, AHL level is altered in response to without or with salt stress in roots, leaf veins, and hypocotyl tissues.
Overexpression of AHL Confers High Tolerance to Salt Stress
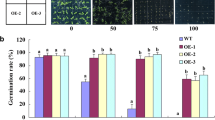
Since AHL expression is upregulated by NaCl treatment (Fig. 1), we examined whether AHL is required for regulation of the tolerance of AHL transgenic plants to salt stress by measuring seed germination and survival rates. The seeds of WT, ahl, AHL-RNAi (ri 2-3, ri 5-2), and AHL-overexpressing (OE 2-2, OE 5-1) plants were germinated on one-half strength MS medium containing 0 or 150 mM NaCl. The germination rates did not differ between WT, ahl, and AHL transgenic plants in the absence of 150 mM NaCl (data not shown). With exposure to 150 mM NaCl at 4 days after seed sowing, 84–92% of the two types of AHL-overexpressing seeds germinated, as compared to 63% of WT, while 53% and 47–49% of the ahl and two individual AHL-RNAi lines, respectively, germinated (Fig. 2A). These findings indicate that seed germination in AHL-overexpressing lines is more likely than that in ahl mutant and AHL-RNAi lines to be tolerant to salt stress. High salt stress also resulted in significant differences in survival rate at 18 days after seed sowing: 65–69% of the two types of AHL-overexpressing seeds produced green leaves, compared with 53% of WT and 23% and 27–36% of ahl and the two AHL-RNAi lines, respectively (Fig. 2B, C). Collectively, these observations suggest that AHL expression is associated with early seedling growth under salt stress conditions.
Physiological responses of ahl mutant and AHL transgenic plants to salt stress. A Comparison of the germination rate of WT, ahl mutant and AHL transgenic plants under salt stress conditions. The seed germination rate on 150 mM NaCl is shown for samples of each genotype on different days. The points represent the mean values (± SD) of three biological replicates. Different letters above the bars indicate a statistically significant difference at P < 0.05 by Duncan’s multiple-range test. B Phenotypes of WT, ahl, AHL-RNAi (ri 2-3, ri 5-2) and AHL-overexpressing (OE 2–2, OE 5–1) seedlings exposed to 150 mM NaCl for 18 d. C Quantification of the survival rates of WT, ahl, AHL-RNAi and AHL-OE seedlings exposed to 150 mM NaCl for 18 d. D Two-week-old seedlings of each sample were subjected to 0 and 150 mM NaCl for 12 h and used to measure proline concentration. The bars represent the mean values (± SD) of three biological replicates. Different letters above the bars in C, D indicate a statistically significant difference at P < 0.05 by Duncan’s multiple-range test.
A previous report demonstrated that accumulation of Pro plays a role as an adaptive mechanism that counteracts salt stress (Hasanuzzaman et al. 2013; Hayat et al. 2012). To measure Pro content in AHL transgenic lines under salt stress conditions, Pro content assays were applied to whole seedlings of WT, ahl, AHL-RNAi (ri 2-3), and AHL-overexpressing (OE 2-2) plants. In the absence of salt stress treatment, the Pro levels in all of these samples were similar (Fig. 2D). However, in the presence of 150 mM NaCl, the Pro content was higher in the AHL-OE line than in WT, ahl, and AHL-RNAi plants, while it was lower in the ahl and AHL-RNAi lines than in WT plants (Fig. 2D). This analysis reveals that AHL is necessary to regulate Pro accumulation under salt stress conditions.
AHL Regulates H2O2 Accumulation and Oxidative Damage During Salt Stress
Salt stress leads to the accumulation of ROS such as H2O2 and oxygen-free radicals in plant tissue (Hasanuzzaman et al. 2013; Ahanger and Agarwal 2017). To evaluate the effects of AHL expression on ROS production under salt stress conditions, whole seedlings of WT, ahl, and AHL transgenic samples were treated with 150 mM NaCl for 12 h, then stained with DAB solution for the H2O2 accumulation assay. In the absence of salt stress, H2O2 levels were similar among the WT, ahl, AHL-RNAi (ri2-3), and AHL-overexpressing (OE2-2) seedlings (Fig. 3A). With salt stress treatment, H2O2 production was higher in ahl and ri2-3 whole seedlings than in WT and OE2-2 seedlings (Fig. 3A). Subsequently, we measured H2O2 production in high salt-treated whole seedlings. As shown in Fig. 3B, H2O2 levels were significantly higher in the ahl and AHL-RNAi lines than in WT and AHL-overexpressing seedlings, while the H2O2 level was significantly lower in the AHL-overexpressing transgenic line than in WT seedlings under salt stress conditions. These observations demonstrate that AHL is involved in the regulation of hydrogen peroxide accumulation under salt stress.
H2O2 and MDA contents in ahl mutant and AHL transgenic plants during salt stress treatment. A Ten-day-old whole seedlings of each sample were treated with H2O (control) and 150 mM NaCl for 12 h and DAB staining was conducted to assess H2O2 accumulation. B, C Quantification of H2O2 (B) and MDA (C) accumulation in 10-day-old seedlings after treatment with H2O (control) or 150 mM NaCl for 12 h. The bars represent the mean values (± SD) of three biological replicates. Different letters above the bars indicate a statistically significant difference at P < 0.05 by Duncan’s multiple-range test
In general, MDA content is considered an indicator of oxidative damage (Sadak et al. 2020). Thus, we evaluated oxidative damage in ahl and AHL-RNAi seedlings subjected to salt stress conditions by analyzing the MDA content of the seedlings. Without salt stress treatment, the basal MDA levels of all sample plants were similar (Fig. 3C). With salt stress treatment, the MDA contents of ahl and AHL-RNAi seedlings were greater than those of WT or AHL-overexpressing transgenic seedlings (Fig. 3C), implying that AHL regulates the salt stress response by participating in ROS-related processes.
Expression of Salt Stress-Responsive Genes in AHL Transgenic Lines
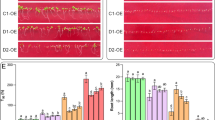
To examine the salt stress response in AHL transgenic lines at the molecular level, we analyzed the transcriptional levels of the ABA-insensitive 5 (ABI5), Arginine Decarboxylase 1 (ADC1), ADC2, Alternative Oxidase 1a (AOX1a), ABA-Responsive Element Binding 3 (AREB3), Arabidopsis thaliana Oxidation-related Zinc Finger 2 (AtOZF2), Delta 1-pyrroline-5-carboxylate synthase 1 (P5CS1), Response to Drought 26 (RD26), and Respiratory burst oxidase homologue-D (RbohD) genes, which are known to be induced by ABA, dehydration, salinity, oxidative, and biotic stresses (Lopez-Molina and Chua 2000; Li et al. 2015; Sánchez-Rangel et al. 2016; Kim et al. 2017; Zhang et al. 2017; Shin et al. 2019). qPCR analyses showed that, in comparison to WT plants, the expression levels of ABI5, ADC1, ADC2, AOX1a, AREB3, AtOZF2, P5CS1, and RD26 were higher in the AHL-overexpressing (OE2-2) lines, whereas they were lower in the ahl and AHL-RNAi (ri2-3) lines under salt stress conditions (Fig. 4A–H). RbohD expression was higher in salt stress-treated ahl and ri2-3 plants than in WT, while it was lower in salt stress-treated OE2-2 plants than in WT (Fig. 4I). These observations indicate that both ahl and AHL-RNAi lines can suppress salt stress-induced ABI5, ADC1, ADC2, AOX1a, AREB3, AtOZF2, P5CS1, and RD26 or enhance salt stress-induced RbohD expression. Hence, it is likely that AHL can act positively or negatively depending on the signal required for modulating the type of salt stress-inducible genes under high salinity condition.
qPCR analysis of salt stress-related genes in ahl mutant and AHL transgenic plants. A–I Expression of ABI5 (A), ADC1 (B), ADC2 (C), AOX1a (D), AREB3 (E), AtOZF2 (F), P5CS1 (G), RD26 (H) and RbohD (I) in WT, ahl, AHL-RNAi (ri 2-3) and AHL-overexpressing (OE 2-2) plants under salt stress. Ten-day-old seedlings were treated with H2O or 150 mM NaCl for 12 h. Total RNA was extracted from each type of seedling and analyzed by qPCR. The bars represent the mean values (± SD) of three biological replicates. Different letters above the bars indicate a statistically significant difference at P < 0.05 by Duncan’s multiple-range test
The ahl Mutation Reduces Cysteine Levels Under Salt Stress Conditions
Cys is a vital organic amino acid that is involved in reduced sulfur synthesis in plants. Cys is also an important precursor for essential components, such as antioxidant glutathione, vitamins, thionins, and glucosinolates (Álvarez et al. 2011; Romero et al. 2014; Wawrzynska et al. 2015). Moreover, Cys acts as a bio-regulator and plays an important role in promoting plant growth and productivity under high glucose, oxidative, and salt stress conditions (Genisel et al. 2015; Romero et al. 2014; Nguyen et al. 2019). To further assess the response to salt stress, the Cys contents in salt stress-treated rosette leaves of WT, ahl, and AHL transgenic plants were estimated. As shown in Fig. 5A, the Cys content was higher in the AHL-overexpressing line than in WT, ahl, and AHL-RNAi plants, while it was lower in the ahl and AHL-RNAi lines than in WT plants in the presence or absence of high NaCl concentration. This result indicates that AHL modulates Cys production in A. thaliana.
Cys concentration and transcript levels of sulfate metabolism-related genes in ahl mutant and AHL transgenic plants under salt stress. A Cys concentration in 10-day-old seedlings after treatment with H2O (control) or 150 mM NaCl for 12 h. The bars represent the mean values (± SD) of three biological replicates. Different letters above the bars indicate a statistically significant difference at P < 0.05 by Duncan’s multiple-range test. B–E qPCR results of several genes related to sulfate metabolism in ahl mutant and AHL transgenic plants under salt stress conditions. Expression of APK1 (B), APR1 (C), OASA1 (D) and SiR (E) in WT, ahl, AHL-RNAi (ri 2-3) and AHL-overexpressing (OE 2-2) plants under salt stress. Ten-day-old seedlings were treated with H2O or 150 mM NaCl for 12 h. Total RNA was extracted from each type of seedling and analyzed by qPCR. The bars represent the mean values (± SD) of three biological replicates. Different letters above the bars indicate a statistically significant difference at P < 0.05 by Duncan’s multiple-range test
The change in Cys content due to AHL expression prompted us to assess genes, including Adenosin-5’-phosphosulfate kinase 1 (APK1), 3'-Phosphoadenosine 5'-phosphosulfate reductase 1 (APR1), O-Acetylserine (thiol), Lyase 1 (OASA1), and Sulfite reductase (SiR), that are known to be responsible for sulfate metabolism. qPCR analyses showed that, compared to WT plants, the expression levels of APK1, APR1, and OASA1 were higher in the AHL-overexpressing (OE2-2) lines and lower in salt stress-treated ahl and AHL-RNAi (ri2-3) lines (Fig. 5B–D). The expression level of SiR in salt stress-treated OE2-2 plants was greater than that in WT, ahl, and ri2-3 plants, whereas transcription of SiR showed similar levels in WT, ahl, and ri2-3 plants during salt stress treatment (Fig. 5E). These results indicate that AHL regulates the expression of these sulfate metabolism-related genes under salt stress, implying that AHL participates in the salt stress response through a sulfate metabolism-mediated pathway.
Cys Modulates Salt Stress-Induced Inhibition of Early Seedling Growth
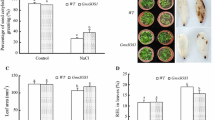
Previous data showed that the accumulation of Cys and the expression of OASA1 (Cys biosynthesis enzyme) were lower in the ahl and AHL-RNAi lines than in WT and AHL-overexpressing plants during salt stress treatment (Fig. 5A, D). In addition, the expression levels of polyamine synthesis-related genes (ADC1 and ADC2) in the cysteine-methionine derivative pathway (Khan et al. 2014) were suppressed to a greater extent in ahl and AHL-RNAi lines compared to WT and AHL-overexpressing plants under salt stress conditions (Fig. 4B, C). Hence, we tested whether the sensitivity to salt stress of the ahl and AHL-RNAi lines could be rescued by application of exogenous Cys. To analyze the effect of Cys on the salt stress response, we estimated the survival rate of WT, ahl, AHL-RNAi (ri2-3, ri5-2), and AHL-overexpressing (OE2-2, OE5-1) seedlings under salt stress conditions in the presence of 40 µM Cys. Under high-NaCl conditions, all sample seedlings treated with Cys showed a higher survival rate than untreated seedlings (Fig. 6). These results indicate that Cys can modulate salt stress-induced inhibition of survival in A. thaliana. As shown in Fig. 6, when treated with Cys, the survival rates of WT, ahl, and AHL-RNAi seedlings were similar under high salt stress conditions, but the survival rates were lower than that of AHL-overexpressing transgenic lines in response to salt stress. This survival data suggests that Cys plays an important role in regulating seedling growth under salt stress conditions.
Comparison of the Cys response of ahl mutant and AHL transgenic plants under salt stress. A Effect of exogenous Cys on growth in WT, ahl, AHL-RNAi (ri 2-3, ri 5-2) and AHL-overexpressing (OE 2-2, OE 5-1) seedlings under salt stress. Seedlings were grown on one-half strength MS plant medium containing 40 μM Cys with 150 mM NaCl for 18 d. B Survival rate of WT, ahl and AHL transgenic seedlings subjected to 150 mM NaCl or 150 mM NaCl plus 40 μM Cys for 18 d. The bars represent the mean values (± SD) of three biological replicates. Different letters above the bars indicate a statistically significant difference at P < 0.05 by Duncan’s multiple-range test
Discussion
Regulation of sulfate metabolism alleviates the adverse effects of salt stress, as sulfate metabolites modulate a wide range of plant growth processes (Khan et al. 2014). Sulfur is an essential component in plants; it is necessary for abiotic stress tolerance and an integral part of important metabolic molecules, such as sulfolipids, non-enzymatic antioxidants (glutathione), and amino acids (cysteine and methionine) (Nocito et al. 2007; Khan et al. 2014). In addition, sulfur-containing compounds are connected with the antioxidant system in plants subjected to salt stress (Khan et al. 2014). Previous studies have reported that AHL, which encodes for a component of secondary sulfate metabolism, PAP phosphatase (Chen et al. 2011), modulates sulfate metabolism. Regulated abiotic stress responses that affect sulfate metabolism include the ABA, drought, high glucose, and osmotic responses (Shin et al. 2019; Nguyen et al. 2019).
In the present study, using AHL transgenic lines (Shin et al. 2019), we further investigated the mechanism through which AHL underlies the sulfate metabolic pathway under high-salt conditions. A quantitative analysis of AHL expression under salt stress condition showed that AHL is up-regulated by NaCl (Fig. 1A). In addition, in the seedling stage, strong AHL activity was detected in hypocotyl tissue under salt stress conditions, but not under normal conditions, implying that AHL may perform a specific function at the spatial level in response to salt stress (Fig. 1B).
Increased expression of sulfate metabolism-responsive genes is related to improved salt tolerance in plants. The activity of O-acetylserine (thiol) lyase (OASTL) in Typha and Phragmites plants increases under salt stress conditions, and this increased activity fulfills the higher demand for cysteine to ensure protection and adaptation in response to salt stress (Fediuc et al. 2005). Additionally, overexpression of the BrECS (for Brassica juncea Glutamylcysteine Synthetase) gene in transgenic rice lines results in increased tolerance to high salt stress conditions by preventing membrane oxidation (Bae et al. 2013). Here, as in the salt stress assay, the germination and survival rates of the ahl and AHL-RNAi lines were lower than those of WT and AHL-overexpressing plants under salt stress conditions (Fig. 2A–C). In addition, the survival rate of AHL-overexpressing transgenic plants was higher than that of WT plants following NaCl treatment (Fig. 2B, C). These data suggest that AHL, which codes for the secondary sulfate metabolism-related enzyme PAP phosphatase, serves as a positive regulator in salt stress tolerance.
In plants, the amino acid Pro functions as an osmolyte or chaperone molecule to maintain cell turgor or stabilize the structure of a protein; it also serves as an antioxidant that regulates ROS levels (Hare et al. 1999). In addition, Pro accumulation is believed to enhance salt stress tolerance in plants (Hasanuzzaman et al. 2013; Hayat et al. 2012). Pro accumulation was higher in AHL-overexpressing lines than in WT, ahl, and AHL-RNAi plants after salt stress treatment (Fig. 2D). Furthermore, the Pro content of ahl and AHL-RNAi lines was lower than that of WT plants under salt stress conditions (Fig. 2D), which suggests that AHL may be responsible for the reduction in salt stress-induced sensitivity via regulation of the levels of ROS and MDA (an indicator of oxidative damage). Consequently, our data showed an apparent difference in H2O2 and MDA levels between AHL-RNAi and AHL-overexpressing transgenic lines during salt stress treatment (Fig. 3). The seedlings of the ahl and AHL-RNAi lines showed increased H2O2 and MDA induction under salt stress conditions compared with those of AHL-overexpressing transgenic lines (Fig. 3). Furthermore, the results shown in Fig. 4 also revealed a distinct difference in the expression levels of salt stress-associated genes between AHL-RNAi and AHL-overexpressing transgenic lines under salt stress conditions. These transcriptional profile changes imply that AHL enhances the capacity for anti-salt stress by modulating salinity-related signaling transduction. Finally, AHL activity may be a component of the oxidative stress or salinity signal that influences the adaptation mechanism.
In plants, Cys acts not only as an amino acid in protein synthesis but also as a precursor in important biological components including antioxidants, fatty acids, vitamins, cofactors, and methionine (Khan et al. 2014; Romero et al. 2014). In addition, conservation of Cys is very important for the function and signaling processes of metabolic enzymes under environmental stressors (Meyer and Hell 2005). Several groups have established a relationship between increased Cys concentration and salt stress tolerance in plants (Barroso et al. 1999; Fediuc et al. 2005; Bae et al. 2013). Consequently, the results in Fig. 5A show an apparent difference in Cys levels between AHL-RNAi and AHL-overexpressing transgenic lines during salt stress treatment. Seedlings of the ahl and AHL-RNAi lines showed a greater reduction in Cys accumulation under salt stress conditions compared with those of AHL-overexpressing transgenic lines (Fig. 5A). Furthermore, the results in Fig. 5B–E also show a distinct difference in transcription of sulfate metabolism pathway-related genes between the ahl mutant and AHL-overexpressing transgenic lines under salt stress conditions. Therefore, these data imply that AHL participates in salt stress tolerance in a Cys-dependent manner or through a sulfate metabolism-mediated pathway. In addition, exogenous application of Cys can rescue the salt stress-induced sensitive phenotype of ahl mutant and AHL-RNAi lines, such that their survival rates were similar to that of WT plants (Fig. 6). These results indicate that AHL-induced salinity tolerance is mainly dependent on Cys concentration.
Collectively, our results demonstrate that AHL-overexpressing transgenic lines exhibit enhanced salt stress tolerance. Under salt stress conditions, AHL positively regulates survival by increasing the expression of salt stress tolerance- and sulfate metabolism-associated genes. Based on Figs. 2B, 3A, B, 5A, the regulation of AHL expression is important for Pro, H2O2, and Cys biosynthesis in response to salt stress. Thus, AHL is linked to the regulation of Cys concentration for mediation of the salt stress response, although its biochemical and molecular functions have not yet been fully investigated. Further functional studies of AHL in Cys metabolism are needed to elucidate salt stress response in plants.
Abbreviations
- Cys:
-
Cysteine
- Pro:
-
Proline
- qPCR:
-
Quantitative real-time polymerase chain reaction
- WT:
-
Wild-type
References
Ahanger MA, Agarwal RM (2017) Salinity stress induced alterations in antioxidant metabolism and nitrogen assimilation in wheat (Triticum aestivum L.) as influenced by potassium supplementation. Plant Physiol Biochem 115:449–460
Ahmad P, Sarwat M, Sharma S (2008) Reactive oxygen species. Antioxidants and signaling in plants. J Plant Biol 51:167–173
Álvarez C, Ángeles BM, Romero LC, Gotor C, García I (2011) Cysteine homeostasis plays an essential role in plant immunity. New Phytol 193:165–177
Bae MJ, Kim YS, Kim IS, Choe YH, Lee EJ, Kim YH, Yoon HS (2013) Transgenic rice overexpressing the Brassica juncea gamma-glutamylcysteine synthetase gene enhances tolerance to abiotic stress and improves grain yield under paddy field conditions. Mol 31:931–945
Barroso JB, Corpas FJ, Carreras A, Sandalio LM, Valderrama R, Palma JM, Lupiáñez JA, del Río LA (1999) Localization of nitric-oxide synthase in plant peroxisomes. J Biol Chem 274:36729–36733
Bates LS, Waldren RP, Teare ID (1973) Rapid determination of free proline for water-stress studies. Plant Soil 39:205–207
Chen H, Zhang B, Hicks LM, Xiong L (2011) A nucleotide metabolite controls stress-responsive gene expression and plant development. PLoS ONE 6:e26661
Daudi A, O’Brien JA (2012) Detection of hydrogen peroxide by DAB staining in Arabidopsis leaves. Bio-Protoc 2:e263
Fatma M, Asgher M, Masood A, Khan NA (2014) Excess sulfur supplementation improves photosynthesis and growth in mustard under salt stress through increased production of glutathione. Environ Exp Bot 107:55–63
Fediuc E, Lips SH, Erdei L (2005) O-acetylserine (thiol) lyase activity in Phragmites and Typha plants under cadmium and NaCl stress conditions and the involvement of ABA in the stress response. J Plant Physiol 162:865–872
Genisel M, Erdal S, Kizilkaya M (2015) The mitigating effect of cysteine on growth inhibition in salt-stressed barley seeds is related to its own reducing capacity rather than its effects on antioxidant system. Plant Growth Regul 75:187–197
Hare PD, Cress WA, Staden JV (1999) Proline synthesis and degradation: a model system for elucidating stress-related signal transduction. J Exp Bot 50:413–434
Hasanuzzaman M, Nahar K, Fujita M (2013) Plant response to salt stress and role of exogenous protectants to mitigate salt-induced damages. In: Ahmad P, Azooz M, Prasad M (eds) Ecophysiology and responses of plants under salt stress. Springer, New York, pp 25–87
Hayat S, Hayat Q, Alyemeni MN, Wani AS, Pichtel J, Ahmad A (2012) Role of proline under changing environments: a review. Plant Signal Behav 7:1456–1466
Iqbal N, Shahid US, Khan NA (2015) Nitrogen availability regulates proline and ethylene production and alleviates salinity stress in mustard (Brassica juncea). J Plant Physiol 178:84–91
Junglee S, Urban L, Sallanon H, Lopez-Lauri F (2014) Optimized assay for hydrogen peroxide determination in plant tissue using potassium iodide. AIDS Patient Care STDs 5:730–736
Khan NA, Khan MIR, Asgher M, Fatma M, Masood A et al (2014) Salinity tolerance in plants: revisiting the role of sulfur metabolites. J Plant Biochem Physiol 2:120
Kim AR, Min JH, Lee KH, Kim CS (2017) PCA22 acts as suppressor of atrzf1 to mediate proline accumulation in response to abiotic stress in Arabidopsis. J Exp Bot 68:1797–1809
Kopriva S, Mugford SG, Baraniecka P, Lee BR, Matthewman CA, Koprivova A (2012) Control of sulfur partitioning between primary and secondary metabolism in Arabidopsis. Front Plant Sci 3:163
Li X, Zhang H, Tian L, Huang L, Liu S, Li D et al (2015) Tomato SIRbohB, a member of the NADPH oxidase family, is required for disease resistance against Botrytis cinerea and tolerance to drought stress. Front Plant Sci 6:463
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-delta delta CT) method. Methods 25:402–408
Lopez-Molina L, Chua NH (2000) A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol 41:541–547
Meyer AJ, Hell R (2005) Glutathione homeostasis and redox-regulation by sulfhydryl groups. Photosynth Res 86:435–457
Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7:405–410
Nguyen TV, Chung MS, Chung JS, Kim CS (2019) proline content alterative 17 (pca17) is involved in glucose response through sulfate metabolism-mediated pathway. Plant Physiol Bioch 143:320–328
Nocito FF, Lancilli C, Giacomini B, Sachi GA (2007) Sulfur metabolism and cadmium stress in higher plants. Plant Stress 1:142–1156
Priya M, Raj AS, Muthukumar PV, Bharathiraja B (2016) Experimental study on biochemical and physiological adaptation of mercury accumulation and tolerance in Clitoria ternatea L. JCHPS 9:298–303
Romero LC, Aroca MA, Laureano-Marín AM, Moreno I, García I, Gotor C (2014) Cysteine and cysteine-related signaling pathways in Arabidopsis thaliana. Mol Plant 7:264–276
Sadak MS, El-Hameid ARA, Zaki FSA, Dawood MG, El-Awadi ME (2020) Physiological and biochemical responses of soybean (Glycine max L.) to cysteine application under sea salt stress. Bull Natl Res Centre 44:1
Sánchez-Rangel D, Chávez-Martíne AI, Rodríguez-Hernández AA, Maruri-López I, Urano K, Shinozaki K, Jiménez-Bremont JF (2016) Simultaneous silencing of two arginine decarboxylase genes alters development in Arabidopsis. Front Plant Sci 7:300
Shin DJ, Min JH, Nguyen TV, Kim YM, Kim CS (2019) Loss of Arabidopsis Halotolerance 2-like (AHL), a 3′- phosphoadenosine-5′-phosphate phosphatase, suppresses insensitive response of Arabidopsis thaliana ring zinc finger 1 (atrzf1) mutant to abiotic stress. Plant Mol Biol 99:363–377
Takahashi H, Kopriva S, Giordano M, Saito K, Hell R (2011) Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu Rev Plant Biol 62:157–184
Wawrzynska A, Moniuszko G, Sirko A (2015) Links between ethylene and sulfur nutrition-a regulatory interplay or just metabolite association? Front Plant Sci 6:1053
Zhang X, Ivanova A, Vandepoele K, Radomiljac J, Van de Velde J, Berkowitz O et al (2017) The transcription factor MYB29 is a regulator of Alternative Oxidase 1a. Plant Physiol 173:1824–1843
Acknowledgements
This work was supported by a National Research Foundation (NRF) of Korea grant funded by the Korean government (Ministry of Science and ICT) (2021R1A2C1009748).
Author information
Authors and Affiliations
Contributions
TVN and CSK designed the experiments; TVN, JIK, CRP, MSC, and CSK performed the experiments and interpreted the data; TVN, MSC, and CSK wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
12374_2021_9320_MOESM1_ESM.tif
Expression level of AHL in WT, ahl, and AHL transgenic plants. A and B mRNA level of AHL was analyzed by RT-PCR (A) and qPCR (B) using total RNA extracted from 10-day-old WT, ahl, AHL-RNAi (ri2-3, ri5-2), and AHL-overexpressing (OE2-2, OE5-1) transgenic plants. Actin 1 (ACT1) was used as internal control. The bars represent the mean values (± SD) of three biological replicates. Different letters above the bars indicate statistically significant differences at P < 0.05 by Duncan’s multiple-range test (TIF 1746 kb)
Rights and permissions
About this article
Cite this article
Van Nguyen, T., Kim, JI., Park, CR. et al. Increased Cysteine Accumulation is Essential for Salt Stress Tolerance in Arabidopsis Halotolerance 2-Like (AHL)-Overexpressing Transgenic Plants. J. Plant Biol. 64, 475–485 (2021). https://doi.org/10.1007/s12374-021-09320-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12374-021-09320-7