Abstract
The complete genome of a double-stranded RNA (dsRNA) mycovirus, Phoma matteuccicola partitivirus 1 (PmPV1) was sequenced. It consists of two dsRNA segments, 1664 bp (dsRNA-1) and 1383 bp (dsRNA-2) in length, each containing a single open reading frame (ORF) potentially encoding a 46.78-kDa protein and a 40.92-kDa protein, respectively. dsRNA-1 encodes a putative polypeptide with a conserved RNA-dependent RNA polymerase (RdRp) domain that shows sequence similarity to the corresponding proteins of partitiviruses. The protein encoded by dsRNA-2 has no significant similarity to the typical coat proteins (CPs) of partitiviruses, but structure analysis nevertheless suggested that it might function as a coat protein. Purified viral particles of PmPV1 were isometric and approximately 29 nm in diameter. Phylogenetic analysis showed that PmPV1 is closely related to members of the genus Gammapartitivirus within the family Partitiviridae but forms a separate branch with Colletotrichum acutatum RNA virus 1 and Ustilaginoidea virens partitivirus 2. This is the first report of the full-length nucleotide sequence of a novel virus of the genus Gammapartitivirus infecting P. matteuccicola strain LG915, the causal agent of leaf blight of Curcuma wenyujin.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mycoviruses (fungal viruses) are ubiquitous in all major important taxonomic groups of filamentous fungi, yeasts and oomycetes [1]. The mycoviruses with dsRNA genomes are now classified into seven families, namely Partitiviridae, Totiviridae, Chrysoviridae, Reoviridae, Endornaviridae, Quadriviridae, and the proposed family “Megaviridae” [2]. Members of the family Partitiviridae are currently subdivided into five genera, of which alphapartitiviruses and betapartitiviruses mainly infect fungi and plants, gammapartitiviruses infect only fungi, deltapartitiviruses have only been isolated from plants, and cryspoviruses only infect protozoa, based on the latest International Committee on Taxonomy of Viruses (ICTV) standards. The genomes of partitiviruses consist of two linear dsRNA segments, ranging in size from 1.4 to 2.4 kb, each containing a single open reading frame (ORF). The larger genome segment often codes for the RNA-dependent RNA polymerase (RdRp), whereas the smaller one usually codes for the coat protein (CP).
Phoma matteuccicola, which causes leaf blight disease in Curcuma wenyujin, is a destructive pathogenic fungus of C. wenyujin, in Hainan, China, that was first reported in our laboratory [3]. Partitiviruses have been extensively studied in many plants and fungi but had not been reported in P. matteuccicola until our present work. Here, we isolated and identified a novel virus from P. matteuccicola strain LG915 and named it “Phoma matteuccicola gammapartitivirus 1” (PmPV1).
Provenance of the virus material
P. matteuccicola strain LG915 was isolated from C. wenyujin showing leaf blight disease in Hainan, China, in 2016 and was maintained on potato dextrose agar (PDA) slants at 4-8 °C. A mycelium plug 5 mm in diameter was cut off from the edge of strain LG915 and cultured on a PDA plate covered with cellophane membrane at 25 °C for 14 days for dsRNA extraction. The dsRNAs of LG915 were extracted from approximately 3.0 g of harvested mycelia according to the protocols of Morris and Dodds, with minor modifications [4]. To improve the yield of dsRNA, a nucleic acid co-precipitator was added before precipitating RNA with ethanol. The dsRNAs were further treated with DNase I and S1 nuclease (Takara, Japan) to eliminate contaminating DNA and single-stranded RNA (ssRNA) and then used to construct a complementary DNA (cDNA) library [5]. The terminal sequences of the dsRNA were determined using a slightly modified protocol of rapid amplification of cDNA ends (RACE) [6]. Each base was identified by sequencing of at least three separate clones in both directions. The full-length cDNA sequences of the two dsRNA segments from P. matteuccicola strain LG915 were deposited in the GenBank database under the accession numbers MK211276 and MK211277 for dsRNA-1 and dsRNA-2, respectively. ORFs were identified using the NCBI ORF Finder program (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Multiple sequence alignments of the RdRp sequences were processed using the Clustal-X program [7]. Phylogenetic trees were constructed using the maximum-likelihood (ML) method in Molecular Evolutionary Genetics Analysis (MEGA) version 6.0 with 1,000 bootstrap replicates [8]. Virus particles were purified using the sucrose density gradient method described previously [5]. Fractions with viral particles were observed under a transmission electron microscope (TEM) after staining with 2% (w/v) phosphotungstic acid solution.
Sequence properties
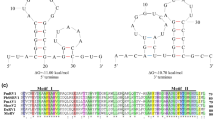
The results indicated that P. matteuccicola strain LG915 was infected by a novel partitivirus, PmPV1. The complete genome has two segments, which are referred to as dsRNA-1 and dsRNA-2. The complete sequences of the two dsRNA segments of PmPV1 were 1664 bp (dsRNA-1) and 1383 bp (dsRNA-2) in length with a GC content of 46% and 48%, respectively (Fig. 1A). The 5’ untranslated region (UTR) and 3’-UTR of PmPV1 were 349 nt and 76 nt long in dsRNA-1 and 136 nt and 116 nt long in dsRNA-2 (Fig. 1A). Comparison of the UTR sequences at the 5’ and 3’ ends of the two dsRNA segments revealed the presence of conserved regions (Fig. 1B).
(A) Schematic representation of the genomic organization of PmPV1. The open reading frame (ORF) and the untranslated regions (UTRs) are indicated by an open bar and single lines, respectively. The gray bar indicates the conserved RdRp domain. The nucleotide positions of the initiation and termination codons and the molecular weights of the predicted proteins are shown above the ORFs. The numbers below the arrows indicate the length of the non-coding sequence. (B) Terminal sequence regions of the PmPV1 genome. Identical sequences of the 5’-UTR and 3’-UTR of the two dsRNAs are reverse highlighted. (C) PmPV1 particles of strain LG915. TEM images (negative staining) of the virus particles of PmPV1. (D) Agarose gel electrophoresis of dsRNA extracted from mycelia of strain LG915 (lane 1) and purified viral particles of PmPV1 (lane 2). M indicates molecular markers (DL15000 DNA marker)
dsRNA-1 contains a single ORF (ORF1) in its plus strand (Fig. 1A) and encodes a 411-amino-acid (aa) protein with a molecular weight of 46.78 kDa. A database search using BLASTp showed that the RdRp sequence of PmPV1 is 66% identical to that of Ustilaginoidea virens partitivirus 2 (UvPV2) [9] and 53% identical to that of Colletotrichum acutatum RNA virus 1 (CaRV1) [10]. A search of the Conserved Domain Database (CDD) and polyprotein sequence alignment showed that the predicted RdRp domain contains six conserved motifs that are typical of members of the family Partitiviridae (Fig. 2A).
(A) Sequence alignment of PmPV1 RdRp motifs with those of selected viruses of the family Partitiviridae. Horizontal lines above the alignment indicate the six motifs. Shaded areas show identical amino acid residues. Asterisks, colons and dots indicate the identical amino acids, conservative differences and semi-conservative differences, respectively. (B) Phylogenetic analysis based on the deduced amino acid sequences of putative RdRps using the maximum-likelihood (ML) method with 1,000 bootstrap replicates. The scale bar represents a genetic distance of 0.2 amino acid substitutions per site. A red circle indicates the novel mycovirus PmPV1. The abbreviations of virus names and GenBank accession numbers are as follows: DdV1, Discula destructiva virus 1 (NP_116716.1); DdV2, Discula destructiva virus 2 (NP_620301.1); BfPV1, Botryotinia fuckeliana partitivirus 1 (CAM33266.1); VdPV1, Verticillium dahliae partitivirus 1 (YP_009164038.1); PaPV1, Penicillium aurantiogriseum partitivirus 1 (YP_009182336.1); GaRVMS1, Gremmeniella abietina RNA virus MS1 (AII16004.1); AoV, Aspergillus ochraceous virus (ABV30675.1); FsV1, Fusarium solani virus 1 (NP_624350.1); PmPv1, Phoma matteuccicola partitivirus 1 (MK211276); UvPV2, Ustilaginoidea virens partitivirus 2 (YP_008327312.1); CaRV1, Colletotrichum acutatum RNA virus 1 (AGL42312.1); CpV1, Cryptosporidium parvum virus 1 (AAC47805.1); BcV1, beet cryptic virus 1 (ACA81389.1); WcCV1, white clover cryptic virus 1 (AAU14888.1); CcV, carrot cryptic virus (ACL93278.1); ScPVS, Sclerotinia sclerotiorum partitivirus S (ACT55329.1); RnPV2, Rosellinia necatrix partitivirus 2 (BAM78602.1); AhV, Atkinsonella hypoxylon virus (AAA61829.1); CrV1, Ceratocystis resinifera virus 1 (AAU26069.1); RsV717, Rhizoctonia solani virus 717 (AAF22160.1); FpV1, Fusarium poae virus 1 (AAC98734.1); PoV1, Pleurotus ostreatus virus 1 (AAT07072.1); DcV2, dill cryptic virus 2 (AGJ83771.1); CcBV, cannabis cryptic virus (AET80948.1); BcV3, beet cryptic virus 3 (AAB27624.1); BrCV, black raspberry cryptic virus (ABU55400.1); RsCV2, Raphanus sativus cryptic virus 2 (ABB04855.1); FcCV, Fragaria chiloensis cryptic virus (AAZ06131.2); The tree was rooted with the RdRp of southern tomato virus (StV; NC_003607) as an outgroup
The sequence of dsRNA-2 also contains a single ORF (ORF2), starting at nt 137 and ending at nt 1267 (Fig. 1A), and encodes a 375-aa protein with a molecular weight of 40.92 kDa. A BLASTp search using the deduced amino acid sequence indicated that ORF2 codes for a hypothetical protein that has 56% identity to a protein from UvPV2 and 27% identity to a protein from CaRV1. The secondary structure of this protein was predicted using PSIPRED software. The results showed that the protein encoded by dsRNA-2 is composed of an alpha helix, beta turn folding patterns, a helix, and an extended strand, which is typical of CPs of partitiviruses (data not shown). In addition, a more sensitive and hidden HHpred analysis based on a Markov model was performed to identify a group of more remote homologs. The results indicated that dsRNA-2 ORF2 encoded protein has many potential distant homologs, including the capsid protein of Rous sarcoma virus (PDB ID 3G21_A), the lipid-binding domain of fowlpox virus (PDB ID 6BR9_A), and the capsid protein of feline immunodeficiency virus (PDB ID 5DCK_B). These results further suggest that this protein has structural similarities to viral capsid proteins. Therefore, we postulate that the protein encoded by dsRNA-2 may have the function of a capsid protein.
Viral particles were purified from P. matteuccicola strain LG915 were found to be isometric and approximately 29 nm in diameter when observed under a TEM (Fig. 1C). These particles contained two dsRNA segments with sizes similar to dsRNA-1 and dsRNA-2 extracted from the mycelia of strain LG915 (Fig. 1D), suggesting that both the viral particles and dsRNAs extracted directly from the mycelia belonged to the same virus, PmPV1.
To analyze the relationship between PmPV1 and other mycoviruses, a phylogenetic tree was constructed based on the amino acid sequence of the RdRp region of PmPV1 and those of related viruses in the family Partitiviridae. The results showed that PmPV1 was closely related to members of the genus Gammapartitivirus in the family Partitiviridae but formed a distinct and independent branch with UvPV2 and CaRV19 (Fig. 2B). Therefore, this dsRNA virus should be considered a new member of the genus Gammapartitivirus, which we have tentatively named “Phoma matteuccicola partitivirus 1” (PmPV1).
References
Ghabrial SA, Jiang DH, Nibert ML, Suzuki N (2015) 50-plus years of fungal viruses. Virology 479:356–368
Zhong J, Zhu JZ, Lei XH, Chen D, Zhu HJ, Gao BD (2014) Complete genome sequence and organization of a novel virus from the rice false smut fungus Ustilaginoidea virens. Arch Virol 48:329–333
Zheng F, Ma R, Xu G, Zheng FQ, Ding XF, Xie CP (2018) Leaf blight on Curcuma wenyujin caused by Phoma matteucciicola in China. Plant Dis 102:2042
Morris TJ, Dodds JA (1979) Isolation and analysis of double stranded RNA from virus-infected plant and fungal tissue. Phytopathology 69:854–858
Zheng L, Liu HQ, Zhang ML, Cao X, Zhou EX (2013) The complete genomic sequence of a novel mycovirus from Rhizoctonia solani AG-1 IA strain B275. Arch Virol 158:1609–1612
Darissa O, Willingmann P, Adam G (2010) Optimized approaches for the sequence determination of double-stranded RNA templates. J Virol Methods 169:397–403
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Zhong J, Zhu JZ, Lei XH, Chen D, Zhu HJ, Gao BD (2014) Complete genome sequence and organization of a novel virus from the rice false smut fungus Ustilaginoidea virens. Virus Genes 48:329–333
Zhong J, Chen D, Lei XH, Zhu HJ, Zhu JZ, Gao BD (2014) Detection and characterization of a novel Gammapartitivirus in the phytopathogenic fungus Colletotrichum acutatum strain HNZJ001. Virus Res 190:104–109
Funding
This study was financially supported by Hainan Province Key R&D Project (ZDYF2019143), the Scientific Research Foundation for Advanced Talents, Hainan University (no. KYQD(ZR)1873), Hainan Major Research Found of Science and Technology (no. ZDKJ201817), and the National Natural Science Foundation of China (no. 31701732).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with animals or human participants performed by any of the authors.
Additional information
Handling Editor: Robert H. A. Coutts.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zheng, F., Xu, G., Zhou, J. et al. Complete genomic sequence and organization of a novel mycovirus from Phoma matteuccicola strain LG915. Arch Virol 164, 2209–2213 (2019). https://doi.org/10.1007/s00705-019-04314-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-019-04314-w