Abstract
Porcine circovirus type 2 (PCV2) is the cause of postweaning multisystemic wasting syndrome (PMWS), which encompasses several distinct symptoms in pigs. PCV2 infection and clinical incidence of PMWS have increased in recent years, possibly due to shifts in viral populations and mutations. In this study, we identified PVC2 strains currently afflicting pig populations in mainland China, because this is a prerequisite for developing a specific vaccine to control the spread of PMWS. We collected 235 tissue samples from 16 provinces between 2014 and 2016. Of these, 152 samples were positive for PCV2. We compared the sequences we obtained for the PVC2 capsid gene, ORF2, to those of the Chinese PCV2 sequences deposited in GenBank between 2002 and 2016 (n = 648). Phylogenetic analyses demonstrated that the PCV2d genotype was the most prevalent strain in the sample population included in GenBank and among the positive samples from this study. We also found one PCV2c strain among the GenBank sequences. Furthermore, PCV2a-2F was the predominant genotype in the PCV2a cluster. Amino acid sequence comparisons demonstrated 70.8–100% identity within PCV ORF2 and several consistent mutations in ORF2. More interestingly, six isolates were classified as recombinant strains. Cumulatively, this study represents the first comprehensive description of PCV2 strains distribution, including recent samples, in Chinese porcine populations. We demonstrate the existence of high genetic variability among PVC2 strains and the ability of this virus to rapidly evolve.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Porcine circovirus type 2 (PCV2) is a highly infectious pathogen that causes immune suppression in pigs. It has resulted in tremendous economic losses in the swine industry. Taxonomically, Porcine circovirus 2 is a member of the genus Circovirus, within the family Circoviridae. PCV2 is one of the smallest DNA viruses infecting mammals, possessing a non-enveloped virion particle of 12–23 nm in diameter [34]. PCV2 is the primary pathogen underlying several syndromes collectively known as porcine circovirus-associated disease (PCVAD), which include postweaning multisystemic wasting syndrome (PMWS) [1, 7] and porcine dermatitis and nephropathy syndrome (PDNS) [2, 35]. The clinical signs of PCV2-infected pigs include weight loss, proliferative and necrotizing pneumonia, enteritis, reproductive disorders in sows, fetal myocarditis, and respiratory disease in weaned and fattening pigs [36].
PCV2 has four major open reading frames (ORFs). These are ORF1–ORF4, which encode the replicase, capsid protein (Cap), viral pathogenesis-associated protein, and apoptosis-suppressing protein, respectively [8, 9, 36]. The Cap protein is the major immunogenic molecule, and virus-like particles of the Cap protein provide effective protection [9]. Due to the lack of a viral envelope, Cap is exposed on the surface of the virion. This has led to remarkable genetic diversity of Cap proteins among viruses. As a result, the Cap protein has proved very useful for epidemiological and phylogenetic studies of PCV2 [5, 25]. Phylogenetic analyses indicated that PCV2 strains could be divided into five genotypes (PCV2a–2e) based on pairwise sequence comparisons of PCV2 isolates [6]. PCV2a could be further subdivided into five clusters (2A–2E), while PCV2b could be subdivided into three clusters (1A–1C) [28]. Previous studies revealed that the majority of Chinese PCV2 strains are of genotype 2a or 2b. A few strains could be classified into genotype 2d. However, no strains of genotype 2c could be isolated [13]. Based on these observations, it has been suggested that the PCV2b genotype has become the dominant viral strain in China in recent years [15, 21, 39].
Although the majority of pigs are vaccinated against PCV2 using killed virus, the incidence of clinical disease in China is still on the rise [37]. In this study, we obtained clinical samples from dead pigs from 16 different provinces. Using sequencing technologies, we analyzed the genetic diversity of these PCV2 strains and identified possible recombination events. Cumulatively, this work helps elucidate important aspects of the molecular genetic evolution of this virus, which is a prerequisite for the future development of effective disease control and prevention strategies.
Materials and methods
Clinical samples
Two hundred and thirty-five tissue samples (lymph nodes) were collected from 235 dead pigs on different farms in 16 provinces in China. Pigs were 4–10 weeks of age, and were suspected to be infected by PMWS and/or PDNS based upon evidence of growth retardation, dyspnea, paleness of the skin, and enlarged lymph nodes upon necropsy (Table 1).
PCR amplification and DNA sequencing of ORF2
Tissues were analysed using published PCR methods to identify samples positive for PCV2 [32], porcine reproductive respiratory syndrome virus (PRRSV) [43], porcine parvovirus [26], classical swine fever virus (CSFV) [18], and Mycoplasma hyopneumoniae [16]. From the PCV2-positive samples, ORF2 was amplified using primers: 5′-TGAGTCTTTTTTATCACTTCGT-3′ (position 993–1014 bp) and 5′-CTTACAGCGCACTTCTTTCGT-3′ (position 1743–1763 bp). Thermal cycling conditions were: 95 °C for 5 min; followed by 35 cycles of 94 °C for 1 min, 52 °C for 1 min and 72 °C for 1 min; and a final elongation step at 72 °C for 10 min. PCR products were run on a 1% agarose gel and imaged under ultraviolet light. The positive PCR products (777 bp) were purified with an E.Z.N.A.™ Gel Extraction Kit (OMEGA, Georgia, USA), and cloned using the pMD18-T Vector System (Takara, Dalian, China). Four individual clones for each insert were sequenced by Sanger sequencing (Life Technologies, Shanghai, China), and the consensus sequence was obtained using Vector NTI Suite 9 (InforMax Inc., Maryland, USA).
Phylogenetic analysis
In addition to the sequences obtained from dead animals, we downloaded 496 ORF2 sequences from GenBank, which were identified in 28 provinces in the principal pig farming areas of China. This gave us a total of 648 Chinese PCV2 strains to examine. Representative sequences for PCV2 and PCV1 (GenBank accession no. FJ475129) were used as references. Sequence alignment was carried out using MEGA software (v. 6.0) (Pennsylvania, USA) and the ClustalW algorithm; the identity among sequences, at the nucleotide or amino acid level, was determined using BioEdit (v7.0.5) (California, USA). A phylogenetic tree was constructed using MEGA software based on the cap nucleotide sequence, using the neighbor-joining (NJ) method with the Kimura two parameter model for nucleotide substitution.
Recombination analysis
To detect putative recombination breakpoints in the PCV2 ORF2 gene of the complete (cumulative) dataset, and to identify sequences that might have originated from a recombination event, six methods (RDP, GeneConv, BootScan, MaxChi, Chimaera, and SiScan) were implemented using the RDP program (v. 4.46) (Cape Town, South Africa) [24]. We employed the following settings in these analyses: window size = 20, highest multiple comparison-corrected P value = 0.01, Bonferroni correction, finding consensus daughter sequences, and polishing breakpoints. Only putative recombination events detected by more than one method were considered. Base to base analysis was used to confirm the recombination events detected by the SimPlot program (v. 3.5.1) (Maryland, USA) when compared to the parental strain sequences, as described previously [22].
Results
Sample screening and identification
We collected 235 tissue samples from dead pigs that were suspected to have suffered from PMWS, and we found that 152 were positive for PCV2 infection. Of these, 25 samples were positive for both PCV2 and PRRSV infections, 20 were positive for both PCV2 and CSFV infections, 17 were positive for both PCV2 and pseudorabies virus (PRV) infections, and five were positive for triple infection with PCV2, PRRSV, and CSFV.
Identification of PCV2 genotypes
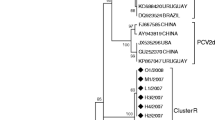
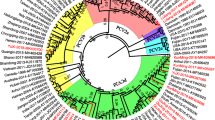
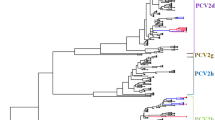
The NJ phylogenetic tree based on ORF2 sequences from the 648 PCV2 strains showed that all of the Chinese PCV2 strains belonged to four genotypes: PCV2a, PCV2b, PCV2c, and PCV2d (Fig. 1). Among the Chinese PCV2 strains obtained from GenBank, 75 strains (11.6%) belonged to the PCV2a genotype, 246 strains (38.0%) belonged to the PCV2b genotype, and 327 strains (50.4%) to the PCV2d genotype. Only one strain, collected in 2010, was genotyped as PCV2c (GenBank accession no. KC823058). The phylogenetic distances between the genotypes ranged from 0.057 (genotype 2b vs. genotype 2d) to 0.098 (genotype 2d vs. genotype 2a). Within each genotype, the average distances ranged from 0.004 (PCV-2a) to 0.021 (PCV-2d).
Phylogenetic tree of Chinese PCV2 isolates. PCV2 strains (n = 648) were used to construct trees based on the ORF2 gene with MEGA software. The neighbor-joining method was used as statistical method, with the Tamura-Nei model and 1000 bootstrap replications, to assess the reliability of the tree. Strains detected in our lab are indicated by triangles. Genotypes and clusters are indicated by square brackets
Mutational analysis of the ORF2-encoded Cap protein
Sequence analysis of the ORF2 gene in the 648 PCV2 strains revealed that nucleotide variation ranged from 89.6–100% and predicted amino acid identity ranged from 70.8–100%. Furthermore, amino acid alignments of the Cap protein encoded by ORF2 indicated that there are five major regions of variation among the PCV2 strains (Fig. 2). These include residues 57–91, 121–151, 181–191, 206–215, and 230–233. The ORF2 amino acid variations at two positions (residue 53: F to I; and residue 68: A to N) were unique to genotype PCV2d. In addition, the amino acid variations at four positions (residue 47: T/A/G to S; residue 130: V/L to F; residue 133: A/V to S; and residue 191: G/A/R/E to K) were unique to genotype PCV2a-2F in the PCV2a cluster.
Alignment of the capsid protein of PCV2 strains. The ORF2 amino acid variations at positions 53 (F to I) and 68 (A to N) were unique to genotype PCV2d. In addition, the amino acid variations at positions 47 (T/A/G to S), 130 (V/L to F), 133 (A/V to S), and 191 (G/A/R/E to K) were unique to genotypes PCV2a–2F in the PCV2a cluster. Mutation of an amino acid is indicated by a block
Detection and analysis of recombinants
Within the 648 PCV2 ORF2 sequences, six were identified as potential recombinant strains (Table 2). The possible breakpoints for recombination were determined (Fig. 3). The nucleotide sequences before the putative breakpoints were very similar to the sequences of the minor parent, while the regions after the breakpoint were most similar to the major parent sequences. Amino acid sequence similarities between recombinants and their parents ranged from 96.4–99.8%.
Recombination analysis of the PCV2 ORF2 gene. Recombination events were analyzed by Simplot software (v. 3.5.1). The similarity among 648 ORF2 genes was calculated using the Kimura-2-parameter method with a transition-transversion ratio of 2. The y-axis indicates the percentage identity within a sliding window of 200 bp centered on the position plotted, with a step size between plots of 20 bp. The red vertical line shows the potential breakpoint
Discussion
In China, PCV2 infection is very common, and PMWS has been a major problem for the swine industry since a 2002 outbreak caused substantial economic losses to farmers [42]. Since then, PMWS outbreaks have occurred frequently, along with an increase in the incidence of other swine diseases. For example, previous studies reported higher rates of PCV2 comorbidity with porcine parvoviruses [30, 37]. Furthermore, numerous studies have identified an increase in cases of coinfection with viruses including PRRSV, porcine epidemic diarrhea virus (PEDV), PRV, and with Mycoplasma hyopneumoniae [29]. Consistent with this, our results show that a variety of pathogens enhanced PCV2 lesions and disease. It is possible that the comorbidity of combinations of PCVADs varies from region to region, but similar mechanisms might underlie the observed enhancement of the disease phenotype.
Based on phylogenetic studies, a classification scheme for PCV2 was proposed, which divides the viral strains into several major groups based on genotypes identified in different countries [41]. In Malaysia, amino acid sequence analysis of the PCV2 capsid protein (ORF2) revealed that the PCV2b genotype constituted a major subgroup of viral strains [19]. Phylogenetic analysis and comparison with reference sequences demonstrated that PCV2b was most prevalent between 2007 and 2014 in northern Italy [11]. In Taiwan, PCV2a was the most common strain isolated in 2001, but, since 2003, PCV2b has become the predominant subgroup found on pig farms [38]. In Brazil, the results revealed remarkable genetic diversity: all four genotypes currently recognized were detected, including PCV2a, PCV2b, PCV2c and PCV2d [10]. In India, the molecular characterization of PCV2 revealed that individual pigs could harbor multiple genotypes simultaneously, including combinations of PCV2a-2D and PCV2d [3]. Between 2004 and 2008, phylogenetic analyses indicated that PCV2 strains isolated in China could be divided into four genotypes (PCV2a, PCV2b, PCV2d, and PCV2e), and PCV2b was the most common [41]. Between 2009 and 2010, PCV2b became the predominant genotype in mainland China. Base-by-base comparisons of the ORF2 gene sequences indicated that PCV2 evolution traced from PCV2a to PCV2b to PCV2d.
PCV2d was initially identified in 1999 in samples collected in Switzerland. This strain now appears to be widespread in China and has been present in North America since 2010 [42]. From 2012 to 2013, 37% of all PCV2 sequences isolated from US pigs were classified as PCV2d. The study of Mu et al. showed that the approximate percentages of genotypes PCV2a, PCV2b and PCV2c in Henan Province, between 2005 and 2011, were 6.5% (2/31), 93.5% (29/31) and 0%, respectively [27]. Five and 61 of the 66 PCV2 strains belonged to genotypes PCV2a and PCV2b, respectively, indicating that PCV2b was the predominant genotype circulating in southern China from 2011 to 2012 [40]. Our results in this study revealed that the PCV2d genotype constituted 50.4% of all collected samples. Cumulatively, the data suggest an ongoing genotype shift from PCV2b to PCV2d is occurring in pig populations in China. There are four major regions (57–91, 121–151, 181–191, and 230–233) of amino acid variation among the PCV2 strains in our study, which were identified as dominant immunoreactive areas [20]. One study reported that a PCV2 vaccine based on genotype PCV2b was more effective in protecting pigs against the effects of PCV2b infection than those based on genotype PCV2a [31]. Commercial PCV2 vaccines in China are mainly based on the PCV2a and PCV2b genotypes. Vaccines may have become less effective in recent years because of the antigenic variability of PCV2.
The genotype PCV2a may be divided into different clusters. Phylogenetic analysis revealed that new PCV2a isolates were not included in clusters 2A-2E. The new cluster was called PCV2a 2F, or, more simply, PCV2e. PCV2e was first reported in 2009 in China, and constituted approximately 50.0% of all isolates in the PCV2a cluster [39]. However, in our study, strains obtained were predominately of the PCV2e genotype (75.6%) within the PCV2a cluster. The amino acid substitutions specific to the genotype PCV2e were mainly localized to positions 47 (T/A/G to S), 130 (V/L to F), 133 (A/V to S), and 191 (G/A/R/E to K) in the PCV2 capsid protein. These amino acid substitutions are more diverse than those observed in 2009.
We demonstrated that recombination in the ORF2 gene occurred between the PCV2a/2b or PCV2a/2d strains, yielding different recombinants. Some examples of recombination identified by Huang et al. (2013) were not found again here, because the sequence analyses were different (full-length genome in Huang et al. and only the ORF2 gene in our study [17]). The diversity of PCV2 is closely related to these virus recombinants. Intergenotype recombination was found in several countries and regions, including South America, China and India [3, 17, 33]. It also occurred between different genotypes. For example, natural recombination was observed among different lineages of PCV2 strains from Hong Kong [23] and between PCV2a and PCV2b [4]. Gagnon et al. found a type 1 and type 2 PCV recombinant genome that contained the ORF1 of PCV1 and the ORF2 of PCV2a [12]. Recombinant mutants may enhance viral replication and alter antigenicity in vitro [14]. Nevertheless, the recombination between PCV2 strains likely contributes to the genetic variation and diversity that we observed for this virus in the field.
Conclusion
We conducted a comprehensive molecular analysis of Chinese PCV2 strains based on the ORF2 gene. The predominant genotype has shifted from PCV2b to PCV2d. This is of great concern, because current vaccines target only PCV2a and PCV2b strains. Furthermore, there have been shifts within the PCV2a subgroup itself, since we found that PCV2a-2F is now the main genotype. These findings highlight the importance of understanding the composition and dynamics of genetic diversity within the Chinese PCV2 strains because this information is critical for developing effective vaccines and control strategies.
References
Allan GM, Mc Neilly F, Meehan BM, Kennedy S, Mackie DP, Ellis JA, Clark EG, Espuna E, Saubi N, Riera P, Botner A, Charreyre CE (1999) Isolation and characterisation of circoviruses from pigs with wasting syndromes in Spain, Denmark and Northern Ireland. Vet Microbiol 66:115–123
Allan GM, McNeilly E, Kennedy S, Meehan B, Moffett D, Malone F, Ellis J, Krakowka S (2000) PCV-2-associated PDNS in Northern Ireland in 1990. Porcine dermatitis and nephropathy syndrome. Vet Rec 146:711–712
Anoopraj R, Rajkhowa TK, Cherian S, Arya RS, Tomar N, Gupta A, Ray PK, Somvanshi R, Saikumar G (2015) Genetic characterisation and phylogenetic analysis of PCV2 isolates from India: indications for emergence of natural inter-genotypic recombinants. Infect Genet Evol 31:25–32
Cai L, Han X, Ni J, Yu X, Zhou Z, Zhai X, Chen X, Tian K (2011) Natural recombinants derived from different patterns of recombination between two PCV2b parental strains. Virus Res 158:281–288
Choi KS, Chae JS (2008) Genetic characterization of porcine circovirus type 2 in Republic of Korea. Res Vet Sci 84:497–501
Cortey M, Olvera A, Grau-Roma L, Segales J (2011) Further comments on porcine circovirus type 2 (PCV2) genotype definition and nomenclature. Vet Microbiol 149:522–523
Ellis J, Hassard L, Clark E, Harding J, Allan G, Willson P, Strokappe J, Martin K, McNeilly F, Meehan B, Todd D, Haines D (1998) Isolation of circovirus from lesions of pigs with postweaning multisystemic wasting syndrome. Can Vet J 39:44–51
Fenaux M, Halbur PG, Gill M, Toth TE, Meng XJ (2000) Genetic characterization of type 2 porcine circovirus (PCV-2) from pigs with postweaning multisystemic wasting syndrome in different geographic regions of North America and development of a differential PCR-restriction fragment length polymorphism assay to detect and differentiate between infections with PCV-1 and PCV-2. J Clin Microbiol 38:2494–2503
Fenaux M, Opriessnig T, Halbur PG, Elvinger F, Meng XJ (2004) Two amino acid mutations in the capsid protein of type 2 porcine circovirus (PCV2) enhanced PCV2 replication in vitro and attenuated the virus in vivo. J Virol 78:13440–13446
Franzo G, Cortey M, de Castro AM, Piovezan U, Szabo MP, Drigo M, Segales J, Richtzenhain LJ (2015) Genetic characterisation of Porcine circovirus type 2 (PCV2) strains from feral pigs in the Brazilian Pantanal: An opportunity to reconstruct the history of PCV2 evolution. Vet Microbiol 178:158–162
Franzo G, Tucciarone CM, Dotto G, Gigli A, Ceglie L, Drigo M (2015) International trades, local spread and viral evolution: the case of porcine circovirus type 2 (PCV2) strains heterogeneity in Italy. Infect Genet Evol 32:409–415
Gagnon CA, Music N, Fontaine G, Tremblay D, Harel J (2010) Emergence of a new type of porcine circovirus in swine (PCV): a type 1 and type 2 PCV recombinant. Vet Microbiol 144:18–23
Ge X, Wang F, Guo X, Yang H (2012) Porcine circovirus type 2 and its associated diseases in China. Virus Res 164:100–106
Guo L, Lu Y, Wei Y, Huang L, Wu H, Liu C (2011) Porcine circovirus genotype 2a (PCV2a) and genotype 2b (PCV2b) recombinant mutants showed significantly enhanced viral replication and altered antigenicity in vitro. Virology 419:57–63
Guo LJ, Lu YH, Wei YW, Huang LP, Liu CM (2010) Porcine circovirus type 2 (PCV2): genetic variation and newly emerging genotypes in China. Virol J 7:273
Haldimann A, Nicolet J, Frey J (1993) DNA sequence determination and biochemical analysis of the immunogenic protein P36, the lactate dehydrogenase (LDH) of Mycoplasma hyopneumoniae. J Gen Microbiol 139:317–323
Huang Y, Shao M, Xu X, Zhang X, Du Q, Zhao X, Zhang W, Lyu Y, Tong D (2013) Evidence for different patterns of natural inter-genotype recombination between two PCV2 parental strains in the field. Virus Res 175:78–86
Hurtado A, Garcia-Perez AL, Aduriz G, Juste RA (2003) Genetic diversity of ruminant pestiviruses from Spain. Virus Res 92:67–73
Jaganathan S, Toung OP, Yee PL, Yew TD, Yoon CP, Keong LB (2011) Genetic characterization of porcine circovirus 2 found in Malaysia. Virol J 8:437
Lekcharoensuk P, Morozov I, Paul PS, Thangthumniyom N, Wajjawalku W, Meng XJ (2004) Epitope mapping of the major capsid protein of type 2 porcine circovirus (PCV2) by using chimeric PCV1 and PCV2. J Virol 78:8135–8145
Li W, Wang X, Ma T, Feng Z, Li Y, Jiang P (2010) Genetic analysis of porcine circovirus type 2 (PCV2) strains isolated between 2001 and 2009: genotype PCV2b predominate in postweaning multisystemic wasting syndrome occurrences in eastern China. Virus Genes 40:244–251
Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC (1999) Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73:152–160
Ma CM, Hon CC, Lam TY, Li VY, Wong CK, de Oliveira T, Leung FC (2007) Evidence for recombination in natural populations of porcine circovirus type 2 in Hong Kong and mainland China. J Gen Virol 88:1733–1737
Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P (2010) RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26:2462–2463
Gomes Martins, de Castro AM, Cortez A, Heinemann MB, Brandao PE, Richtzenhain LJ (2007) Genetic diversity of Brazilian strains of porcine circovirus type 2 (PCV-2) revealed by analysis of the cap gene (ORF-2). Arch Virol 152:1435–1445
Molitor TW, Oraveerakul K, Zhang QQ, Choi CS, Ludemann LR (1991) Polymerase chain reaction (PCR) amplification for the detection of porcine parvovirus. J Virol Methods 32:201–211
Mu CL, Yang QY, Zhang YN, Zhou YH, Zhang JX, Martin DP, Xia PA, Cui BA (2012) Genetic variation and phylogenetic analysis of porcine circovirus type 2 infections in central China. Virus Genes 45:463–473
Olvera A, Cortey M, Segales J (2007) Molecular evolution of porcine circovirus type 2 genomes: phylogeny and clonality. Virology 357:175–185
Opriessnig T, Halbur PG (2012) Concurrent infections are important for expression of porcine circovirus associated disease. Virus Res 164:20–32
Opriessnig T, Xiao CT, Gerber PF, Halbur PG (2014) Identification of recently described porcine parvoviruses in archived North American samples from 1996 and association with porcine circovirus associated disease. Vet Microbiol 173:9–16
Opriessnig T, O’Neill KGP, de Castro AM, Gimenéz-Lirola LG, Beach NM, Zhou L, Meng XJ, Wang C, Halbur PG (2013) A PCV2 vaccine based on genotype 2b is more effective than a 2a-based vaccine to protect against PCV2b or combined PCV2a/2b viremia in pigs with concurrent PCV2, PRRSV and PPV infection. Vaccine 31:487–494
Pogranichnyy RM, Yoon KJ, Harms PA, Swenson SL, Zimmerman JJ, Sorden SD (2000) Characterization of immune response of young pigs to porcine circovirus type 2 infection. Viral Immunol 13:143–153
Ramos N, Mirazo S, Castro G, Arbiza J (2013) Molecular analysis of Porcine Circovirus Type 2 strains from Uruguay: evidence for natural occurring recombination. Infect Genet Evol 19:23–31
Rodriguez-Carino C, Segales J (2009) Ultrastructural findings in lymph nodes from pigs suffering from naturally occurring postweaning multisystemic wasting syndrome. Vet Pathol 46:729–735
Rosell C, Segales J, Ramos-Vara JA, Folch JM, Rodriguez-Arrioja GM, Duran CO, Balasch M, Plana-Duran J, Domingo M (2000) Identification of porcine circovirus in tissues of pigs with porcine dermatitis and nephropathy syndrome. Vet Rec 146:40–43
Stevenson GW, Kiupel M, Mittal SK, Choi J, Latimer KS, Kanitz CL (2001) Tissue distribution and genetic typing of porcine circoviruses in pigs with naturally occurring congenital tremors. J Vet Diagn Invest 13:57–62
Sun J, Huang L, Wei Y, Wang Y, Chen D, Du W, Wu H, Liu C (2015) Prevalence of emerging porcine parvoviruses and their co-infections with porcine circovirus type 2 in China. Arch Virol 160:1339–1344
Wang C, Huang TS, Huang CC, Tu C, Jong MH, Lin SY, Lai SS (2004) Characterization of porcine circovirus type 2 in Taiwan. J Vet Med Sci 66:469–475
Wang F, Guo X, Ge X, Wang Z, Chen Y, Cha Z, Yang H (2009) Genetic variation analysis of Chinese strains of porcine circovirus type 2. Virus Res 145:151–156
Wei CY, Zhang MZ, Chen Y, Xie JX, Huang Z, Zhu WJ, Xu TC, Cao ZP, Zhou P, Shuo SuS, Zhang GH (2013) Genetic evolution and phylogenetic analysis of porcine circovirus type 2 infections in southern China from 2011 to 2012. Infect Genet Evol 17:87–92
Xiao CT, Halbur PG, Opriessnig T (2015) Global molecular genetic analysis of porcine circovirus type 2 (PCV2) sequences confirms the presence of four main PCV2 genotypes and reveals a rapid increase of PCV2d. J Gen Virol 96:1830–1841
Yang HC (2004) Epidemical characterizations and control strategies of swine immunosuppressive diseases (PRRS and PMWS). Chin J Anim Husband Vet Med 31:41–43
Yoon KJ, Zimmerman JJ, Chang CC, Cancel-Tirado S, Harmon KM, McGinley MJ (1999) Effect of challenge dose and route on porcine reproductive and respiratory syndrome virus (PRRSV) infection in young swine. Vet Res 30:629–638
Acknowledgements
This work was supported by a grant from the National Natural Science Foundation of China (No. 31270045).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
None of the authors of this paper has a financial or personal relationship with other people or organizations that could inappropriately influence or bias the content of this paper.
Rights and permissions
About this article
Cite this article
Jiang, CG., Wang, G., Tu, YB. et al. Genetic analysis of porcine circovirus type 2 in China. Arch Virol 162, 2715–2726 (2017). https://doi.org/10.1007/s00705-017-3414-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-017-3414-1