Abstract
Porcine circovirus type 2 (PCV2) infection causes postweaning multisystemic wasting syndrome and porcine circovirus-associated diseases in many regions. A total of 77 sequences, including 31 sampled from Henan province of China, were retrieved from GenBank and subjected to amino acid variation and phylogenetic analyses. The two PCV genotypes prevailing in Henan were PCV-2a and PCV-2b with PCV-2b accounting for 93.5 % (29/31) of the Henan isolates. The 31 Henan isolates all shared between 92.7 and 100 % sequence similarity. Amino acid variation analysis of the capsid protein revealed that Henan PCV2 strains tended to accumulate more substitutions within epitopic regions—a substitution pattern consistent with host immune system-mediated selection of virus immune escape variants. The analysed PCV sequences carry evidence of at least six unique recombination events. Selective pressure analysis of the relative recombination-free ORF2 sequences of these viruses revealed evidence of sites that are likely evolving in response to host-driven immune pressures—a finding that coupled with information on the prevalent diversity in Henan PCV2 isolates of known immunoreactive genomic loci will aid in future studies aiming to assess the evolutionary responses of PCV2 in China to the widespread deployment of anti-PCV vaccines in the country.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Porcine circovirus (PCV) is a spherical, small, non-enveloped virus of the family Circoviridae that contains a single-stranded circular DNA genome. There are two known main PCV lineages known as PCV types 1 and 2, which are antigenically and genetically distinct from one other [1]. As a non-cytopathic contaminant from the porcine kidney cell line PK-15, PCV type 1 is non-pathogenic and is incapable of causing clinical disease in swine [2]. In contrast, porcine circovirus type 2 (PCV2) is the causative agent of postweaning multisystemic wasting syndrome (PMWS), porcine dermatitis and nephropathy syndrome (PDNS) and other porcine circovirus-associated diseases (PCVAD) [3–5]. Co-infection of PCV2 with porcine reproductive and respiratory syndrome virus and Haemophilus parasuis could aggravate the disease phenotypes of these other pathogens via immunosuppression [6]. Currently, PCV2 seriously affects the swine industry worldwide.
PCV2 genomes tend to be either 1,767 or 1,768 bp in length and contain two major open reading frames (ORFs). ORF1 encodes two replication-associated proteins (Rep and Rep’) and ORF2 encodes the capsid protein (Cap). Besides the capsid protein being the primary elicitor of protective host immune responses [7, 8], it also plays a pivotal role in viral attachment to target cells such as monocytes or macrophages via the heparin sulphate and chondroitin sulphate B glycosaminoglycan receptors [9].
Globally, when compared to other circoviruses such as beak and feather disease virus, PCV2 diversity is relatively low. Nevertheless, whatever PCV2 diversity does exist has been used to classify different PCV2 isolates into genotype groupings [5]. The currently accepted methodology for classifying PCV2 lineages into genotype groupings is based on pairwise sequence comparison (PASC) analyses of ORF2 gene sequences with a genotype demarcation threshold of 0.035 [10–12]. By means of this analysis methodology and threshold, only three PCV2 genotypes are currently accepted: PCV-2a, PCV-2b and PCV-2c.
Previous analyses of Chinese PCV2 diversity has indicated that Chinese isolates on a genome-wide scale tend to be >94.6 % identical to one another and have, overall, an only slightly lower range of total diversity than PCV2 sequences sampled world-wide (which all >93.4 % identical) [13–17]. For these past analyses, it is apparent that the majority of Chinese PCV2 strains belong to genotypes PCV-2a and PCV-2b, with PCV-2b presently being the dominant Chinese genotype [13, 14, 16]. Slightly confounding the simple pairwise sequence similarity-based classification of PCV2s is the fact that many PCV2 isolates are known to be recombinants, either of different PCV2 genotypes or of PCV2 genotypes and currently unknown divergent PCV-like viruses [18–21]. In addition, there likely exist recombination hotspots within the ORF1 gene [19, 21, 22].
Little is currently known about the genetic variation and evolutionary dynamics of PCV2 within China’s main swine husbandry hub in the province of Henan. Throughout this province, PCV2 infections are a major emerging threat to pig farming and for this reason, vaccination against PCV2 is likely to be the most effective ways of mitigating losses to this virus. The main objective of this study was, therefore, to catalogue the genetic variation and phylogenetic characteristics of Henan PCV2 populations prior to widespread vaccine deployment, both to inform the choice of vaccines that are ultimately used and to provide a baseline from which to monitor the molecular epidemiological impacts of these vaccines on future Chinese PCV2 diversity and evolution.
Materials and methods
PCV2 genome sequence data set
A total of 31 PCV2 full genome sequences, all derived from Henan, were retrieved from GenBank as of April, 2012. We downloaded full genome sequences of an additional 46 PCV2 isolates, originating outside Henan Province, available in GenBank that contained information on sample collection dates and geographical locations. All these sequences, deposited between 1997 and 2011, were downloaded, linearized at the same point and aligned with the CLUSTALW [23] component of the MegAlign program (DNASTAR, version 7.10, Madison, WI, USA). The isolates are listed in Tables 1 and 2.
Genetic variation and phylogenetic analyses of PCV2 sequences
A phylogenetic tree of the ORF2 gene was generated by the neighbor-joining method by means of MEGA 5 [24] with 1,000 bootstrap replicates and the Kimura 2-parameter nucleotide substitution model (identified as the best-fitting substitution model implemented in MEGA 5.0). Deduced Cap protein amino acid sequences were also aligned to identify potentially immunologically important residue substitutions amongst the Henan isolates. PCV2 genotyping was based on the currently accepted pairwise sequence comparison methods outlined above in the introduction.
Recombination between PCV2 sequences
To detect putative recombination breakpoints in the PCV2 genomes and identify likely parental sequences, six methods implemented in the Recombination Detection Program (RDP) were used [25]. To minimise the number of tests performed (so as to reduce the severity of the multiple testing correction needed to ensure an acceptably low rate of false positives), the 'auto mask for optimal recombination detection' setting was chosen. The six recombination detection methods used the following general settings: window size = 30, highest acceptable P value = 0.01 and Bonferroni multiple comparison correction.
Selection pressure
Selective pressure along the PCV2 ORF2 gene was assessed (this gene is relatively free of recombination—a process that can lead to false positive inferences of positive selection). Amino acid entropy scores per site were calculated by means of DAMBE [26] and plotted versus the difference between the non-synonymous (dN) and synonymous (dS) substitution rates (dN−dS) for sites along the ORF2 gene. The difference was calculated by means of the Tamura–Nei method implemented in the MEGA 5.0 software [27, 28]. dN−dS > 0, dN−dS = 0 and dN−dS < 0 suggested, respectively, positive selection (adaptive molecular evolution), neutral selection and negative selection (purifying selection) [29].
Results
Sequence similarity analysis
The sequence analysis showed the pairwise similarities of the 31 Henan isolates ranged between 92.6 and 100 % (Table 3). Compared with the representative strain first confirmed in Canada (AF027217, isolates PMWS PCV) and the strains BF, HR, BX, which were first isolated in mainland China in 2001, the identity ranged from 92.9 to 96.9 % and 92.6 to 96.9 %.
Phylogenetic relationships of PCV2 isolated in Henan
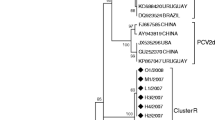
Phylogenetic analysis indicated that PCV-2a and PCV-2b (Fig. 1) are the only PCV2 genotypes prevalent in Henan. While only the PCV-2a isolates (EF524531 and JF928003) from this province cluster most closely with one another relative to PCV-2a isolates from China and from Canada, the 29 PCV-2b isolates were interspersed with PCV-2b isolates from elsewhere in the country. The genetic distances between the Henan PCV2 isolates ranged from 0.003 to 0.055.
The linearized phylograms constructed by the neighbor-joining method for the nucleotide sequences of PCV2 ORF2 gene sequences. Filled circle Henan isolates in genotype PCV2a and PCV2b; filled triangle reference strains first isolated in China; filled square reference strains first confirmed in Canada
Analysis of amino acid substitutions of ORF2 genes
To investigate variation in the deduced amino acid sequences of ORF2 gene products, the amino acid sequences of 35 PCV2 isolates including the 31 Henan isolates were aligned. As shown in Fig. 2 the ORF2 amino acid variations at seven positions, 49 (leucine to serine), 59 (arginine to alanine), 72 (methionine to leucine), 77 (asparagine to aspartic acid), 151 (threonine to proline), 206 (isoleucine to lysine) and 232 (asparagine to lysine), were unique to the two Henan genotype PCV-2a isolates (EF524531 and JF928003). Besides, ten out of 29 isolates in PCV-2b, which had 234 amino acids for Cap protein, all carried substitutions at positions 8 (tyrosine to phenylalanine), 53 (phenylalanine to isoleucine), 59 (arginine to lysine), 68 (alanine to asparagine), 89 (arginine to leucine), 90 (serine to threonine), 121 (serine to threonine), 134 (threonine to asparagine), 169 (serine to glycine or arginine), 190 (alanine to threonine), 210 (glutamic acid to aspartic acid) and 215 (valine to isoleucine). In addition, there were several major regions of variation within the Henan PCV2 ORF2 sequences including residues 57–81, 86–91, 130–133, 185–191, 206–215 and 230–233.
Recombination analysis
By running the RDP software, 28 potential recombination events were detected amongst all the analysed PCV2 genomes (including all the reference and Henan isolates). Only the six most statistically significant events are reported here (Table 4). One example is shown in Fig. 3. Although no evidence of direct genetic exchange between PCV2a and PCV2b sequences was detected, four of these recombination events likely involved exchanges between PCV2b sequences. However, in two cases, PCV2b sequences (EU418627 and JN639857) carried evidence of having acquired sequences from PCV-like viruses related to PCV2a. Five out of the six recombination events involved recombination transfers of large fragments of the ORF1 gene. The detected recombination events, except for event 5, had likely breakpoint positions within 100 nucleotides of the PCV virion strand origin of replication: both known recombination hot-spots in all circoviruses [21]. The ORF2 gene was, conversely, relatively free of recombination (only 1/12 detected breakpoints were within this gene), an observation which is again consistent with Cap protein genes of single-stranded DNA viruses in general including circoviruses such as beak and feather disease virus, but has not so far been identified as a recombination cold spot for PCV [21].
Identification of the recombination events. One of the recombinations between AY146992 and AF381176 was shown by means of the BootScan method. Recombination was detected between sequences sharing 70–100 % sequence identify. The left and right bounds of the pink tract region indicate breakpoint positions. The analysis based on the pairwise distance and modeled with a window size 30. The x-axis refers to the positions of informative sites and y-axis represents bootstrap support
Selective pressure analysis of the PCV genome
To find positions under positive and negative selection in the relatively recombination-free ORF2 genes of the Henan PCV2 sequences, the difference between non-synonymous and synonymous substitution rates (dN−dS) for every codon was evaluated (Fig. 4). According to the previous report, we designated the antigenic regions of this gene between positions 51–84, 113–136, 166–208, 228–234 as epitopes A, B, C, D, respectively [8, 30, 31]. Epitope A, B and C contained more positions under positive selection (5, 5 and 4 sites, respectively, with dN−dS scores > 0) than under negative selection (3, 1 and 3 sites, respectively, with dN−dS scores < 0). In epitope D, five amino acid positions were apparently evolving under neutral selection, one under negative selection and one under positive selection. The immunodominant decoy epitope (169–180) within epitope C contained one amino acid position (178) under positive selection. There was one amino acid site at position 91 within the heparin sulphate binding receptor domain (positions 91 and 113) that was possibly evolving under positive selection.
Discussion
Since the recognition of porcine circovirus type 2 in 2001, a considerable number of PCV2 sequences from various areas of China have been deposited in Genbank. These include multiple deposits of isolates from the Henan province, the first of which was only deposited in 2005. During the past 11 years, there have been several investigations that have analysed the distribution and prevalence of PCV2 infections in China [13–17]. To give a more detailed description of amino acid sequence variations and the phylogenetic characteristics of Henan PCV2 sequences, a total of 77 PCV2 sequences from Henan and elsewhere were retrieved from Genbank and analysed.
The lowest degree of identity between any two Henan isolates was 92.7 %. Although this implies slightly higher diversity amongst Chinese PCV2 isolates than has been reported in other studies (for example 94.6 % in [13]), it is still apparent that the over-all diversity of PCV2 isolates is low. A combination of this low diversity and the fact that recombination is a prominent feature of PCV2 evolution could seriously undermine the power of phylogenetic and other genetic variation analyses seeking to illuminate the evolution and spread of PCV2 genomes.
As reported previously, the ORF2 gene is reasonably free from the impact of recombination, so that it will generally yield the same tree as the whole viral genome [30]. Therefore, the ORF2 gene was employed for phylogenetic analysis in this study which showed that the Henan PCV2 isolates could be divided into two major genotypes and that isolates in PCV-2b belonged to several distinct clusters. Our study showed the approximate percentages of genotypes PCV-2a, PCV-2b and PCV-2c in Henan between 2005 and 2011 were 6.5 % (2/31), 93.5 % (29/31) and 0 %, respectively. Over the past 7 years, genotype PCV-2b has, therefore, probably been the overwhelmingly predominant PCV2 genotype in this province. Besides, finding no evidence of PCV-2c in Henan, we also found no evidence of two rare genotypes, PCV-2d and PCV-2e, that have previously been detected in China [13]. We noted, however, that the Henan isolate, EF524531 (strain HN0601), which has a clearly PCV-2a-derived ORF2 sequence, has a full genome sequence that has been previously identified as predominantly PCV-2e-like.
The two identified Henan PCV-2a isolates were obtained five years apart in 2006 and 2011 from different parts of the province. Nevertheless, the phylogenetic analysis revealed that these two isolates are clearly more closely related to one another than they are to any other PCV-2a isolates and that they may therefore represent a Henan specific PCV-2a lineage. It will, however, require a substantially more dense sampling of Chinese PCV-2a isolates to determine whether this Henan PCV-2a lineage originated in and is endemic to Henan or whether these PCV-2a isolates were carried into the province from elsewhere in China.
At least some of the amino acid variation in the Cap protein could be associated with the pathogenicity and/or immunogenicity of PCV2. The most variable of these regions amongst the predicted expressed amino acid sequences of the Henan PCV2 isolates include the known immunoreactive residues in the Cap protein (57–91, 121–134 185–191, 206–215, 230–234 [13, 32, 33]), the hypothetical ORF1 protein (amino acid residues 81–100 and 201–220) [34].
It has previously been demonstrated that PCV Cap proteins contain a highly conserved putative N-glycosylation site at amino acids 143–145 (N143YS) [35]. Deletion of this site could heighten specific immune responses, improving the immunogenicity of Cap-based DNA vaccines and inducing higher Cap-specific T lymphocyte proliferative activity and levels of interferon-γ. This potential N-glycosylation site was absolutely conserved in all of the analysed PCV2 sequences, implying that all of these sequences might, via this site, weaken anti-PCV2 immune responses in the animals that they infect.
It has additionally been reported that the alanine at position 59 in the Cap protein of PCV-2a strains is a crucial determinant of a conformational neutralizing antibody epitope in this virus lineage [36]. While the two Henan PCV-2a isolates are likely to express Cap proteins with an alanine at position 59, the PCV-2b isolates contained a variety of other amino acids at this site (threonine, lysine and arginine). This difference between the PCV-2a and PCV-2b isolates may reduce the recognition of this epitope and decrease the antibody neutralizing ability of PCV-2b infected animals.
The oligopeptide 169-STIDYFQPNNKR-180 serves as an immunodominant epitope associated with disease [8]. Through Alanine scanning, it has been determined that Y-173, F-174, Q-175 and K-179 are crucial for antibody recognition. However, this peptide may also serve as a decoy, diverting the humoral response away from some other potentially protective epitope [8]. Amongst the ORF2 genes of the 31 Henan isolates, this potential decoy epitope was conserved in both the PCV-2a isolates and 19 of the PCV-2b isolates. In the ten other PCV-2b isolates, either a glycine or an arginine was observed at position 169 and an isoleucine was observed at position 178. Since these substitutions are not within the four sites previously identified as being crucial for immune recognition, it is unknown what effect these substitutions might have on the immunogenicity of this epitope.
Recent studies have demonstrated that an antigenic epitope, 26-RPWLVHPRHRY-36, in the nuclear localization signal region of the PCV2 Cap protein could elicit a neutralizing anti-PCV2 host immune response [37]. As is shown here, this epitope is conserved in both the Henan PCV-2a isolates and 24 of the Henan PCV-2b isolates. While five of Henan PCV-2b isolates expressed a leucine at position 30, one of these five isolates (EU656143) also expressed a histidine at position 35. While the change at position 30 was from a polar amino acid (valine) to a hydrophobic amino acid (leucine) with the same isoelectric points, that at position 35 was from one alkaline amino acid (arginine) to another (histidine), but with different isoelectric points. The effect of these apparently conservative substitutions on the immunogenicity of these PCV-2a proteins should be further explored.
Overall, the various amino acid substitutions that we have detected within immunoreactive protein domains could potentially impact the ease with which PCV2 could be managed by a vaccination programme.
Genetic recombination is known to occur frequently between and within different PCV lineages [19] and is an important evolutionary mechanism whereby PCV2 diversity is generated. Also, given that co-infections of individual cells with genetically distinct virus strains were an obvious pre-condition for recombination [38], the pervasiveness of detectable recombinant PCV genotype suggests that an appreciable proportion of PCV infection involves multiple PCV lineages. For example, Cai et al. [19] identified a novel PCV genotype that had apparently arisen through recombination between PCV-2a and PCV-2b strains within the ORF2 gene. Our analysis demonstrated that recombination has likely also played a significant role in the generation of PCV2 genetic diversity in Henan. Specifically, we found that Henan PCV2 isolates have been involved in both inter- and intra-genotype recombination events. Consistent with previous reports on recombination in PCV [18], the genome regions observed to be most frequently transferred in the Henan isolates were portions of the replication-associated protein gene and ORF2. Recombinants, displaying evidence of ORF1 and ORF2 genes' transfers between different PCV2 genotypes, have previously been shown to display significantly enhanced viral replication and altered antigenicity in vitro [39]. Although we did not detect obvious evidence of recombination between Chinese PCV-2a and PCV-2b genotypes, it is probable that some of the Henan PCV-2b isolates contain fragments of sequence inherited from divergent PCV lineages more distantly related to PCV-2b than PCV-2a (see events 1 and 6 in Table 3). Our results also indicated that recombination between different PCV-2b lineages can occur in the ORF1 and ORF2 genes and that such recombination likely contributes to the genetic diversity of this lineage.
Our analysis of selective pressures acting on ORF2 of the Henan PCV2 strains showed that the four ORF2 epitopes (A, B, C and D) contained a variety of positively and negatively selected positions. Specifically, the few codon sites within these epitopes that were not conserved were mostly detected to be evolving under positive rather than negative selection. This observation corroborates a previous report based on analysis of a far larger PCV2 dataset (including many of the same Chinese sequences analysed here) [30]. In this report, it was demonstrated that although epitope C was mainly evolving under negative selection, position 170, located in decoy epitope, exhibited evidence of positive selection. In our Henan PCV2 dataset, we found that position 178 of this decoy epitope (within the receptor binding domain of the Cap protein) is likely also evolving under positive selection. Such evidence of positive selection within epitopes is generally considered to be evidence of host-driven immune evasion [40]. However, very little experimental evidence exists to corroborate the role of immune evasion in driving the fixation of non-synonymous substitutions in this region of the PCV2 Cap gene.
So far, series commercial recombinant baculovirus expressed PCV-2a capsid protein-based vaccines are available for application in the field. Such proteins have been shown to be protective against PCV-2b infection in the field [41, 42]. Given the capacity for PCV2 to evolve via recombination and mutation in response to the widespread deployment of such vaccines, continual epidemiology surveillance and extra laboratory work is needed to both monitor the changing landscape of PCV2 diversity and determine the impact of this diversity of control strategies. In summary, the current study indicated that a diverse range of recombinogenic PCV2 genotypes is likely endemic in Henan province and these may be capable of rapidly evolving novel lineages capable of overcoming future vaccines as and when they are widely deployed within this region of China.
References
G.M. Allan, F. Mc Neilly, B.M. Meehan, S. Kennedy, D.P. Mackie, J.A. Ellis, E.G. Clark, E. Espuna, N. Saubi, P. Riera, A. Botner, C.E. Charreyre, Vet. Microbiol. 66, 115–123 (1999)
B.M. Meehan, F. McNeilly, D. Todd, S. Kennedy, V.A. Jewhurst, J.A. Ellis, L.E. Hassard, E.G. Clark, D.M. Haines, G.M. Allan, J. Gen. Virol. 79(Pt 9), 2171–2179 (1998)
G.M. Allan, J.A. Ellis, J. Vet. Diagn. Invest. 12, 3–14 (2000)
S. Krakowka, J.A. Ellis, F. McNeilly, S. Ringler, D.M. Rings, G. Allan, Vet. Pathol. 38, 31–42 (2001)
X. Ge, F. Wang, X. Guo, H. Yang, Virus Res. 164, 100–106 (2012)
L. Grau-Roma, L. Fraile, J. Segales, Vet. J. 187, 23–32 (2011)
P. Nawagitgul, I. Morozov, S.R. Bolin, P.A. Harms, S.D. Sorden, P.S. Paul, J. Gen. Virol. 81, 2281–2287 (2000)
B.R. Trible, R.R. Rowland, Virus Res. 164, 68–77 (2012)
G. Misinzo, P.L. Delputte, P. Meerts, D.J. Lefebvre, H.J. Nauwynck, J. Virol. 80, 3487–3494 (2006)
L. Grau-Roma, E. Crisci, M. Sibila, S. Lopez-Soria, M. Nofrarias, M. Cortey, L. Fraile, A. Olvera, J. Segales, Vet. Microbiol. 128, 23–35 (2008)
M. Cortey, A. Olvera, L. Grau-Roma, J. Segales, Vet. Microbiol. 149, 522–523 (2011)
J. Segales, A. Olvera, L. Grau-Roma, C. Charreyre, H. Nauwynck, L. Larsen, K. Dupont, K. McCullough, J. Ellis, S. Krakowka, A. Mankertz, M. Fredholm, C. Fossum, S. Timmusk, N. Stockhofe-Zurwieden, V. Beattie, D. Armstrong, B. Grassland, P. Baekbo, G. Allan, Vet. Rec. 162, 867–868 (2008)
F. Wang, X. Guo, X. Ge, Z. Wang, Y. Chen, Z. Cha, H. Yang, Virus Res. 145, 151–156 (2009)
L.J. Guo, Y.H. Lu, Y.W. Wei, L.P. Huang, C.M. Liu, Virol. J. 7, 273 (2010)
L. Wen, X. Guo, H. Yang, Vet. Microbiol. 110, 141–146 (2005)
W. Li, X. Wang, T. Ma, Z. Feng, Y. Li, P. Jiang, Virus Genes 40, 244–251 (2010)
J. Shuai, W. Wei, X. Li, N. Chen, Z. Zhang, X. Chen, W. Fang, Virus Genes 35, 619–627 (2007)
L. Cai, X. Han, J. Ni, X. Yu, Z. Zhou, X. Zhai, X. Chen, K. Tian, Virus Res. 158, 281–288 (2011)
L. Cai, J. Ni, Y. Xia, Z. Zi, K. Ning, P. Qiu, X. Li, B. Wang, Q. Liu, D. Hu, X. Yu, Z. Zhou, X. Zhai, X. Han, K. Tian, Virus Res. 165, 95–102 (2012)
C.M. Ma, C.C. Hon, T.Y. Lam, V.Y. Li, C.K. Wong, T. de Oliveira, F.C. Leung, J. Gen. Virol. 88, 1733–1737 (2007)
P. Lefeuvre, J.M. Lett, A. Varsani, D.P. Martin, J. Virol. 83, 2697–2707 (2009)
R. Hesse, M. Kerrigan, R.R. Rowland, Virus Res. 132, 201–207 (2008)
J.D. Thompson, T.J. Gibson, D.G. Higgins, Curr. Protoc. Bioinformatics. Chapter 2, Unit 2.3 (2002)
K. Tamura, D. Peterson, N. Peterson, G. Stecher, M. Nei, S. Kumar, Mol. Biol. Evol. 28, 2731–2739 (2011)
D. Martin, E. Rybicki, Bioinformatics 16, 562–563 (2000)
X. Xia, Z. Xie, J. Hered. 92, 371–373 (2001)
Y. Suzuki, T. Gojobori, Mol. Biol. Evol. 16, 1315–1328 (1999)
S.L. Pond, S.D. Frost, S.V. Muse, Bioinformatics 21, 676–679 (2005)
Z. Yang, R. Nielsen, N. Goldman, A.M. Pedersen, Genetics 155, 431–449 (2000)
A. Olvera, M. Cortey, J. Segales, Virology 357, 175–185 (2007)
L.J. Perez, H.D. de Arce, M. Cortey, P. Dominguez, M.I. Percedo, C.L. Perera, J. Tarradas, M.T. Frias, J. Segales, L. Ganges, J.I. Nunez, Vet. Microbiol. 151, 245–254 (2011)
P. Lekcharoensuk, I. Morozov, P.S. Paul, N. Thangthumniyom, W. Wajjawalku, X.J. Meng, J. Virol. 78, 8135–8145 (2004)
D. Mahe, P. Blanchard, C. Truong, C. Arnauld, P. Le Cann, R. Cariolet, F. Madec, E. Albina, A. Jestin, J. Gen. Virol. 81, 1815–1824 (2000)
L.S. Stevenson, D.F. Gilpin, A. Douglas, F. McNeilly, I. McNair, B.M. Adair, G.M. Allan, Viral Immunol. 20, 389–398 (2007)
J. Gu, R. Cao, Y. Zhang, X. Lian, H. Ishag, P. Chen, Vet. J. 192, 385–389 (2012)
L.P. Huang, Y.H. Lu, Y.W. Wei, L.J. Guo, C.M. Liu, BMC Microbiol. 11, 188 (2011)
L. Guo, Y. Lu, L. Huang, Y. Wei, C. Liu, Intervirology 54, 156–163 (2011)
H.K. Kim, Y. Luo, H.J. Moon, S.J. Park, H.O. Keum, S. Rho, B.K. Park, Virus Genes 39, 352–358 (2009)
L. Guo, Y. Lu, Y. Wei, L. Huang, H. Wu, C. Liu, Virology 419, 57–63 (2011)
B.T. Grenfell, O.G. Pybus, J.R. Gog, J.L. Wood, J.M. Daly, J.A. Mumford, E.C. Holmes, Science 303, 327–332 (2004)
M. Fort, M. Sibila, A. Allepuz, E. Mateu, F. Roerink, J. Segales, Vaccine 26, 1063–1071 (2008)
Y. Takahagi, S. Toki, Y. Nishiyama, F. Morimatsu, H. Murakami, J. Vet. Med. Sci. 72, 35–41 (2010)
Acknowledgements
This study was supported by the National Nature Science Foundation of China (No. 31172346).We are grateful to Professor Darren P. Martin in University of Cape Town for technical support. We would like to thank the reference sequence submitters for their original works.
Conflict of interest
None of the authors has any financial or personal relationships that could inappropriately influence or bias the content of the paper
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Mu, C., Yang, Q., Zhang, Y. et al. Genetic variation and phylogenetic analysis of porcine circovirus type 2 infections in central China. Virus Genes 45, 463–473 (2012). https://doi.org/10.1007/s11262-012-0789-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-012-0789-7