Abstract
Mosquitoes of the genus Aedes are known vectors of pathogenic flaviviruses, and insect-specific flaviviruses (ISFs) have been detected in members of this genus in numerous parts of the world. In order to gain insight into whether Aedes mosquitoes in Greece are infected by flaviviruses, 1173 Aedes spp. mosquitoes collected in 2010 and 2012 were grouped in 53 pools and tested by RT nested PCR using flavivirus generic primers. Eight pools (15.09 %) were found to be PCR positive: five pools (5/53, 9.4 %) contained RNA sequences related to Ochlerotatus caspius flavivirus (OCFV), an ISF previously detected in the Iberian peninsula, two pools (2/53, 3.8 %) contained sequences related to a mosquito flavivirus detected in Aedes vexans (AeveV) in Italy and the Czech Republic, and one pool contained a DNA sequence that was too short to identify accurately. The highest OCFV prevalence (12.9 %) was observed in August 2010 in the regional unit of Thessaloniki. Similar sequences were later obtained from two Culex spp. pools collected in 2013 in the same regions. A genetic difference of 0.2-1.4 % was seen among the Greek OCFV strains, which differed by 2.2-4.1 % from the Iberian strains and by 6.2-11.1 % from the Finnish Hanko virus. The genetic distances among strains varied depending on the genome region (genes for E, NS3 and NS5 proteins), with NS3 being the most variable. The present study shows no evidence of infection of Aedes mosquitoes with known pathogenic flaviviruses, but it expands the geographic distribution of OCFV in the eastern Mediterranean area. Any implication of ISFs for public health (either directly or through interactions with other flaviviruses in the mosquitoes) remains to be elucidated.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Aedes mosquitoes are known vectors of pathogenic flaviviruses (genus Flavivirus, family Flaviviridae), including dengue virus and yellow fever virus. Furthermore, insect-specific flaviviruses (ISFs) have been detected in these mosquito species in numerous regions of the world, including European countries [2, 7, 14, 16, 24, 25, 31]. ISFs comprise a distinct group of flaviviruses that do not infect mammalian cells, replicate only in mosquitoes or mosquito cell lines, and are not associated with disease in humans. They are probably primordial flaviviruses [8, 27]. Some ISFs have been shown to produce DNA forms of their genomic RNA; integrated sequences related to ISFs have been detected in A. aegypti and A. albopictus mosquitoes [9, 31].
In 2010, West Nile virus (WNV) emerged in Greece and caused large outbreaks of human infections [20]. During entomological surveys conducted in the summer months of 2010-2014, WNV (lineage 2) was detected every year in Culex spp. mosquitoes [11, 18, 19, 21–23]. Furthermore, Culex theileri flavivirus, an ISF, has been detected in Culex spp. mosquitoes in Greece [23]. There is increasing interest in the interactions of ISFs with pathogenic flaviviruses (including WNV) in mosquitoes, and studies have produced contradictory results. It appears that these interactions depend on several factors, including the species of mosquitoes, the flaviviruses involved, the timing of infection, and the load of each virus [4, 5, 13, 15]. Concerning the role of Aedes spp. in the WNV life cycle, vector competence studies have shown that Aedes (Ochlerotatus) caspius is an inefficient vector of WNV in the laboratory [3], while Aedes vexans may play a role in WNV enzootic cycles [12, 30]. The vast majority of Aedes spp. mosquitoes in the rice fields in Thessaloniki, northern Greece, are Aedes caspius, representing 28.5 % of the trapped mosquitoes. The other mosquito species are Anopheles pseudopictus (21 %), Culex modestus (19.5 %), C. pipiens (16.8 %), Anopheles hyrcanus (9.6 %) and Anopheles sacharovi (4.3 %) [8]. Since Aedes mosquitoes in Greece have never been tested for flaviviruses, the aim of the present study was to check whether Aedes spp. mosquitoes collected during WNV outbreaks were infected by the virus and to investigate whether they were carrying any other pathogenic flavivirus or ISF.
Materials and methods
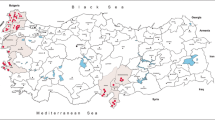
Following the emergence of WNV in 2010 in Greece, mosquito surveillance studies were conducted every year, targeting mainly Culex spp. mosquitoes. During 2010 and 2012, 1173 Aedes spp. mosquitoes were transported in dry ice to the National Reference Centre for Arboviruses in order to be tested for WNV infection (in 2011, only Culex spp. mosquitoes were studied). The mosquitoes were collected using CO2–baited light traps and were identified at the genus level using dichotomous determination keys [10]. Of these, 469 were collected in August and September 2010 (soon after the initiation of the WNV outbreak) at 33 sites in six regional units (Nomenclature of Territorial Units for Statistics 3 level: Imathia, Kilkis, Larisa, Pella, Pieria, Thessaloniki), and 704 were collected in October 2012 at nine sites in five regional units (Chalkidiki, Imathia, Pieria, Serres, Thessaloniki) (Table 1, Fig. 1).
The Aedes spp. mosquitoes were grouped into 53 pools of up to 100 individuals that were sorted according to the collection site and date. RNA was extracted using the RNeasy Mini Kit (QIAGEN, Hilden, Germany), and a reverse transcriptase (RT)-nested PCR using flavivirus generic primers was applied [26]. Positive samples were tested by PCR with three additional sets of primers (primer sequences are available upon request) to obtain sequences of a 520-bp fragment of the envelope (E) protein gene and 500-bp and 650-bp fragments of the genes for the non-structural (NS) proteins NS3 and NS5, respectively. All flavivirus-positive pools were re-tested without the reverse transcription (RT) step in order to check for possible DNA forms. Sequencing of the PCR products was performed using a BigDye Terminator v3.1 Cycle Sequencing Kit in a 3130 ABI Genetic Analyzer (Life Technologies, Carlsbad, CA, USA). Multiple alignment of the sequences was performed using Clustal W, and phylogenetic trees were constructed by the neighbor-joining method with 1,000 bootstrap replicates in MEGA6 [29]. Virus isolation was attempted in Vero E6 cells.
Results
Eight (8/53, 15.1 %) pools of Aedes spp. mosquitoes were found to contain flavivirus sequences. Using the NCBI BLAST search tool [1], five sequences showed the highest similarity to Ochlerotatus caspius flavivirus (OCFV), which was previously detected in Portugal [14] and Italy [6], two sequences were 100 % identical to Aedes vexans flavivirus (AeVeV), previously detected in Italy (GQ476997) [6] and the Czech Republic (JN802280) [7], and one sequence was too short (<60 bp) to be indentified accurately (Table 1). When the eight positive pools were retested by PCR without the reverse transcription step, the five OCFV-positive pools were negative, suggesting that they were RNA forms, while the remaining three pools were positive, suggesting that they included DNA forms. However, it cannot be excluded that RNA forms were also present in the samples. Virus isolation attempted in Vero E6 cells from two OCFV-positive pools was not successful. The Aedes albopictus C6/36 cell line, which is suitable for the replication of ISFs, was not available.
The percentage of OCFV-positive mosquito pools was 9.43 % (5/53 pools), with the highest percentage observed in 2010 in the regional unit of Thessaloniki (4/31, 12.9 %) (Table 1, Fig. 1). The exact collection sites and dates of the OCFV-positive pools are shown in Table 2. Two pools of Culex spp. mosquitoes (100 mosquitoes per pool) collected in 2013 in which OCFV sequences were detected are also shown in Table 2 (marked with an asterisk).
Most of the OCFV-positive Aedes mosquitoes (4/5 pools) were collected in August 2010 in the regional unit of Thessaloniki, while all Aedes mosquitoes (23 pools) collected in September 2010 at the same sites were negative. Additional sequences from the partial E, NS3 and NS5 genes were obtained from three of the five OCFV-positive samples. The mean genetic differences among the Greek OCFV flaviviruses in partial fragments of the E, NS3 and NS5 genes at the nucleotide level were 0.4 %, 1.4 % and 0.2 %, respectively, and 0 %, 0.2 % and 0 % at the amino acid level. In all three phylogenetic trees, the Greek OCFV strains constitute a distinct subclade, clustering with related sequences obtained from Portugal [14], Spain (referred to as Mediterranean Ochlerotatus flavivirus) [31] and Finland (Hanko virus, HANKV) [16] (Fig. 2A, B, and C). The genetic differences at the nucleotide and amino acid levels between the Greek OCFV strains and the related viruses from the Iberian Peninsula (Spain and Portugal) and Finland are shown in Table 3. In all fragments, the nucleotide sequence identity is >84 %, which is the threshold value for flaviviruses to be classified within the same species [17]. The nucleotide sequences obtained from the present study have been deposited in the GenBank database under the accession numbers KM245094-KM245102.
Neighbor-joining phylogenetic trees based on partial fragments of the genes encoding the A) E (507 nt), B) NS3 (461 nt), and C) NS5 (460 nt) proteins of OCFV. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The percentage of replicate trees in which the sequences clustered together in the bootstrap test (1000 replicates) is shown next to the branches. Sequences from the present study are indicated. Evolutionary analysis was conducted in MEGA6 [29]
Discussion
In order to determine whether Aedes mosquitoes in Greece were infected by WNV and/or other flaviviruses during WNV outbreaks, a generic flavivirus RT nested PCR was applied to test genetic material extracted from Aedes spp. mosquitoes collected in various locations in northern Greece. Of the 53 Aedes spp. pools tested, eight (15.09 %) were found to be flavivirus positive. Five of the eight sequences obtained from the generic PCR showed highest similarity (>90 %) to OCFV detected in Portugal [14]. This was confirmed by phylogenetic analysis of partial genome fragments corresponding to the E, NS3 and NS5 genes: the Greek OCFV sequences clustered together with OCFV sequences from Spain [31] and Portugal [14], with HANKV being included in the same clade [16].
The genetic difference among the Greek OCFV strains was 0.2-1.4 %, and these strains differed by 2.2-4.1 % from the Iberian strains, and by 6.2-11.1 % from HANKV. The differences among strains varied depending on the genome region, with the NS3 gene being the most variable; however, the threshold value of 84 % [17] was not overcome even in the NS3 gene. Nucleotide sequence differences were observed even between sequences detected in pools from mosquitoes collected in the same area. For example, the pools Thessaloniki 23/10 and Thessaloniki 25/10 consisted of mosquitoes collected on the same night in two locations in the Thessaloniki region only 10 km apart. According to Kuno et al. (1998), all these viruses should be considered members of one virus species. Blitvich and Firth [4] suggested HANKV as the name of these related viruses.
There was a drastic difference in the prevalence between 2010 and 2012, since all of the OCFV-positive pools were collected in 2010 (5 of 44 pools, 11.36 %), while all mosquitoes collected in 2012 were OCFV negative. It has to be mentioned that the collection of 2010 was performed from mid-August to late September, while the collection of 2012 was conducted from late September to early October. A seasonality of activity was observed, since all OCFV-positive Aedes mosquitoes were collected in August 2010, while all (23 pools) Aedes mosquitoes collected in September 2010 at the same sites were negative. Furthermore, OCFV-positive Culex spp. mosquitoes were collected in the same season (late July and mid-August of 2013 (Table 3). The fact that similar OCFV sequences were taken from Culex mosquitoes suggests that Culex spp. can also be infected by OCFV. Similar findings were reported also in other countries in southern Europe [7].
Since 1975, when the first ISF (cell fusing agent virus, CFAV) was isolated [28], the number of ISFs has increased dramatically [4], mainly through entomological surveys in which the detection of pathogenic flaviviruses was attempted using PCR protocols with generic primers. So far, ISFs of the OCFV and HANKV group have been detected in Portugal [14], Spain [31], Italy [6], Finland [16] and Greece, suggesting that they are widely distributed in Europe. There are several issues concerning ISFs that need to be clarified, such as their spatial and temporal distribution, factors associated with their prevalence, their complete genome sequences, and their possible effect on human health, either directly or through their interactions with other flaviviruses in the mosquitoes.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Aranda C, Sanchez-Seco MP, Caceres F, Escosa R, Galvez JC, Masia M, Marques E, Ruiz S, Alba A, Busquets N, Vazquez A, Castella J, Tenorio A (2009) Detection and monitoring of mosquito flaviviruses in Spain between 2001 and 2005. Vector Borne Zoonotic Dis 9:171–178
Barrera R, MacKay A, Amador M, Vasquez J, Smith J, Diaz A, Acevedo V, Caban B, Hunsperger EA, Munoz-Jordan JL (2010) Mosquito vectors of West Nile virus during an epizootic outbreak in Puerto Rico. J Med Entomol 47:1185–1195
Blitvich BJ, Firth AE (2015) Insect-specific flaviviruses: a systematic review of their discovery, host range, mode of transmission, superinfection exclusion potential and genomic organization. Viruses 7:1927–1959
Bolling BG, Olea-Popelka FJ, Eisen L, Moore CG, Blair CD (2012) Transmission dynamics of an insect-specific flavivirus in a naturally infected Culex pipiens laboratory colony and effects of co-infection on vector competence for West Nile virus. Virology 427:90–97
Calzolari M, Bonilauri P, Bellini R, Caimi M, Defilippo F, Maioli G, Albieri A, Medici A, Veronesi R, Pilani R, Gelati A, Angelini P, Parco V, Fabbi M, Barbieri I, Lelli D, Lavazza A, Cordioli P, Dottori M (2010) Arboviral survey of mosquitoes in two northern Italian regions in 2007 and 2008. Vector Borne Zoonotic Dis 10:875–884
Calzolari M, Ze-Ze L, Ruzek D, Vazquez A, Jeffries C, Defilippo F, Osorio HC, Kilian P, Ruiz S, Fooks AR, Maioli G, Amaro F, Tlusty M, Figuerola J, Medlock JM, Bonilauri P, Alves MJ, Sebesta O, Tenorio A, Vaux AG, Bellini R, Gelbic I, Sanchez-Seco MP, Johnson N, Dottori M (2012) Detection of mosquito-only flaviviruses in Europe. J Gen Virol 93:1215–1225
Cammisa-Parks H, Cisar LA, Kane A, Stollar V (1992) The complete nucleotide sequence of cell fusing agent (CFA): homology between the nonstructural proteins encoded by CFA and the nonstructural proteins encoded by arthropod-borne flaviviruses. Virology 189:511–524
Crochu S, Cook S, Attoui H, Charrel RN, De Chesse R, Belhouchet M, Lemasson JJ, de Micco P, de Lamballerie X (2004) Sequences of flavivirus-related RNA viruses persist in DNA form integrated in the genome of Aedes spp. mosquitoes. J Gen Virol 85:1971–1980
Darsie RF, Samanidou-Voyadjoglou A (1997) Keys for the identification of the mosquitoes of Greece. J Am Mosq Control Assoc 13:247–254
Engler O, Savini G, Papa A, Figuerola J, Groschup MH, Kampen H, Medlock J, Vaux A, Wilson AJ, Werner D, Jost H, Goffredo M, Capelli G, Federici V, Tonolla M, Patocchi N, Flacio E, Portmann J, Rossi-Pedruzzi A, Mourelatos S, Ruiz S, Vazquez A, Calzolari M, Bonilauri P, Dottori M, Schaffner F, Mathis A, Johnson N (2013) European surveillance for West Nile virus in mosquito populations. Int J Environ Res Public Health 10:4869–4895
Fall AG, Diaite A, Etter E, Bouyer J, Ndiaye TD, Konate L (2012) The mosquito Aedes (Aedimorphus) vexans arabiensis as a probable vector bridging the West Nile virus between birds and horses in Barkedji (Ferlo, Senegal). Med Vet Entomol 26:106–111
Farfan-Ale JA, Lorono-Pino MA, Garcia-Rejon JE, Hovav E, Powers AM, Lin M, Dorman KS, Platt KB, Bartholomay LC, Soto V, Beaty BJ, Lanciotti RS, Blitvich BJ (2009) Detection of RNA from a novel West Nile-like virus and high prevalence of an insect-specific flavivirus in mosquitoes in the Yucatan Peninsula of Mexico. Am J Trop Med Hyg 80:85–95
Ferreira DD, Cook S, Lopes A, de Matos AP, Esteves A, Abecasis A, de Almeida AP, Piedade J, Parreira R (2013) Characterization of an insect-specific flavivirus (OCFVPT) co-isolated from Ochlerotatus caspius collected in southern Portugal along with a putative new Negev-like virus. Virus Genes 47:532–545
Hobson-Peters J, Yam AW, Lu JW, Setoh YX, May FJ, Kurucz N, Walsh S, Prow NA, Davis SS, Weir R, Melville L, Hunt N, Webb RI, Blitvich BJ, Whelan P, Hall RA (2013) A new insect-specific flavivirus from northern Australia suppresses replication of West Nile virus and Murray Valley encephalitis virus in co-infected mosquito cells. PLoS One 8:e56534
Huhtamo E, Moureau G, Cook S, Julkunen O, Putkuri N, Kurkela S, Uzcategui NY, Harbach RE, Gould EA, Vapalahti O, de Lamballerie X (2012) Novel insect-specific flavivirus isolated from northern Europe. Virol 433:471–478
Kuno G, Chang GJ, Tsuchiya KR, Karabatsos N, Cropp CB (1998) Phylogeny of the genus Flavivirus. J Virol 72:73–83
Papa A, Bakonyi T, Xanthopoulou K, Vazquez A, Tenorio A, Nowotny N (2011) Genetic characterization of West Nile virus lineage 2, Greece, 2010. Emerg Infect Dis 17:920–922
Papa A, Xanthopoulou K, Gewehr S, Mourelatos S (2011) Detection of West Nile virus lineage 2 in mosquitoes during a human outbreak in Greece. Clin Microbiol Infect 17:1176–1180
Papa A (2013) West Nile virus infections in humans-Focus on Greece. J Clin Virol 58:351–353
Papa A, Papadopoulou E, Gavana E, Kalaitzopoulou S, Mourelatos S (2013) Detection of West Nile virus lineage 2 in Culex mosquitoes, Greece, 2012. Vector Borne Zoonotic Dis 13:682–684
Papa A, Xanthopoulou K, Tsioka A, Kalaitzopoulou S, Mourelatos S (2013) West Nile virus in mosquitoes in Greece. Parasitol Res 112:1551–1555
Papa A, Papadopoulou E, Kalaitzopoulou S, Tsioka K, Mourelatos S (2014) Detection of West Nile virus and insect-specific flavivirus RNA in Culex mosquitoes, central Macedonia, Greece. Trans R Soc Trop Med Hyg 108:555–559
Roiz D, Vazquez A, Rosso F, Arnoldi D, Girardi M, Cuevas L, Perez-Pastrana E, Sanchez-Seco MP, Tenorio A, Rizzoli A (2012) Detection of a new insect flavivirus and isolation of Aedes flavivirus in Northern Italy. Parasit Vectors 5:223
Roiz D, Vazquez A, Seco MP, Tenorio A, Rizzoli A (2009) Detection of novel insect flavivirus sequences integrated in Aedes albopictus (Diptera: Culicidae) in Northern Italy. Virol J 6:93
Sanchez-Seco MP, Rosario D, Domingo C, Hernandez L, Valdes K, Guzman MG, Tenorio A (2005) Generic RT-nested-PCR for detection of flaviviruses using degenerated primers and internal control followed by sequencing for specific identification. J Virol Methods 126:101–109
Schlesinger RW (1971) New opportunities in biological research offered by arthropod cell cultures. I. some speculations on the possible role of arthropods in the evolution of arboviruses. Curr Top Microbiol Immunol 55:241–245
Stollar V, Thomas VL (1975) An agent in the Aedes aegypti cell line (Peleg) which causes fusion of Aedes albopictus cells. Virol 64:367–377
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tiawsirisup S, Kinley JR, Tucker BJ, Evans RB, Rowley WA, Platt KB (2008) Vector competence of Aedes vexans (Diptera: Culicidae) for West Nile virus and potential as an enzootic vector. J Med Entomol 45:452–457
Vazquez A, Sanchez-Seco MP, Palacios G, Molero F, Reyes N, Ruiz S, Aranda C, Marques E, Escosa R, Moreno J, Figuerola J, Tenorio A (2012) Novel flaviviruses detected in different species of mosquitoes in Spain. Vector Borne Zoonotic Dis 12:223–229
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
The study was funded by the Development Agency of Thessaloniki, SA. The collaboration between Aristotle University of Thessaloniki and Robert Koch Institute was supported by IKYDA 2013 (granted by the State Scholarships Foundation in Greece and German Academic Exchange Service-DAAD).
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
Not needed.
Rights and permissions
About this article
Cite this article
Papa, A., Papadopoulou, E., Paliwal, R. et al. Insect-specific flaviviruses in Aedes mosquitoes in Greece. Arch Virol 161, 2183–2188 (2016). https://doi.org/10.1007/s00705-016-2877-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-016-2877-9