Abstract
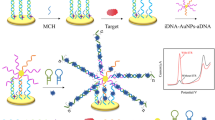
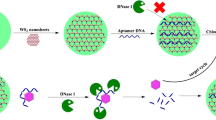
An electrochemical aptasensor, including the polyethyleneimine-graphite-like carbon nitride/Au nanowire nanocomposite (PEI-C3N4/AuNWs) and exonuclease-assisted signal amplification strategy was constructed for the determination of chloramphenicol (CAP). Initially, a nanocomposite with substantial electrocatalytic property was synthesized by PEI-C3N4/AuNWs. This improves the conductivity and specific surface area of the PEI-C3N4/AuNW–modified gold electrode. Next, a DNA with a complementary sequence to a CAP aptamer (cDNA) was immobilized on the PEI-C3N4/AuNW–modified electrode, followed by the CAP aptamer hybridized with cDNA. The lower signal at this time is due to the negatively charged phosphate group of the oligonucleotide and [Fe (CN)6]3−/4− electrostatically repelling each other. The presence of the CAP would cause aptamer on the electrode surface to fall off and be digested by Recjf exonuclease, which resulted in target recycling, and a significant increase in DPV signal can be observed at a potential of 0.176 V (vs. Ag/AgCl). Under optimal conditions, there is a linear relationship between the peak current and the logarithm of CAP concentration in the range 100 fM–1 μM, and the detection limit of this aptasensor is 2.96 fM (S/N = 3). Furthermore, the resultant aptasensor has excellent specificity, reproducibility, and long-term stability, and has been applied to the detection of CAP in milk samples.

The detection principle of the electrochemical aptasensor for CAP detection was based on PEI-C3N4/AuNWs and exonuclease-assistant signal amplification. It is based on the fact that PEI-C3N4/AuNWs nanocomposites on the surface of the electrode can effectively improve the performance of the aptasensor, and Recjf exonuclease initiates the target recycling process, causes signal amplification.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As the first mass-produced antibiotic, chloramphenicol (CAP) is used to treat human and animal diseases in the breeding industry worldwide [1,2,3]. Nevertheless, some side effects may be caused by the enrichment of the food chain in the human body [4]. Excessive intake of CAP has been found to be harmful to human health, such as bone marrow suppression, leukemia, and fatal irreversible aplastic anemia on human beings [5]. Thus, it is crucial to establish a rapid and efficient method to realize the sensitive detection of CAP residues in foodstuffs.
Traditionally, CAP has been detected using a variety of techniques such as high-performance liquid chromatography (HPLC) [6], gas chromatography-mass spectrometry (GC-MS) [7], and liquid chromatography-tandem mass spectrometry [8]. Although these methods are reliable and stable, there are still some restrictions in their practical application, such as expensive instruments, high expense, and time-consuming. Electrochemical analysis has attracted researchers’ attention due to its advantages such as easy to operation, rapid response, and excellent sensitivity [9, 10]. As an identification element in the electrochemical sensor, aptamer (Apt) has been used in the development of various biosensors and is widely applied in CAP analysis [11,12,13]. Compared with other recognition elements such as antibody, aptamers exhibit excellent stability, low cost, and target diversity [14]. Therefore, we conducted CAP detection studies based on electrochemical aptasensors.
Nanomaterials, such as nanoparticles [15, 16], graphene [17], metal-organic frameworks (MOFs) [18], and other carbon-based nanomaterials [19, 20], are widely used in the construction of electrochemical sensors to amplify signals. They can provide high specific surface area to enhance electron transfer, and improve sensors’ performance [10]. Graphite-like carbon nitride (g-C3N4) has the advantages of large specific surface area, stable chemical properties, and unique electronic, mechanical properties [21], which makes it become an ideal material for constructing biosensors [22]. Polyethyleneimine (PEI), a liquid crystal polymer, is widely used to enhance the dispersibility and stability of nanomaterials by functionalizing them [23]. It can bind g-C3N4 to the gold electrode (AuE) surface and further bind it to Au nanomaterials based on its amino group. Therefore, to further improve the conductivity of nanomaterials, we introduced Au nanowires to prepare composite nanomaterials. Au nanowires (AuNWs) have good conductivity, promote electron transfer, and improve analytical sensitivity. It also has the advantages of a unique quantum size effect, a large number of exposed surface atoms, high biocompatibility, and high flexibility [24, 25]. Besides, AuNWs can provide binding sites for aptamer through the Au-N bond, which allows for specific recognition of sensors. For the above reasons, PEI-C3N4/AuNWs is considered a suitable choice for fabricating electrochemical aptasensor, which has been rarely reported in the field aptasensor for the detection of chloramphenicol.
On the other hand, the sensitivity and specificity of electrochemical aptasensor depend mainly on the signal changes before and after the specific-recognition. Exonuclease-assisted target recycling offered a feasible strategy for signal amplification. As an exonuclease acting exclusively on single-stranded DNA (ssDNA), RecJf exonuclease can catalyze the removal of deoxynucleotide monophosphates from ssDNA in the 5′-3′ direction [14]. In particular, RecJf exonuclease is expected to selectively cleave the target-aptamer complex and liberate the target, which allows the target to further complex with a remaining aptamer on the electrode surface. In this way, the availability of aptamer is improved and the target recycling is realized.
Inspired by the as-mentioned items, in this work, a sensitive electrochemical aptasensor based on PEI-C3N4/AuNWs and RecJf exonuclease was developed determination of chloramphenicol (CAP). The as-prepared PEI-C3N4/AuNWs can be a suitable nanomaterial to improve the conductivity and specific surface area of the gold electrode. Then, a complementary DNA (cDNA) and CAP aptamer were hybridized on the PEI-C3N4/AuNW–modified electrode. The corresponding current was low attributable to the electrostatic repulsion between [Fe (CN)6]3−/4− and the aptamer. In the presence of CAP and Recjf exonuclease, the CAP-aptamer complex was cleaved from the electrode surface, RecJf exonuclease release CAP from the CAP-aptamer complex, allowing it to initiate the next hybridization process. Furthermore, the remaining cDNA on the electrode surface is also digested by Recjf exonuclease. Eventually, a significantly amplified current can be obtained owing to the endeavor of the RecJf exonuclease and PEI-C3N4/AuNWs. Moreover, the resultant aptasensor has excellent specificity, reproducibility, and long-term stability, and has been applied to the detection of CAP in milk samples.
Experimental section
Reagent, materials, and apparatus
All reagents, materials, and apparatus used in the experiments are shown in the “Supplementary information” section.
Synthesis of PEI-C3N4/AuNWs
In a typical synthesis of ultrathin AuNWs [24], 4.5 mL of 0.50 M α-naphthol ethanol solution was added into 4.5 mL of 0.05 M HAuCl4 aqueous solution at 60 °C. The solution color changed immediately from yellow to black, indicating the rapid reduction of Au (III) to Au (0). After aging for a short while (~ 1 min), the resultant product was collected by centrifugation and washed with ethanol.
Carboxylated g-C3N4 was prepared by a thermal oxidation method [26]. First, 10 g white melamine powers were annealed at 550 °C for 4 h. After that, a yellow bulk C3N4 (b-C3N4) was obtained. To improve the solubility of g-C3N4 in water,1 g above obtained products ground to powder was put into 100 mL of HNO3 (5 mol/L) and the mixture was subjected to ultrasound for 1 h. Subsequently, the solution was refluxed for 24 h at 125 °C. Until it cooled, centrifuge at 5000 rpm to remove residual unpeeled C3N4 and collect the supernatant. The supernatant was filtered through a 45 μm water-phase microporous membrane filter and washed with secondary distilled water to neutrality, and carboxylated g-C3N4 nanosheets were separated. Finally, the final product was dried in a vacuum oven at 35 °C for 12 h.
After preparing carboxylated g-C3N4 and AuNWs separately, the synthesis of PEI-C3N4/AuNWs was performed. First, to increase the amount of amino groups in carboxylated g-C3N4 to load more AuNWs, PEI functionalization was performed on it. The obtained carboxylated g-C3N4 (6 mg) was dissolved in ultrapure water (6 mL) containing 60 μL of PEI (2%, 20 mg/mL), and stirred at room temperature overnight (13 h). Centrifuge at 12000 rpm for 5 min to remove unreacted PEI and resuspend the obtained PEI-C3N4 precipitate in 2 mL of twice-distilled water. After that, 3 mL of the prepared AuNWs was added to the obtained PEI-C3N4 solution under magnetic stirring (magnetic stirring for 12 h). Subsequently, the PEI-C3N4/AuNWs composite obtained by centrifugation was collected and then dispersed in 1 mL of secondary distilled water.
Fabrication of the aptasensor
The fabrication process of the aptasensor was exhibited in Scheme 1. The gold electrodes (AuE) need pretreatment before the fabrication of the aptasensor. Briefly, 0.05 μm Al2O3 was used to polish AuE, followed by ultrasonically washing in ethanol and distilled water for 5 min, in turn. After that, soak the AuE in the freshly prepared piranha solution (H2SO4: 30% H2O2 = 7:3) for 10 min, and then, it was rinsed with double-distilled water. After drying at room temperature, the prepared AuE was modified.
Determination of CAP
For the detection of CAP, 10 μL mixture containing 8 μL of different CAP concentrations and 2 μL of 2 U/μL RecJf exonuclease was added to the surface of the electrodes and then incubated for 2 h at 37 °C for an adequate reaction. Subsequently, the electrode was gently rinsed with Tris-HCL and the response of the electrochemical signal was recorded in 5 mM [Fe (CN)6]3−/4− solution. The concentration of CAP was quantified according to the difference of the DPV signal (ΔI). The calculation formula is ΔI = I1 − I0, where I0 is the DPV signal value after incubation of the blank solution, I1 is the DPV signal value in the presence of CAP.
Real sample preparation
First, a certain amount of CAP was added to the milk; 2 mL of spiked milk and 3 mL of water were mixed, and then, 1 mL of 10% trichloroacetic acid and 1 mL of chloroform were added to the solution, which was then vortex-mixed for 1 min to precipitate proteins and dissolve organic substances such as fat in the milk. Furthermore, the mixture was treated with ultrasound at 20 °C for 15 min, the solution was centrifuged at 12,000 rpm for 20 min, and the supernatant was retained. Finally, the supernatant was analyzed according to the procedure in the “Determination of CAP” section. Each experiment was analyzed three times. The data points were acquired from the current value at working voltage 0.176 V (vs. Ag/AgCl).
Results and discussion
Characterization of materials
The morphology of the as-prepared product is illustrated by TEM. The sheet structure with curled edges of PEI-C3N4 can be seen in Fig. 1a, which providing a large surface area to load more AuNWs. Figure 1b and c show that the as-prepared AuNWs showed a linear structure with the length of hundreds of nanometers and the average diameter of merely 10 nm, it interlaced to form a tight network structure. Figure 1d shows the TEM images of PEI-C3N4/AuNWs. Many AuNWs are connected to the surface, and the morphology of each material is the same as before the composite, no impurities appear.
Figure 1e shows the FT-IR spectrum of as-prepared carboxylated g-C3N4 in the wavelength range of 500–4000 cm−1. The band at 812 cm−1 is a characteristic of tristriazine. Also, the band at 1247 cm−1 corresponds to the C–N stretching of g-C3N4. Furthermore, the band at 3127 cm−1 may be attributed to the intermolecular hydrogen bonding of primary and secondary amines and the stretching vibration of O–H in water molecules. Moreover, the band at 1414 cm−1, 1636 cm−1, 1575 cm−1, and 3249 cm−1 may correspond to bending vibration of C=O, stretching vibration of C–O, -COO, and O–H. This result is consistent with the literature reported previously [27], and the above results indicate that carboxylated g-C3N4 was successfully prepared.
Moreover, the prepared PEI-C3N4 and PEI-C3N4/AuNWs structure was recorded by X-ray diffraction (XRD). As shown in curve a of Fig. 1f, the strong and sharp peak at 27.3° is attributed to characteristic (002) plane diffraction of PEI-C3N4 as a layered nanosheet material. After incorporating with AuNWs, the obtained PEI-C3N4/AuNWs maintain the XRD characteristic peaks of the PEI-C3N4, while the new diffraction peaks (curves b, Fig. 1f) emerge at 37.9°, 44.8°, 64.4°, 77.4°, and 81.7°, which are attributed to (111), (200), (220), (311), and (222) planes of AuNWs [24]. These results indicate that the AuNWs has been incorporated with the PEI-C3N4 successfully, which was consistent with the reported literature results [28].
To investigate the electroactive surface area of PEI-C3N4/AuNWs, we measured and compared the corresponding current response of the bare AuE and the AuE modified with PEI-C3N4/AuNWs in 5 mM [Fe (CN)6]3−/4-. According to the Randles-Sevcik equation, electroactive surface area of two electrodes was further quantitated. The detailed calculation process was given in the “Supplementary information” section.
Electrochemical characterization of the constructed aptasensor
EIS monitored the stepwise modifications of the sensing electrode surface. In the EIS spectrum, the diameter of the semicircle represents the electron transfer resistance (Ret). When different materials are modified onto the electrode surface, the value of Ret changes accordingly. As shown in Fig. 2, the bare AuE shows a large semicircle diameter (curve a, 187 Ω). When the PEI-C3N4/AuNWs is bonded to the electrode surface, the Ret of PEI-C3N4/AuNWs/AuE decreased dramatically (curve b, 58 Ω). This shows that PEI-C3N4/AuNWs can facilitate electron transfer and improves electrode performance effectively. With the modification of cDNA on the electrode, a large semicircle diameter appears (curve c, 220 Ω), suggesting that cDNA binds to the electrode surface and prevents electron transfer. After the introduction of MCH (curve d, 303 Ω) and Apt (curve e, 425 Ω), the Ret increased gradually owing to their non-electroactive property. After incubation with the reaction mixture containing 1 μM CAP and Recjf exonuclease, the semicircle diameter decreases (curve f, 149 Ω), since the Recjf exonuclease initiates the hybridization cycle process consequently reduce immobilized DNA strands on the electrode surface. These results are in line with expectations and demonstrate the successful construction of the aptasensor.
EIS of different modified steps in 5 mM [Fe (CN)6]3−/4- solution: (a) AuE, (b) PEI-C3N4/AuNWs/AuE, (c) cDNA/PEI-C3N4/AuNWs/AuE, (d) MCH/cDNA/PEI-C3N4/AuNWs/AuE, (e) Apt/MCH/cDNA/PEI-C3N4/AuNWs/AuE, and (f) incubated with the 10 μL mixture containing 8 μL of 1 μM CAP, 2 μL of 2 U/μL RecJf exonuclease. EIS was tested in 5 mM [Fe (CN)6]3−/4− solution including 0.1 M KCl
Feasibility of the sensing strategy
We used DPV to study the effects of Recjf exonuclease and PEI-C3N4/AuNWs on signal amplification in this strategy. As shown in Fig. 3a, before the target’s participation, the double-strand DNA (dsDNA) structure formed by Apt and cDNA hinders the Recjf exonuclease reaction, the corresponding DPV signal was low attributable to the hindrance from the biomolecules (curve a). Also, it exhibited a weak enhancement of the DPV signal when CAP existed alone (curve b); this is mainly due to the specific recognition between CAP and aptamer leading to the CAP-aptamer complex was cleaved from the electrode surface, thereby reducing the biomolecules on the surface of the electrode. While the CAP and Recjf exonuclease were both exist, the DPV signal increases significantly (curve c). This indicates that RecJf exonuclease releases CAP from the CAP-aptamer complex in the presence of the target, allowing it to initiate the target recycling process resulting in signal amplification. Compared with the case without Recjf exonuclease (curve b), the exonuclease-assisted signal amplification strategy increased the DPV signal by 188% (curve c).
Research on signal amplification strategy: (A) DPV of (a) Apt/MCH/cDNA/PEI-C3N4/AuNWs/AuE in the absence of CAP, (b) Apt/MCH/cDNA/PEI-C3N4/AuNWs/AuE after incubation with 1 μM CAP in the (b) absence and (c) presence of RecJf exonuclease, respectively; (B) DPV of different modified electrodes: (a) Apt/MCH/cDNA/AuE and (b) Apt/MCH/cDNA/PEI-C3N4/AuNWs/AuE after incubating with a 10 μL mixture containing 8 μL of 1 μM CAP, 2 μL of 2 U/μL RecJf exonuclease, respectively
Figure 3b compares the DPV signals after Apt/MCH/cDNA/AuE/ and Apt/MCH/cDNA/PEI-C3N4/AuNWs/AuE incubating with a mixture containing CAP and Recjf exonuclease, respectively. The peak current value increases by about 35% after the PEI-C3N4/AuNW–modified electrode. This indicates that PEI-C3N4/AuNWs can facilitate the electron transfer to increase the performance of the modified electrode. Thus, the proposed signal amplification strategy was demonstrated to be feasible.
Moreover, gel electrophoresis was employed to verify the target recycling induced by Recjf exonuclease. Characterization figures of gel electrophoresis were given in the “Supplementary information” section.
Optimization of experimental conditions
The following parameters were optimized: (a) PEI-C3N4/AuNWs volume; (b) the concentration of RecJf exonuclease; (c) cDNA concentration; (d) reaction time. Detailed optimized data and figures are shown in the “Supplementary information” section. Subsequent experimental conditions were based on the optimized results: (a) optimal volume: 2 μL; (b) optimal Recjf exonuclease concentration: 2.0 U/μL; (c) optimal cDNA concentration: 2 μM; and (d) optimal reaction time: 90 min.
Performance of the detection method
The DPV signal response of the electrochemical aptasensor to CAP standard was studied under optimal conditions. Figure 4a depicts the DPV recorded by analyzing CAP at different concentrations. Figure 4b shows the ΔI value corresponding to different concentration of CAP; there is a linear relationship between the ΔI value and the logarithmic of CAP concentration in the range of 100 fM–1 μM. The equation was ΔI (μA) = 21.3 lgCCAP (M) + 312.3 (R2 = 0.995) and according to the formula S/N = 3, LOD was calculated to be 2.96 fM, and the electrochemical sensitivity was calculated to be 380.36 μA·μM−1·cm−2. The electrochemical response value of the blank solution was described in Table S1. Several recently reported methods for CAP detection are compared with the aptasensor in this work. The data is shown in Table 1 after unit conversion. We found that the constructed aptasensor performed better with a lower LOD than others.
a The DPV signal of the aptasensor to different concentrations of CAP (1 μM, 100 nM, 10 nM, 1 nM, 100 pM, 10 pM, 1 pM, 100 fM); b linear relationship between DPV signal and lgCCAP. The data points were acquired from the current value at working voltage 0.176 V (vs. Ag/AgCl) in the inserted DPV graph
Specificity, interference study, stability, and reproducibility of the aptasensor
The specificity of the aptasensor is an essential index for the practical implementation of CAP detection. To assess the specificity of our CAP detection method, four other antibiotics, including kanamycin, doxycycline, oxytetracycline, and tetracycline, were also detected. As shown in Fig. 5a, the current of the other antibiotics (10 μM) is close to that of the blank test; when CAP exists and the concentration is lower than that of other control molecules, the DPV signal increases significantly. Besides, incubation of mixed solution containing all the above antibiotics and pure CAP solution at the same concentration showed little change in DPV signal, which reflects the high binding specificity of the CAP aptamer to the CAP target. In other words, other interfering molecules have little effect on the detection performance of aptasensor. In addition, we selected several other substances (mercury ions (Hg+), methylene blue (MB), and ascorbic acid (AA)) to perform the interference study. Figure 5b shows that none of these substances will significantly affect the sensitivity of the aptasensor.
a The specificity of the aptasensor for CAP (10 nM) compared with blank buffer and other interferents for the same concentration (10 μM): KANA, DOX, OTC, TET, and their mixture containing with 100 nM CAP. b The interference study of the aptasensor for CAP (1 nM) compared with other interferents for the same concentration (1 μM): Hg+, MB, AA, and their mixture containing with 1 nM CAP
Reproducibility and long-term stability are also two essential indicators of the aptasensor. Therefore, seven identical aptasensor with the same fabricated process were used to determine 1 μM CAP. By comparing their DPV signal values, the relative standard deviation was 1.77%, verifying that the aptasensor has good reproducibility. Finally, the sensor was stored at 4 °C to carry out its stability study. After 14 days, the signal value decreased by 5.89%, which may be due to the aptamer’s falling off on the electrode surface during storage.
Real sample analysis
The aptasensor is used to detect CAP in milk samples to examine its potential application in real samples analysis. A series of CAP concentrations include 0.8 μM, 100 nM, and 20 nM prepared by adding CAP standards into milk samples and assayed via the proposed method. As shown in Table 2, recovery was from 93.38 to 111.54% and RSD was from 0.91 to 3.36%. Simultaneously, milk samples containing the same CAP concentration were tested by LC-MS/MS (the Chinese National Standard GB/T 29688- 2013), and the results showed the recovery from 102.16 to 112.69%. The recovery results of the two methods are close, indicating that the sensor has good accuracy and has the potential to be applied in real samples.
Conclusions
In conclusion, an electrochemical aptasensor based on PEI-C3N4/AuNWs and RecJf exonuclease was developed to determine chloramphenicol (CAP). The PEI-C3N4/AuNWs exhibited a large specific surface and excellent electrocatalytic, which considerably amplified the detection signal. The presence of CAP causes RecJf exonuclease to initiate a target recycling mechanism, leading to an enhancement of the peak current. On the basis of the above signal amplification mechanism, this aptasensor has shown excellent performance in detecting CAP, with a detection limit as low as 2.96 fM. In addition, the constructed aptasensor is low-cost and can simplify the operation without the need for electroactive labeling, and realize flexible detection of other targets by replacing the corresponding aptamer. Therefore, the performance of this aptasensor has the potential to become a powerful tool and reference in the field of antibiotic analysis in the future.
References
Ehrlich J, Bartz QR, Smith RM, Joslyn DA, Burkholder PR (1947) Chloromycetin, a new antibiotic from a soil actinomycete. Ence 106(2757):417–417
Yan L, Luo CH, Cheng W, Mao W, Zhang DC, Ding SJ (2012) A simple and sensitive electrochemical aptasensor for determination of chloramphenicol in honey based on target-induced strand release. J Electroanal Chem 687:89–94
Hanekamp JC, Bast A (2015) Antibiotics exposure and health risks: chloramphenicol. Environ Toxicol Pharmacol 39(1):213–220
Smith AG, Dovey GJ, Cartwright RA (2015) Topical chloramphenicol and the risk of acute leukaemia in adults. Pharmacoepidemiol Drug Saf 9(3):215–219
Noa ER, Adi L, Yaara LW, Michal E, Mical P, Leonard L (2015) Efficacy and safety of chloramphenicol: joining the revival of old antibiotics? Systematic review and meta-analysis of randomized controlled trials. J Antimicrob Chemother 4:979–996
Wang LL, Yang H, Zhang CW, Mo YL, Lu XH (2008) Determination of oxytetracycline, tetracycline and chloramphenicol antibiotics in animal feeds using subcritical water extraction and high performance liquid chromatography. Anal Chim Acta 619(1):54–58
Li P, Qiu YM, Cai HX, Kong Y, Tang YZ, Wang DN, Xie MX (2006) Simultaneous determination of chloramphenicol, thiamphenicol, and florfenicol residues in animal tissues by gas chromatography/mass spectrometry. Se Pu 24(1):14–18
Ottonello G, Ferrari A, Magi E (2014) Determination of polychlorinated biphenyls in fish: optimisation and validation of a method based on accelerated solvent extraction and gas chromatography–mass spectrometry. Food Chem 142:327–333
Yan ZD, Gan N, Li TH, Cao YT, Chen YJ (2016) A sensitive electrochemical aptasensor for multiplex antibiotics detection based on high-capacity magnetic hollow porous nanotracers coupling exonuclease-assisted cascade target recycling. Biosens Bioelectron 78:51–57
Huang KJ, Liu YJ, Zhang JZ (2015) Aptamer-based electrochemical assay of 17β-estradiol using a glassy carbon electrode modified with copper sulfide nanosheets and gold nanoparticles, and applying enzyme-based signal amplification. Microchim Acta 182:409–417
Liu S, Lai GS, Zhang HL, Yu AM (2017) Amperometric aptasensing of chloramphenicol at a glassy carbon electrode modified with a nanocomposite consisting of graphene and silver nanoparticles. Microchim Acta 184(5):1445–1451
Pilehvar S, Dierckx T, Blust R, Breugelmans T, De Wael K (2014) An electrochemical impedimetric aptasensing platform for sensitive and selective detection of small molecules such as chloramphenicol. Sensors 14(7):12059–12069
Yadav SK, Agrawal B, Chandra P, Goyal RN (2014) In vitro chloramphenicol detection in a Haemophilus influenza model using an aptamer-polymer based electrochemical biosensor. Biosens Bioelectron 55:337–342
Yan MM, Bai WH, Zhu C, Huang YF, Yan J, Chen AL (2016) Design of nuclease-based target recycling signal amplification in aptasensors. Biosens Bioelectron 77:613–623
Wang YH, Xia H, Huang KJ et al (2018) Ultrasensitive determination of thrombin by using an electrode modified with WSe2 and gold nanoparticles, aptamer-thrombin-aptamer sandwiching, redox cycling, and signal enhancement by alkaline phosphatase. Microchim Acta 185:1–7
Zhu ZH, Qu LN, Niu QJ, Zeng Y, Sun W, Huang XT (2011) Urchinlike MnO2 nanoparticles for the direct electrochemistry of hemoglobin with carbon ionic liquid electrode. Biosens Bioelectron 26:2119–2124
Huang KJ, Yu S, Li J, Wu ZW, Wei CY (2012) Extraction of neurotransmitters from rat brain using graphene as a solid-phase sorbent, and their fluorescent detection by HPLC. Microchim Acta 176(3–4):327–335
Liu CS, Li J, Pang H (2020) Metal-organic framework-based materials as an emerging platform for advanced electrochemical sensing. Coord Chem Rev 410:213222
Hou H, Banks CE, Jing M, Zhang Y, Ji X (2015) Carbon quantum dots and their derivative 3d porous carbon frameworks for sodium-ion batteries with ultralong cycle life. Adv Mater 27(47):7861–7866
Zhu LM, Wang ZH, Wang LL, Li JJ, Cao XY (2019) ZnSe embedded in N-doped carbon nanocubes as anode materials for high-performance Li-ion batteries. Chem Eng J 364:503–513
Sui CJ, Zhou YL, Wang MY, Yin HS, Wang P, Ai SY (2018) Aptamer-based photoelectrochemical biosensor for antibiotic detection using ferrocene modified DNA as both aptamer and electron donor. Sensors Actuators B Chem 266:514–521
Liu YT, Wang QB, Lei JP, Hao Q, Wang W, Ju HX (2014) Anodic electrochemiluminescence of graphitic-phase C3N4 nanosheets for sensitive biosensing. Talanta 122:130–134
Chen YH, Liu XZ, Guo SL, Cao J, Zhou J, Zuo JL, Bai LJ (2019) A sandwich-type electrochemical aptasensor for Mycobacterium tuberculosis MPT64 antigen detection using C60NPs decorated N-CNTs/GO nanocomposite coupled with conductive PEI-functionalized metal-organic framework. Biomaterials 216:119253
Jiang X, Qiu XY, Fu GT, Sun JZ, Huang ZN, Sun DM, Xu L, Zhou JC, Tang YW (2018) Highly simple and rapid synthesis of ultrathin gold nanowires with (111)-dominant facets and enhanced electrocatalytic properties. J Mater Chem A 6(36):17682–17687
Xue SY, Yi HY, Jing P, Xu WJ (2015) Dendritic Pt@ Au nanowires as nanocarriers and signal enhancers for sensitive electrochemical detection of carcinoembryonic antigen. RSC Adv 5(94):77454–77459
Fu XL, Hou F, Liu FR, Ren SW, Cao JT, Liu YM (2019) Electrochemiluminescence energy resonance transfer in 2D/2D heterostructured g-C3N4/MnO2 for glutathione detection. Biosens Bioelectron 129:72–78
Li JS, Wang H, Guo ZK, Wang YG, Ma HM, Ren X, Du B, Wei Q (2017) A “turn-off” fluorescent biosensor for the detection of mercury (II) based on graphite carbon nitride. Talanta 162:46–51
Sun B, Dong J, Cui L, Feng TT, Zhu JJ, Liu XH, Ai SY (2019) A dual signal-on photoelectrochemical immunosensor for sensitively detecting target avian viruses based on AuNPs/g-C3N4 coupling with CdTe quantum dots and in situ enzymatic generation of electron donor. Biosens Bioelectron 124–125:1–7
Yalikun N, Mamat X, Li Y, Hu X, Wågberg T, Dong YM, Hu GZ (2018) Synthesis of an iron-nitrogen co-doped ordered mesoporous carbon-silicon nanocomposite as an enhanced electrochemical sensor for sensitive and selective determination of chloramphenicol. Colloids Surf B 172:98–104
Roushani M, Rahmati Z, Hoseini SJ, Fath RH (2019) Impedimetric ultrasensitive detection of chloramphenicol based on aptamer MIP using a glassy carbon electrode modified by 3-ampy-RGO and silver nanoparticle. Colloids Surf B 183:110451
Xie Y, Huang Y, Tang D, Cui H, Cao H (2018) A competitive colorimetric chloramphenicol assay based on the non-cross-linking deaggregation of gold nanoparticles coated with a polyadenine-modified aptamer. Microchim Acta 185:534
Qin X, Wang Q, Geng L, Shu X, Wang Y (2019) A “signal-on” photoelectrochemical aptasensor based on graphene quantum dots-sensitized TiO2 nanotube arrays for sensitive detection of chloramphenicol. Talanta 197:28–35
Chu TX, Vu VP, Tran HT, Tran TL, Tran QT, Le Manh T (2020) Molecularly imprinted polyaniline nanowire-based electrochemical biosensor for chloramphenicol detection: a kinetic study of aniline electropolymerization. J Electrochem Soc 167(2):027527
Funding
This study was financially supported by the National Natural Science Foundation of China (Grant No. 61301037), the Program for Science and Technology Innovation Talents in Universities of Henan Province (No. 20HASTIT002), the Cultivation Plan for Young Core Teachers in Universities of Henan Province (No. 2017GGJS072), and the National Engineering Laboratory for Wheat & Corn Further Processing, Henan University of Technology (No. NL2018004).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 385 kb)
Rights and permissions
About this article
Cite this article
He, B., Wang, S. An electrochemical aptasensor based on PEI-C3N4/AuNWs for determination of chloramphenicol via exonuclease-assisted signal amplification. Microchim Acta 188, 22 (2021). https://doi.org/10.1007/s00604-020-04688-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-020-04688-8