Abstract
The authors describe a fluorometric method for the determination of the activity of the enzyme alkaline phosphatase (ALP). It is based on the finding that the formation of ssDNA-templated fluorescent silver nanoclusters (AgNCs) is inhibited by enzyme-assisted silver reduction. On addition of ALP, ascorbic acid 2-phosphate (AAP) is hydrolyzed to form ascorbic acid (AA). The generated AA reduces silver ion to form metallic silver which hampers the formation of fluorescent silver nanoclusters, this leading to low fluorescence. Conversely, in the absence of ALP, AAP cannot reduce Ag(I). As a result, fluorescent AgNCs are being formed. Fluorescence intensity correlates linearly in the 1–100 U·L−1 ALP concentration range, with a detection limit of 0.63 U·L−1. The method was applied to the determination of ALP in spiked fetal calf serum samples. It is cost-effective and convenient, does not require labels, or probes, or complicated operations.

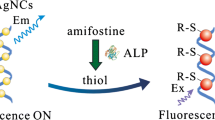
Schematic of a fluorometric method for alkaline phosphatase (ALP) activity assay based on the inhibition of ssDNA-templated fluorescent silver nanoclusters by enzymatic reaction-assisted silver reduction.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As an important enzyme, alkaline phosphatase (ALP) can catalyze the dephosphorylation process of proteins, nucleic acids, and small molecules [1]. Under-expressed levels of serum ALP is a rare form of rickets, and excessive level of ALP in serum are closely related to several diseases of liver and bone, including Paget’s disease, osteomalacia, hepatitis and obstructive jaundice [2, 3]. Therefore, the detection of ALP is highly important for diagnostics. A number of techniques have been developed to quantify ALP activity on the basis of its ability to catalyze the dephosphorylation process of various substrates including nucleic acids, proteins, some small molecules and so on [1]. Among these substrates, small molecules have been intensively employed to detect ALP activity. For example, Ito et al. developed a novel electrochemical method for ALP detection using a mushroom tyrosinase (TN)-embedded carbon paste electrode. This method is high sensitive based on the ALP-catalyzed hydrolysis of phenyl phosphate to accumulate phenol as a product [4]. To further improve the sensitivity, Peng et al. demonstrated an amplification platform based on copper sulfide nanoparticle-decorated graphene nanocomposites, which exhibited excellent electro catalytic activity toward the oxidation of ALP hydrolyzed products. A detection limit of 0.02 U·L−1 was obtained [5]. However, these electrochemical methods always need steps such as preparing of modified electrode, which add to the complexity, cost, and overall assay time. Colorimetric methods employing small molecules as the substrates for ALP activity assay have also been reported lately. Li et al. developed a label-free colorimetric method for ALP detection based on the conjugated gold nanoparticle/adenosine triphosphate (AuNP/ATP) system. The presence of ALP can be visually observed from gray-to-red color change [6]. Later, Gao et al. developed a much higher resolution colorimetric assay for rapid visual readout of ALP activity based on enzymatic reaction-assisted silver deposition on gold nanorod to generate significant color change. With ALP, the color of gold nanorod was changed from red to orange to yellow to green to cyan to blue and to violet [7]. Fluorescence-based strategies for ALP detection also attract much attention due to their high sensitivity. A label-free fluorescent method, using ATP as the substrate, and perylene probe as the signal read-out probe shows promising application for ALP detection [8]. Besides, aggregation-induced emission (AIE) probe was used as the signal read-out probe for ALP detection as well [9]. Although promising, the disadvantages of these assays include complex synthesis procedures and low sensitivity, limiting the scope of its practical applications. Fluorescent methods based on DNA-templated metal nanomaterials using a pyrophosphate (PPi) substrate have been reported [10, 11]. These methods relied on the strong interaction between PPi and Cu2+, which would mediate the formation of fluorescent metal nanomaterials, leading to the fluorescence response to the concentration of ALP. These strategies provide the advantages of simple design, convenient operation, and low experimental cost without need of chemical modification, organic dyes, or separation procedures. These properties suggest the great potential of DNA-templated nanomaterials for facilitating fluorescent methods in ALP detection.
Therefore, the results mentioned above are an excellent motivation for designing a new fluorescent assay system based on DNA-templated nanomaterials. Among the various reported DNA-templated nanomaterials, single stranded-DNA (ssDNA) templated silver nanoclusters (AgNCs) are of special interest due to their ease of preparation and low toxicity [12]. DNA molecules present nitrogen and oxygen functional groups that coordinate with metallic nanomaterials. Besides, they can form and stabilize the nanoparticles via base pairing interactions. Therefore, the synthesis of fluorescent AgNCs using DNA as a scaffold in aqueous solution has attracted extensive attention due to their outstanding spectral and photophysical properties [13,14,15]. AgNCs display highly fluorescent, excellent photostability, sub-nanometer size, and cost-effective to synthesize, and thus are well-suited as a fascinating option to produce label-free fluorescence signals for bioassay and cellular imaging design [11, 16,17,18,19,20,21,22,23,24,25].
Herein, by using ssDNA-templated AgNCs as signal reporter, we developed a novel strategy for simply and accurately determining ALP activity. An ssDNA with a sequence of 5′-CCCTATATACTCCCCGG-3′, is adopted here as a template to stabilize highly fluorescent ssDNA/AgNCs [19]. In the absence of ALP, the fluorescent AgNCs were formed and exhibited strong fluorescence. In the presence of ALP, ascorbic acid 2-phosphate was hydrolyzed to form ascorbic acid. Then, the generated ascorbic acid reduced silver ion to metal silver. The formation of the AgNCs is prohibited due to the lack of silver ion, resulting in weak fluorescence intensity. As a result, the fluorescent response of AgNCs is dependent on the concentration of ALP. This method does not require sophisticated design, which reduces complexity and cost.
Experimental section
Chemicals and materials
The ssDNA with a sequence of 5′-CCCTATATACTCCCCGG-3′ was synthesized by Shanghai Sangon Biotechnology Co., Ltd. (Shanghai, China) (website: https://www.sangon.com/). Ascorbic acid 2-phosphate and ascorbic acid were purchased from Sigma-Aldrich, Inc. (Shanghai, China) (website: http://www.sigmaaldrich.com/china-mainland.html). Silver nitrate (AgNO3) and sodium borohydride (NaBH4) were of analytical grade. All chemicals used were of analytical reagent, obtained from commercial sources, and directly used without additional purification. The Tris-HNO3 reaction buffer used in this experiment consisted of 20 mM Tris–HNO3 (pH 7.4). The hydrolysis buffer consisted of 67 mM glycine-KOH (pH 9.5), and 6.7 mM Mg(NO3)2. Milli-Q purified water was used to prepare all the solutions.
Apparatus
Fluorescent emission spectra were performed on Varian cary eclipse fluorescence spectrophotometer, Varian Medical Systems, Inc. (Palo Alto, American). The sample cell is a 700-μL quartz cuvette. The luminescence intensity was monitored by exciting the sample at 565 nm and measuring the emission at 615 nm. The slits for excitation and emission were set at 5 nm, 10 nm respectively. The fitting of the experimental data was accomplished using the software Origin 8.0.
Optimization of the concentration of HNO3
To optimize the concentration of HNO3, 15 μL hydrolysis buffer, and 6 μL AgNO3 (600 μM) were mixed. Subsequently, 1, 2, 3, 4, 5, 7 and 10 μL of HNO3 (160 mM) and 5 μL probe DNA (100 μM) were added. Then Tris-HNO3 buffer was added to the mixture to make the volume of the reaction solution to be 94 μL. The mixed solution was kept in the dark for 20 min at room temperature. Then, 6 μL of freshly prepared NaBH4 solution (600 μM) was added, followed by vigorous stirring in the dark. The mixture was stored in the dark at room temperature for 60 min, followed by fluorescence measurements. The final concentration of AgNO3, probe DNA, and NaBH4 is 36 μM, 5 μM, and 36 μM, respectively. All the experiments were repeated for three times.
Optimization of the concentration of AgNO3
To optimize the concentration of AgNO3, 15 μL hydrolysis buffer, 5 μL of HNO3 (160 mM) and 2, 4, 5, 6, 8, 10, 12 and 14 μL AgNO3 (600 μM) were mixed. Subsequently, 5 μL probe DNA (100 μM) was added. Then Tris-HNO3 buffer was added to the mixture to make the volume of the reaction solution to be 94 μL. The mixed solution was kept in the dark for 20 min at room temperature. Then, 6 μL of freshly prepared NaBH4 solution (600 μM) was added, followed by vigorous stirring in the dark. The mixture was stored in the dark at room temperature for 60 min, followed by fluorescence measurements. The final concentration of HNO3, probe DNA and NaBH4 is 8 mM, 5 μM and 36 μM, respectively. All the experiments were repeated for three times.
Optimization of the concentration of NaBH4
To optimize the concentration of NaBH4, 15 μL hydrolysis buffer, 5 μL of HNO3 (160 mM), 6 μL AgNO3 (600 μM) and 59 μL Tris-HNO3 buffer were mixed. Subsequently, 5 μL of probe DNA (100 μM) was added. The mixed solution was kept in the dark for 20 min at room temperature. Then, 2, 4, 5, 6, 8, 10, 12 and 14 μL of freshly prepared NaBH4 solution (600 μM) was added, followed by vigorous stirring in the dark. The mixture was stored in the dark at room temperature for 60 min, followed by fluorescence measurements. The final concentration of HNO3, AgNO3, and probe DNA, is 8 mM, 36 μM, and 5 μM, respectively. All the experiments were repeated for three times.
Optimization of the concentration of ascorbic acid 2-phosphate
To optimize the concentration of ascorbic acid 2-phosphate (AAP), different concentrations of AAP were added to the ALP solution (103 U·L−1) with a final volume 15 μL and shaken gently for 30 min at 37 °C. Then, 59 μL Tris-HNO3 buffer, 5 μL of HNO3 (160 mM) and 6 μL AgNO3 (600 μM) were added into the solution and shaken gently for 30 min at 37 °C. Subsequently, 5 μL probe DNA (100 μM) was added. The mixed solution was kept in the dark for 20 min at room temperature. Then, 6 μL of freshly prepared NaBH4 solution (600 μM) was added, followed by vigorous stirring in the dark. The mixture was stored in the dark at room temperature for 60 min, followed by fluorescence measurements. The final concentration of ALP, HNO3, AgNO3, probe DNA and NaBH4 is 103 U·L−1, 8 mM, 36 μM, 5 μM and 36 μM, respectively. All the experiments were repeated for three times.
ALP-catalyzed dephosphorylation
For quantitative measurement of ALP, different concentrations of ALP were added to the AAP solution (200 μM) with a final volume 15 μL and shaken gently for 30 min at 37 °C. Then, 59 μL Tris-HNO3 buffer, 5 μL of HNO3 (160 mM) and 6 μL AgNO3 (600 μM) were added into the solution and shaken gently for 30 min at 37 °C. Subsequently, 5 μL probe DNA (100 μM) was added. The mixed solution was kept in the dark for 20 min at room temperature. Then, 6 μL of freshly prepared NaBH4 solution (600 μM) was added, followed by vigorous stirring in the dark. The mixture was stored in the dark at room temperature for 60 min, followed by fluorescence measurements. The final concentration of AAP, HNO3, AgNO3, probe DNA and NaBH4 is 200 μM, 8 mM, 36 μM, 5 μM and 36 μM, respectively. All the experiments were repeated for three times.
For the ALP activity assay in biological fluids, the fetal calf serum was diluted 100 times with hydrolysis buffer, and different concentrations of ALP was then added to the diluted fetal calf serum to prepare the spiked samples.
Results and discussion
Design strategy
Figure 1 outlines the design principle of the ssDNA-templated AgNCs-based fluorescent method for alkaline phosphatase (ALP) assay. In the absence of ALP, the fluorescence intensity is high, because ascorbic acid 2-phosphate (AAP) does not disrupt the synthesis of ssDNA-templated AgNCs. Upon the addition of ALP, due to the strong hydrolysis ability of ALP toward AAP, ascorbic acid (AA) was formed. And then AA, which has reducing ability toward metal ion, reduced silver ion to metal silver. So, few AgNCs formed and a weak fluorescence signal was recorded. Since the degree of decrease of ssDNA-templated AgNCs is directly related with the addition ALP. Therefore, ALP activity detection can be easily realized by monitoring the change of the fluorescence signal.
Feasibility validation of this strategy
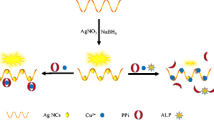
In order to realize the detection of ALP using this approach, firstly we validated the feasibility. The ssDNA-templated AgNCs were synthesized according to the previous reported methods [26, 27]. The fluorescence properties of the AgNCs have a maximum emission wavelength of 615 nm and a maximum excitation wavelength of 565 nm, as seen in Fig. S1. The effect of ALP, AAP and AA on the formation of DNA-templated AgNCs was explored firstly to verify the design principle. To study their effects, Ag+ (36 μM) was pretreated with ALP, AAP, and AA, respectively, at 37 °C for 30 min, then ssDNA and NaBH4 were added to synthesize AgNCs. The fluorescence intensity of AgNCs was used to investigate the effects on AgNCs formation. ALP and AAP affected the fluorescence intensity only slightly (Fig. S2, Fig. S3). However, when Ag+ was pretreated with AA, an obvious fluorescence decrease was observed (Fig. S4). These results indicate that AA can strongly react with Ag+, whereas ALP and AAP has no chemical reaction with Ag+. Additionally, after the hydrolysis of AAP by ALP, a decrease of fluorescence intensity was clearly observed (Fig. 2). On the basis of the observed different fluorescence inhibition ability of AAP and AA, a cost-effective and convenient fluorescent assay for ALP based on the AA-induced inhibition of AgNCs formation can be achieved.
Optimization of assay conditions
The following parameters were optimized: (a) concentration of HNO3; (b) Ag+; (c) BH4 −; (d) AAP and (e) ALP-catalyzed hydrolysis time. Respective disussion and Figures are given in the Electronic Supporting Material. We found the following experimental conditions to give best results: (a) An HNO3 concentration of 8 mM; (b) A Ag+ concentration of 36 μM; (c) A BH4 − concentration of 36 μM; (d) An AAP concentration of 200 μM and (e) ALP-catalyzed hydrolysis time of 30 min.
Fluorescence measurement of ALP activity
Figure 3a depicts the typical fluorescence responses at different ALP concentrations. The fluorescence signals decreased gradually with increasing ALP concentration. There is a linear relationship between the fluorescence intensities and the concentration of ALP in the range 1–100 U·L−1 (Fig. 3b), with a detection limit of 0.63 U·L−1 (3S0/S, in which S0 is the standard deviation for the blank solution, n = 11, and S is the slope of the calibration curve). This detection limit is comparable to or much lower than that of some detection strategies reported in literature (Table 1). Besides, comparing with the previously reported ALP assays, this sensor shows a few advantages. Firstly, our assay does not need nucleic acid covalent immobilization. Secondly, this method is convenient operation comparing with the complicated synthesize of polymer probes [28, 29], perylene probes [8] and aggregation-induced emission (AIE) probes [30, 31]. Thirdly, fewer reagents were needed compared to the other AgNCs-based method [32]. In addition, to verify the repeatability of this method, for each concentration of ALP, the measurement has been repeated for at least three times independently. The average relative standard deviation (RSD) is 4.73% (Table S1), indicating that the precision and reproducibility of the method are acceptable. It should be noted that the AgNCs based method is time consuming, with the testing can be performed at around 2 h.
Fluorescence emission spectra of ssDNA-templated AgNCs-based assay method in the presence of increasing amount of ALP (a) and calibration curve for ALP detection (b). AAP, 200 μM; AgNO3, 36 μM; probe DNA, 5 μM; NaBH4, 36 μM. Excitation, 565 nm; Excitation slit, 5 nm; Emission slit, 10 nm. The error bars illustrate the standard deviations of three replicate measurements
ALP activity assay in biological fluids
As this AgNCs-based method for ALP detection is based on the consumption of silver ion by the product of AA. Therefore, the presence of any other compounds with strong affinity for silver ions or any other reducing agents may affect the target detection. To find out, we investigate the effect of sodium chloride, L-cysteine, D-(+)-glucose and sodium citrate on the formation of ssDNA-templated AgNCs. As we can see from Fig. S7, when Ag+ was pretreated with Cl− and thiols, an obvious fluorescence decrease was observed. However, reducing agents such as glucose and sodium citrate show little influence on the fluorescence of AgNCs. A set of more meaningful ALP determination with this AgNCs-based method was conducted in spiked fetal calf serum, which was aimed at investigating the ability of the AgNCs system to overcome interference from the presence of any other compounds with strong affinity for silver ions. Various concentrations of ALP were added to the sample solution containing AAP and 1% fetal calf serum. The changes in fluorescence intensity are shown in Fig. 4. The results show that with the incremental introduction of ALP, fluorescence decrease can also be observed in the presence of fetal calf serum. This result indicates that although serum containing compounds with strong affinity for silver ions, they may have a very low concentration. Therefore, no obvious influence on the ALP detection in 1% serum was observed. These results have shown high robustness of the AgNCs-based platform in the complex matrix, suggesting the potential of this method for applications in complicated real samples.
Fluorescence intensity of ssDNA-templated AgNCs-based assay method in the presence of increasing amount of ALP in 1% serum solution. AAP, 200 μM; AgNO3, 36 μM; probe DNA, 5 μM; NaBH4, 36 μM. Excitation, 565 nm; Excitation slit, 5 nm; Emission slit, 10 nm. The error bars illustrate the standard deviations of three replicate measurements
Selectivity study
Additional assays were performed to inspect the selectivity of the assay to other proteins and molecules such as human serum albumin (HSA), lysozyme (Lyz), hemoglobin (HGB), ATP, glucose and glycine. In a typical experiment, AAP was incubated with HSA (2500 nM), Lyz (2500 nM), HGB (2500 nM), ATP (2500 nM), glucose (2500 nM), glycine (2500 nM) and ALP (100 U·L−1, 2253.8 nM), respectively. As shown in Fig. 5, the fluorescence of this ssDNA-templated AgNCs-based method changed little for HSA, Lyz, HGB, ATP, glucose and glycine, while a significant fluorescence decrease was observed for ALP. The results clearly show high selectivity of the assay and it is a specific ALP assay method.
Fluorescence intensity of this assay method in the presence of different proteins and small molecules: blank control (without ALP); HSA (2500 nM); Lyz (2500 nM); HGB (2500 nM); ATP (2500 nM); glucose (2500 nM); glycine (2500 nM) and ALP (100 U·L−1, 2253.8 nM). AAP, 200 μM; AgNO3, 36 μM; probe DNA, 5 μM; NaBH4, 36 μM. Excitation, 565 nm; Excitation slit, 5 nm; Emission slit, 10 nm. The error bars illustrate the standard deviations of three replicate measurements
Conclusions
In summary, based on ssDNA-templated AgNCs, a novel fluorescent method for ALP assay has been developed. A small molecule, ascorbic acid 2-phosphate (AAP), has been chosen as the enzyme substrate. This study relies on the strong reduction ability of the product of ALP-catalyzed AAP hydrolysis, which would reduce silver ion to metal silver, disabling the effective formation of fluorescent AgNCs, resulting in low fluorescence intensity. The detection limit of this method is comparable to or much lower than that of some detection strategies reported in literature. Additionally, this method is cost-effective and convenient without complicated operations. However, we should note that it is a little time consuming to synthesis the AgNCs. Nevertheless, on the basis of this excellent performance, as well as the simple-to-implement features, this method using AAP as substrate may hold great potential in the diagnosis of ALP-related diseases.
References
Coleman JE (1992) Structure and mechanism of alkaline-phosphatase. Annu Rev Bioph Biom 21:441–483
Millan JL, Whyte MP (2016) Alkaline phosphatase and hypophosphatasia. Calcif Tissue Int 98(4):398–416
Saif MW, Alexander D, Wicox CM (2005) Serum alkaline phosphatase level as a prognostic tool in colorectal cancer: a study of 105 patients. J Appl Res 5(1):88–95
Ito S, Yamazaki S, Kano K, Ikeda T (2000) Highly sensitive electrochemical detection of alkaline phosphatase. Anal Chim Acta 424(1):57–63
Peng J, Han XX, Zhang QC, Yao HQ, Gao ZN (2015) Copper sulfide nanoparticle-decorated graphene as a catalytic amplification platform for electrochemical detection of alkaline phosphatase activity. Anal Chim Acta 878:87–94
Li CM, Zhen SJ, Wang J, Li YF, Huang CZ (2013) A gold nanoparticles-based colorimetric assay for alkaline phosphatase detection with tunable dynamic range. Biosens Bioelectron 43:366–371
Gao ZQ, Deng KC, Wang XD, Miro M, Tang DP (2014) High-resolution colorimetric assay for rapid visual readout of phosphatase activity based on gold/silver core/shell nanorod. ACS Appl Mater Inter 6 6(20):18243–18250
Chen J, Jiao HP, Li WY, Liao DL, Zhou HP, Yu C (2013) Real-time fluorescence turn-on detection of alkaline phosphatase activity with a novel perylene probe. Chem-Asian J 8(1):276–281
Liang J, Kwok RTK, Shi HB, Tang BZ, Liu B (2013) Fluorescent light-up probe with aggregation-induced emission characteristics for alkaline phosphatase sensing and activity study. ACS Appl Mater Inter 5(17):8784–8789
Zhang LL, Zhao JJ, Duan M, Zhang H, Jiang JH, Yu RQ (2013) Inhibition of dsDNA-templated copper nanoparticles by pyrophosphate as a label-free fluorescent strategy for alkaline phosphatase assay. Anal Chem 85(8):3797–3801
Ma J-L, Yin B-C, Wu X, Ye B-C (2016) Copper-mediated DNA-scaffolded silver nanocluster on-off switch for detection of pyrophosphate and alkaline phosphatase. Anal Chem 88(18):9219–9225
Diez I, Ras RHA (2011) Fluorescent silver nanoclusters. Nano 3(5):1963–1970
Zhang M, Guo S-M, Li Y-R, Zuo P, Ye B-C (2012) A label-free fluorescent molecular beacon based on DNA-templated silver nanoclusters for detection of adenosine and adenosine deaminase. Chem Commun 48(44):5488–5490
Zhang K, Wang K, Xie M, Zhu X, Xu L, Yang R, Huang B, Zhu X (2014) DNA-templated silver nanoclusters based label-free fluorescent molecular beacon for the detection of adenosine deaminase. Biosens Bioelectron 52:124–128
Liu X, Wang F, Aizen R, Yehezkeli O, Willner I (2013) Graphene oxide/nucleic-acid-stabilized silver nanoclusters: functional hybrid materials for optical aptamer sensing and multiplexed analysis of pathogenic DNAs. J Am Chem Soc 135(32):11832–11839
Yang SW, Vosch T (2011) Rapid detection of microRNA by a silver nanocluster DNA probe. Anal Chem 83(18):6935–6939
Zhang JP, Li C, Zhi X, Ramon GA, Liu YL, Zhang CL, Pan F, Cui DX (2016) Hairpin DNA-templated silver nanoclusters as novel beacons in strand displacement amplification for microRNA detection. Anal Chem 88(2):1294–1302
Zhang YD, Cai YA, Qi ZL, Lu L, Qian YX (2013) DNA-templated silver nanoclusters for fluorescence turn-on assay of acetylcholinesterase activity. Anal Chem 85(17):8455–8461
Peng YL, Jiang JH, Yu RQ (2014) Label-free and sensitive detection of micrococcal nuclease activity using DNA-scaffolded silver nanoclusters as a fluorescence indicator. Anal Methods-Uk 6(12):4090–4094
Lee CY, Park KS, Jung YK, Park HG (2016) A label-free fluorescent assay for deoxyribonuclease I activity based on DNA-templated silver nanocluster/graphene oxide nanocomposite. Biosens Bioelectron 93:293–297
Deng L, Zhou ZX, Li J, Li T, Dong SJ (2011) Fluorescent silver nanoclusters in hybridized DNA duplexes for the turn-on detection of Hg2+ ions. Chem Commun 47(39):11065–11067
Lan GY, Huang CC, Chang HC (2010) Silver nanoclusters as fluorescent probes for selective and sensitive detection of copper ions. Chem Commun 46(8):1257–1259
Sharma J, Yeh HC, Yoo Y, Werner JH, Martinez JS (2011) Silver nanocluster aptamers: in situ generation of intrinsically fluorescent recognition ligands for protein detection. Chem Commun 47(8):2294–2296
Yeh HC, Sharma J, Han JJ, Martinez JS, Werner JH (2010) A DNA-silver nanocluster probe that fluoresces upon hybridization. Nano Lett 10(8):3106
Han GM, Jia ZZ, Zhu YJ, Jiao JJ, Kong DM, Feng XZ (2016) Biostable L-DNA-templated aptamer-silver nanoclusters for cell-type-specific imaging at physiological temperature. Anal Chem 88(22):10800–10804
Ganguly M, Bradsher C, Goodwin P, Petty JT (2015) DNA-directed fluorescence switching of silver clusters. J Phys Chem C 119(49):27829–27837
Sengupta B, Ritchie CM, Buckman JG, Johnsen KR, Goodwin PM, Petty JT (2008) Base-directed formation of fluorescent silver clusters. J Phys Chem C 112(48):18776–18782
Li Y, Li YA, Liu ZG, Su XG (2014) Sensitive fluorometric detection of alkaline phosphatase using a water-soluble conjugated polymer. RSC Adv 4(81):42825–42830
Deng JJ, Yu P, Wang YX, Mao LQ (2015) Real-time ratiometric fluorescent assay for alkaline phosphatase activity with stimulus responsive infinite coordination polymer nanoparticles. Anal Chem 87(5):3080–3086
Chen Q, Bian N, Cao C, Qiu XL, Qi AD, Han BH (2010) Glucosamine hydrochloride functionalized tetraphenylethylene: a novel fluorescent probe for alkaline phosphatase based on the aggregation-induced emission. Chem Commun 46(23):4067–4069
Cao FY, Long Y, Wang SB, Li B, Fan JX, Zeng X, Zhang XZ (2016) Fluorescence light-up AIE probe for monitoring cellular alkaline phosphatase activity and detecting osteogenic differentiation. J Mater Chem B 4(26):4534–4541
Guo LY, Chen DL, Yang MH (2017) DNA-templated silver nanoclusters for fluorometric determination of the activity and inhibition of alkaline phosphatase. Microchim Acta 184(7):2165–2170
Zeng WN, Wang LY, Liu SP, Lin SM, Beuerman R, Cao DR (2012) Cationic conjugated polyelectrolyte-based sensitive fluorescence assay for adenosinetriphosphate and alkaline phosphatase. Sensor Actuat B-Chem 171:652–657
Li YA, Li Y, Wang XY, Su XG (2014) A label-free conjugated polymer-based fluorescence assay for the determination of adenosine triphosphate and alkaline phosphatase. New J Chem 38(9):4574–4579
Liu HS, Ma CB, Wang J, Wang KM, Wu KF (2017) A turn-on fluorescent method for determination of the activity of alkaline phosphatase based on dsDNA-templated copper nanoparticles and exonuclease based amplification. Microchim Acta 184(7):2483–2488
Kang WJ, Ding YY, Zhou H, Liao QY, Yang X, Yang YG, Jiang JS, Yang MH (2015) Monitoring the activity and inhibition of alkaline phosphatase via quenching and restoration of the fluorescence of carbon dots. Microchim Acta 182(5–6):1161–1167
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 21405125), Natural Science Foundation Project of CQ (No. CSTC2014JCYJA80041), Fundamental Research Funds for the Central Universities (No. XDJK2015C090).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The author(s) declare that they have no competing interests.
Electronic supplementary material
ESM 1
(DOCX 278 kb)
Rights and permissions
About this article
Cite this article
He, Y., Jiao, B. Determination of the activity of alkaline phosphatase based on the use of ssDNA-templated fluorescent silver nanoclusters and on enzyme-triggered silver reduction. Microchim Acta 184, 4167–4173 (2017). https://doi.org/10.1007/s00604-017-2459-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00604-017-2459-x