Abstract
In colorectal cancer (CRC), dysregulation of noncoding RNA expression is a distinguishing factor. Owing to the conflicting results and the insufficient studies on serum microRNA-378 (miR-378) and long intragenic noncoding RNA00641 (LINC00641) expression patterns, we aimed to explore their expression profiles and diagnostic ability in colorectal cancer. Blood samples were collected from 30 healthy controls and 70 CRC patients. miR-378 and LINC00641 expression levels were determined using quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR), carcinoembryonic antigen (CEA) levels assessed by the enzyme-linked immunosorbent assay (ELISA) method. The expression levels of the studied miR-378 and LINC00641 correlated with patients’ CEA levels. LINC00641 expression was dramatically upregulated, and miR-378 expression was significantly downregulated in colorectal cancer compared to the healthy controls. The differential expression of miR-378 and LINC00641 was inversely correlated, whereas the expression folds of both LINC00641 and CEA directly correlated with the advanced stages of colorectal cancer. Receiver operator characteristic (ROC) curve analysis disclosed the highest diagnostic potential for LINC00641 to discriminate colorectal cancer patients from the control and the highest diagnostic potential for discrimination between colorectal cancer stages.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) has been reported as the most prevalent malignancy worldwide. Many studies have shown that the incidence of CRC has been increasing significantly in recent years (Stoffel and Murphy 2020; Ibrahim et al. 2014). In Egypt, the sixth cancer was CRC. The most important risk factors were metabolic disorders, precancerous bowel injury, and a family history of CRC (Siegel et al. 2019). As a result, there is a pressing need to improve the early molecular diagnosis of colorectal cancer.
In recent years, the underlying molecular mechanisms of the occurrence and progression of CRC have not been fully understood. Previous studies have reported that CRC develops through the accumulation of genetic mutants and epigenetic modifications (Picard et al. 2020).
Noncoding RNAs (ncRNAs) can be classified as long ncRNAs (lncRNAs) and small ncRNAs (sncRNA), depending on the length of nucleotides. Noncoding RNA over 200 nucleotides losing the ability of biological function in the protein translation pathway is known as lncRNA. Positive associations between lncRNA expression and cell growth and differentiation indicate their impact on various diseases (Kanduri 2016; Jarroux et al. 2017). Schmitt and Chang (2016) and Tang et al. (2017) indicated the significant potential of lncRNAs as new tumor-related biomarkers for the early detection of tumors and the prediction of clinical outcomes in patients with tumors.
Recent research has revealed that regulatory sncRNAs (miRNA, piRNA, tRF, and snoRNA) can act as significant gene regulators and play a role in a variety of physiological and pathological processes (Catela et al. 2017). Also, the abnormal expression of sncRNAs is implicated in various human disorders, including cancers (Romano et al. 2017; Elshafei et al. 2017).
The long intragenic noncoding RNA00641 (LINC00641) is a new noncoding RNA located at 14q11.2 on the chromosome. Recent studies have shown that LINC00641 regulates autophagy and disc degeneration under nutritional stress (Wang et al. 2018).
This lncRNA has been suggested as a potential prognostic biomarker for patients with glioblastoma. Recently, Wang et al. (2019) demonstrated that LINC00641 modulates autophagy and is implicated in the pathogenesis of degenerative intervertebral disc. LINC00641 is abnormally expressed in bladder tumor tissues and cell lines and is considered a cancer suppressor in bladder cancer (Li et al. 2018a).
MicroRNA-378a (miR-378a, formerly called miR-378) is a small noncoding RNA molecule that can guide gene expression post-transcriptionally. The two mature strands, miR-378a3p and miR-378a5p, are derived from the first intron of the coactivator-1-beta gene, a peroxisome growth factor-activated gamma that encodes PGC1β. The embedding of this transcriptional regulator in the sequence of oxidative energy metabolism implies the involvement of miR-378a in the metabolic pathway, the expression of miR-378 improves cell survival by altering caspase-3 activity and enhancing angiogenesis and the growth of the tumor (Li et al. 2018b).
Because of the incoherence of the findings and the inadequacy of the studies on the differential expression schemes of miR-378 and LINC00641, we aimed to explore the biochemical contribution of LINC00641 and miR-378 expression in CRC subjects and correlate these data with clinical-pathological characteristics to clarify their role in the early diagnosis of CRC.
Subjects and methods
This study was a pilot retrospective observational study carried out on 100 participants; 70 of them were CRC patients recruited from the Gastrointestinal Endoscopy Unit in Al-Kasr Al-Ainy Hospital, Cairo University, who attended the colonoscopy unit for general colorectal screening and patients suffering from variable colonic symptoms, including CRC alarming symptoms.
The patient’s diagnosis and cancer staging are based on the colonoscopic findings, abdominal radio-imaging, pathological findings, and clinical decisions in three stages:
-
1.
Stage I/II: local cancer.
-
2.
Stage III: involvement of lymph nodes.
-
3.
Stage IV: distant metastasis.
In addition, 30 control subjects were selected as apparently healthy volunteers without gastrointestinal symptoms, chronic disease history, cancer, or family history of CRC. All enrolled subjects’ demographics, such as age, gender, and family history, were gathered. Body mass index (BMI) was calculated for all subjects as an index of the weight in kilograms divided by the square of the height in meters. The Helsinki Declaration was followed in the current research. All study participants provided their knowledge of consent.
Venous blood samples (10 ml) were withdrawn from enrolled participants by trained laboratory technicians. Each sample was divided into three portions for processing, total RNA extraction, determination of biochemical markers, and cell blood count (CBC).
The following biochemical tests were done for all involved subjects: alanine aminotransferase (ALT), aspartate aminotransferase (AST), total bilirubin (T. bilirubin), albumin, urea, and creatinine were assayed using the routine biochemical technique. The level of CEA in the blood was tested using ELISA with a kit for CEA (Pointe Scientific, Inc., 4559 Research Drive, Canton, MI 48,188, USA).
Purification of total RNA from serum, which includes small RNAs such as miRs, was performed using the miRNeasy Kit and the reverse transcription of the total RNA was conducted using the MicroRNA easy kit as previously described (Khalil et al. 2022). For real-time polymerase chain reaction (RT-PCR), 5 μl of diluted RT products (cDNA template) was combined with SYBR Green Master Mix (Qiagen) and primer specific for miR-378 and LINC00641 following the settings for the real-time PCR technique. The melting curve analysis revealed a single peak, confirming the specificity of the target PCR product. qRT-PCR data analysis is accomplished by the 2-ΔΔct comparative expression method (Livak and Schmittgen 2001).
Statistical analysis
Data analysis was performed using the statistical software SPSS 18.0 (SPSS Inc., Chicago, IL, USA). The data was reported as a mean ± standard error mean. Statistical analyses were performed using one-way ANOVA. When the ANOVA was significant, the Duncan method was used to assess the specific differences between the groups. The analysis of the ROC was used to determine the optimum cut-off value for miR-378 and LINC00641. The correlation coefficient (r) was calculated to determine the relationship between LINC00641, miR-378, and the CEA level of the patients using Pearson product-moment correlation. P < 0.001 was considered statistically significant.
Results
This study was carried out on 70 CRC patients aged 34–65 years. Male patients were 36 (51.43%), while female patients were 34 (48.57%). Based on the colonoscopy and radio-imaging findings, the most common CRC locations detected were recto-sigmoid (21.4%), hepatic flexure (17.1%), rectal (12.9%), ascending colon (12.9%), transverse (11.4%), and less commonly sigmoid (10%), cecal (10%), and splenic flexure (4.3%). The recruited CRC patients were grouped into three stages: stage I/II (30), stage III (17), and stage IV (23). Table 1 shows the demographics and clinical findings.
Tables 2, 3, and 4 show a remarkable, statistically significant difference between CRC and healthy control groups regarding hemoglobin, leukocytes, INR, albumin, ALT, AST, total bilirubin, creatinine, and CEA levels with higher levels of ALT, AST, total bilirubin, creatinine, and CEA. There was also a decline in hemoglobin and albumin levels among CRC patients.
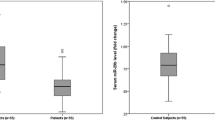
LINC00641 is upregulated in CRC patients compared to healthy participants. Here, LINC00641 shows a highly significant fold elevation in its expression level in CRC patients compared to healthy subjects, confirming that it can be used for early detection of colorectal cancer, as shown in Table 5. A notable and statistically significant elevation in the differential expression folds of LINC00641 was observed among CRC patients along with stages. The highest expression pattern was in stage IV patients (8.89 ± 0.62) with remarkable significance from stages I/II and III, as shown in Table 5.
Our study shows a remarkable statistically significant fold decrease in miR-378 levels in CRC patients compared to healthy subjects, confirming that it can be used as LINC00641 for early detection of CRC, as shown in Table 5. Regarding the gradual decrease in the expression pattern of miR-378 among CRC stages, there was a statistical difference in the expression between stages I/II and III, while stage IV was the most downregulated group (0.34 ± 0.05) with great statistical significance from stage I/II (p < 0.001) and no statistical significance from the stage III group, as shown in Table 5.
The diagnostic validities of the targeted miR-378 and LINC00641 to differentiate between CRC patients and normal subjects were calculated using ROC curve analysis; ROC analysis revealed that serum LINC00641 discriminated CRC from healthy controls with a best cutoff value = 1.09, area under curve (AUC) = 0.996, 95% confidence interval (CI) = 0.988–1.005, and P < 0.001, with sensitivity = 98.57% and specificity = 100% (Fig. 1b). LINC00641 also distinguished late stage of CRC patients with best cutoff = 5.125, AUC = 0.956, 95% CI = 0.900–1.011, and P < 0.001, with sensitivity = 91.3% and specificity = 94.80% (Fig. 1d). ROC analysis revealed that serum miR-378 discriminated CRC from healthy controls with a best cutoff value = 0.90, an AUC = 0.957, 95% CI = 0.915–0.999, and P < 0.001, with sensitivity = 90% and specificity = 100% (Fig. 1a). miR-378 also distinguished the late stage of CRC with a best cutoff = 0.375, an AUC = 0.894, a 95% CI = 0.819–0.969, and P < 0.001, with sensitivity = 73.91% and specificity = 90.90% (Fig. 1c).
miR-378 expression was negatively correlated with the expression of LINC00641 (r = − 0.689, P < 0.001), and serum CEA level (r = − 0.747, P < 0.001) in CRC patients. However, serum expression of LINC00641 was positively correlated with serum CEA level (r = 0.719, P < 0.001), as shown in Table 6 and Fig. 2.
Discussion
Inadequate management and control of colorectal cancer is the most common gastrointestinal malignancy and is considered third cancer in mortality rates viewed as a life-threatening disorder worldwide (Ferlay et al. 2021). In Egypt, CRC has a high incidence among patients > 50 years after colonoscopic screening (Elkeleny et al. 2021). The high prevalence of early-onset colorectal cancer is due to a lack of awareness of contributing factors. So, in the current study, we aimed to explore the expression behavior of LINC00641 and miR-378, their association with cancer progression, and their diagnostic power in CRC early identification and staging. To the best of our knowledge, this is the first serum-based study to investigate the expression level of both LINC00641 and miR-378 in the serum of 70 CRC (grades I/II, III, and IV) Egyptian patients and 30 healthy participants.
The mean age of CRC enrolled patients were 51.79 years, and the major location for the diagnosed cases was colonic, but regarding the colorectal anatomy, most of the cases were recto-sigmoid and hepatic flexure carcinomas, which is consistent with other Egyptian studies (Ramzy et al. 2015; Elkeleny et al. 2021).
Non-coding RNA possesses a remarkable role in cell processes such as proliferation, differentiation, and metastasis. lncRNA and miRNA have emerged as gene expression regulators that target more than 30% of the human genome (Wallace et al. 2016; Parikh et al. 2014), opening up new avenues for therapeutic and detectable biomarkers.
LINC00641 has shown to serve as an important regulator in several cancers, including bladder cancer (Li et al. 2018a), glioblastoma (Liang et al. 2019), lung cancer (Li et al. 2019), prostate cancer (Sajjadi et al. 2021), and nasopharyngeal carcinoma (Ren et al. 2021).
Herein, our results revealed a remarkable, significant upregulation of the LINC00641 expression level in CRC patients compared to healthy controls. The LINC00641 expression levels were elevated and associated with TNM stage and metastasis. Our results are consistent with the previous studies of Zhang et al. (2021) and Hu et al. (2020), who reported that LINC00641 expression is upregulated in various cancers and pointed to LINC00641 as a novel prognostic oncogenic biomarker.
Our study revealed that the LINC00641 expression was elevated significantly in CRC patients with stage IV, which is compatible with the findings of Xue et al. (2021), who reported that elevated LINC00641 expression positively correlated with lymph node metastasis and other clinical-pathological variables of CRC patients. Remarkably, our results showed a positive correlation between CEA and LINC00641 expression.
miR-378 have implicated in the development of a variety of cancers. Reduced miR-378 expression level in gliomas implies significant tumor invasiveness and a poor prognosis (Li et al. 2015). In hepatocellular carcinoma, miR-378 suppresses tumor cell growth and proliferation (Niu et al. 2015). However, there are limited investigations on the link between miR-378 and colon cancer development.
Our study found that the expression fold of miRNA-378 was lower in CRC patients than in healthy control groups. The relative expression of miR-378 in serum could effectively distinguish CRC patients and normal populations, which is consistent with Faltejskova et al. (2012) and Peng et al. (2015). Additionally, Zeng et al. (2017) demonstrated that blocking the Wnt/-catenin pathway with miR-378 expression reduced the malignant behaviors of colon cancer cells. Furthermore, Li et al. (2013) stated that miRNA-378 suppresses the growth features of HBV-related HCC by directly targeting the down expression of IGF 1R 3′-UTR.
Finally, the data obtained from the ROC analysis shows that LINC00641 and miR-378 can successfully discriminate between CRC patients and healthy controls. LINC00641 presented the best diagnostic potential because it showed the highest diagnostic accuracy for CRC patients from control subjects (AUC = 0.996, 98.57% sensitivity) and the highest ability to discriminate late-stage (IV) colorectal cancer (AUC = 0.956, 91.30% sensitivity, and 94.8% specificity). However, serum miR-378 discriminated CRC from healthy controls with an AUC of 0.957, 90% sensitivity, and 100% specificity. Moreover, miR-378 distinguished the late stage of CRC with an AUC of 0.894, with 73.91% sensitivity, and 90.90% specificity. Hence, LINC00641 and miR-378 may be promising circulating biomarkers for early CRC diagnosis and disease prognosis, respectively. However, some limitations exist, such as the study’s small sample size and the evaluation of inflammatory biomarkers. As a result, more large-scale studies and clinical trials are required, as well as an assessment of their functional impacts on putative genes and target pathways.
Conclusion
Our findings suggest that LINC00641 and miR-378 can be used as simple and promising biological biomarkers for the diagnosis of CRC and for predicting the progression of complications. Overall, our results suggested that LINC00641/miR-378 axis might serve as a novel therapeutic target for CRC.
References
Catela IT, Voss G, Cornella H et al (2017) microRNAs as cancer therapeutics: a step closer to clinical application. Cancer Lett 407:113–122
Elkeleny MR, Abdelbaki TN, Sabry AA et al (2021) Colonoscopic screening in early detection of colorectal cancer in high-risk groups: a prospective study. Egypt J Surg 40(1):3–10
Elshafei A, Shaker O, Abd El-motaal O, Salman T (2017) The expression profiling of serum miR-92a, miR-375, and miR-760 in colorectal cancer: an Egyptian study. Tumor Biol 39(6):1–14
Faltejskova P, Svoboda M, Srutova K et al (2012) Identification and functional screening of microRNAs highly deregulated in colorectal cancer. J Cell Mol Med 2655–66
Ferlay J, Colombet M, Soerjomataram I et al (2021) Cancer statistics for the year 2020: An overview. Int J Cancer 149:778–789
Hu Y, Su Y, Lei X et al (2020) LINC00641/miR-582-5p mediate oxaliplatin resistance by activating autophagy in gastric adenocarcinoma. Sci Rep 10(1):14981
Ibrahim AS, Khaled HM, Mikhail NN et al ( 2014). Cancer incidence in Egypt: results of the national population-based cancer registry program. J Cancer Epidemiol 437971
Jarroux J, Morillon A, Pinskay M (2017) History, discover and classification of lncRNAs. Adv Exp Med Biol 1008:1–46
Kanduri C (2016) Long noncoding RNAs: lessons from genomic imprinting. Biochim Biophys Acta 1859:102–111
Khalil EH, Shaker OG, Hasona NA (2022) Impact of rs2107425 Polymorphism and Expression of lncH19 and miR-200a on the susceptibility of colorectal cancer. Ind J Clin Biochem. https://doi.org/10.1007/s12291-022-01052-w
Li Y, Jiang J, Liu W, et al (2018b) microRNA-378 promotes autophagy and inhibits apoptosis in skeletal muscle. Proc Natl Acad Sci USA 115(46):E10849–E10858
Liang R, Zhi Y, Zheng G et al (2019) Analysis of long non-coding RNAs in glioblastoma for prognosis prediction using weighted gene co-expression network analysis, Cox regression and L1-LASSO penalization. Onco Targets Ther 12:157–168
Li B, Wang Y, Li S et al (2015) Decreased expression of miR-378 correlates with tumor invasiveness and poor prognosis of patients with glioma. Int J Clin Exp Pathol 8:7016–7021
Li LH, Gao Q, Wang XY et al (2013) miR-378 suppresses HBV-related hepatocellular carcinoma tumor growth by directly targeting the insulin-like growth factor 1 receptor. Zhonghua Gan Zang Bing Za Zhi 21:609–613
Li L, Zhao P, Zhao Z et al (2019) Long non-coding RNA LINC00641 suppresses non-small-cell lung cancer by sponging miR-424-5p to upregulate PLSCR4. Cancer Biomark 26:79–91
Li Z, Hong S, Liu Z (2018a) LncRNA LINC00641 predicts prognosis and inhibits bladder cancer progression through miR-197-3p/KLF10/ PTEN/PI3K/AKT cascade. Biochem Biophys Res Commun 503(3):1825–1829
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods (san Diego Calif) 25(4):402–408
Niu JX, Meng XK, Ren JJ (2015) Studied microRNA gene expression in human hepatocellular carcinoma by microRNA microarray techniques. World J Gastroenterol 21:12605–12611
Parikh A, Lee C, Joseph P et al (2014) microRNA-181a has a critical role in ovarian cancer progression through the regulation of the epithelial–mesenchymal transition. Nat Commun 5:2977
Peng J, Xie Z, Cheng L et al (2015) Paired design study by real-time PCR: miR-378 and miR-145 are potent early diagnostic biomarkers of human colorectal cancer. BMC Cancer 15:158
Picard E, Verschoor CP, Grace WM et al (2020) Relationships between immune landscapes, genetic subtypes and responses to immunotherapy in colorectal cancer. Front Immunol 6(11):369
Ramzy I, Hasaballah M, Marzaban R et al (2015) Evaluation of microRNAs-29a, 92a and 145 in colorectal carcinoma as candidate diagnostic markers: an Egyptian pilot study. Clin Res Hepatol Gastroenterol 39:508–515
Ren D, Jinlong Lu, Han X et al (2021) LINC00641 contributes to nasopharyngeal carcinoma cell malignancy through FOXD1 upregulation at the post-transcriptional level. Biochem Cell Biol 99(6):750–758
Romano G, Veneziano D, Acunzo M et al (2017) Small non-coding RNA and cancer. Carcinogenesis 38(5):485–491
Sajjadi RS, Modarressi MH, Tabatabaiefar MA (2021) JPX and LINC00641 ncRNAs expression in prostate tissue: a case-control study. Res Pharm Sci 16(5):493–504
Siegel RL, Miller KD, Jemal A (2019) Cancer statistics. CA Cancer J Clin 69:7–34
Stoffel EM, Murphy CC (2020) Epidemiology and mechanisms of the increasing incidence of colon and rectal cancers in young adults. Gastroenterology 158(2):341–353
Schmitt AM, Chang HY (2016) Long noncoding RNAs in cancer pathways. Cancer Cell 29:452–463
Tang Y, Cheung BB, Atmadi Brata B et al (2017) The regulatory role of long noncoding RNAs in cancer. Cancer Lett 391:12–19
Wallace J, Hu R, Mosbruger TL et al (2016) Genome-wide CRISPR-Cas9 screen identifies microRNAs that regulate myeloid leukemia cell growth. PLoS ONE 11(4):e0153689
Wang J, Liu Z-H, Yu L-J (2019) Long non-coding RNA LINC00641 promotes cell growth and migration through modulating miR-378a/ZBTB20 axis in acute myeloid leukemia. Eur Rev Med Pharmacol Sci 23(17):7498–7509
Wang XB, Wang H, Long HQ et al (2018) LINC00641 regulates autophagy and intervertebral disc degeneration by acting as a competitive endogenous RNA of miR-153-3p under nutrition deprivation stress. J Cell Physiol 234(5):7115–7127
Xue D, Xue YF, Zhang LJ et al (2021) LINC00641 induces the malignant progression of colorectal carcinoma through the miRNA-424 5p/PLSCR4 feedback loop. Eur Rev Med Pharmacol Sci 25(2):749–757
Zeng M, Zhu L, Li L et al (2017) miR-378 suppresses the proliferation, migration and invasion of colon cancer cells by inhibiting SDAD1. Cell Mol Biol Lett 22:12
Zhang J, Jin S, Xiao W et al (2021) Long noncoding RNA LINC00641 promotes renal cell carcinoma progression via sponging microRNA-340-5p. Cancer Cell Int 21(1):210
Funding
This work was supported by a project funded by Cairo University entitled “long noncoding RNA signature in CRC.”
Author information
Authors and Affiliations
Contributions
N.H. and O.G. conceived and designed the research study; N.A. and O.G. contributed reagents/materials/analysis tools; N.A. and N.H. analyzed the data; N.H. and N.A. wrote the manuscript. Finally, all authors reviewed the manuscript.
Corresponding author
Ethics declarations
Ethical approval
The study was performed with the approval of the ethics committee of Al-Kasr Al-Ainy Hospital, Cairo University, Egypt, and The Helsinki Declaration was followed in the current research. All of the participants in this study gave their informed consent.
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Abdel Hameed, N.A., Shaker, O.G. & Hasona, N.A. Significance of LINC00641 and miR-378 as a potential biomarker for colorectal cancer. Comp Clin Pathol 31, 807–814 (2022). https://doi.org/10.1007/s00580-022-03384-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00580-022-03384-8