Abstract
Segmentation of the liver from abdominal CT images is an essential step for computer-aided diagnosis and surgery planning. The U-Net architecture is one of the most well-known CNN architectures which achieved remarkable successes in both medical and biological image segmentation domain. However, it does not perform well when the target area is small or partitioned. In this paper, we propose a novel architecture, called dense feature selection U-Net (DFS U-Net), which addresses this challenging problem. Specifically, The Hounsfield unit values were windowed in a range to exclude irrelevant organs, and then use the pre-processed data to train our proposed DFS U-Net model. To further improve the segmentation accuracy of the small region and disconnected regions of interests with limited training datasets, we improve the loss function by adding a parameter to the formula. With respect to the ground truth, the Dice score ratio can reach over 94.9% for the liver. Our experimental results demonstrate its potential in clinical usage with high effectiveness, robustness and efficiency.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
It is greatly significant to delineate the area of an organ of computed tomography-based clinical diagnosis. The accuracy of segmentation not only facilitates the subsequent quantitative assessment of the regions of interest but also benefits precise diagnosis, prediction of prognosis, and surgical planning and intra-operative guidance [1]. Recently, the convolutional neural networks (CNNs) have been utilized to solve high-level tasks in medical fields, such as organ recognition and tissues detection [2]. The salient features learned by end-to-end approaches are the major advantage of CNNs, which are more representative and effective than traditional handcrafted features with heuristically tuned parameters for the final task [3]. Similarly, CNNs demonstrate their outstanding achievements and promising performance for pixel-level classification and labeling problems e.g., fully convolutional neural networks (FCN) [4], DeepLab [5] and U-Net [6]. These methods have all garnered significant improvements in performance over previous approaches by applying the state-of-the-art CNN-based image classifiers and representation to the semantic segmentation problem in both domains [7]. CNNs-based applications are very wide, e.g., semantic segmentation in recent computer vision and medical imaging analysis work. The ASDNet [8] showed good results in medical image segmentation and adopted semi-supervised learning. The deep adversarial network [9] introduced the adversarial method to reduce the demand of labeling image data and could segment a part of the biological image. This same idea is applied in [10] with an images to images neural network for 3D automatic segmentation.
Semantic segmentation for a target organ involves assigning a label contour to each pixel in a medical image. On one hand, features of single pixels (or patches) play a major role in classification. On the other hand, factors such as edges, i.e., organ boundaries, spatial consistency, and appearance consistency, could greatly impact the overall performance [11]. Furthermore, there are indications of semantic vision tasks requiring hierarchical levels of visual perception and abstraction [12, 13]. Therefore, generating rich feature hierarchies for both interior and boundary of organs will provide important “mid-level visual cues” [14] for semantic segmentation. Subsequent spatial aggregation of these mid-level cues then has the prospect of improving semantic segmentation by enhancing the accuracy and consistency of pixel-level labeling.
Liver disease is widely endangering the health of people worldwide. According to statistics, liver disease is one of the main causes of premature human death. From this point of view, liver surgery treatment is particularly important. Liver surgery is one of the main treatment methods for common benign and malignant liver diseases. Liver segmentation, as a fundamental and essential step of computer-assisted liver disease [15], plays a crucial role in liver analysis and clinical surgical planning. Traditional manual segmentation is very time consuming and poorly reproducible due to high similarity between liver tissue and its adjacent organs as well as the difference between the livers and lesion, as Fig. 1 shows. Therefore, it is essential to develop a high-quality automatic liver segmentation method.
Medical image segmentation is more and more applied in assistant diagnosis and has drawn more and more attention in the field of the enhancement of the efficiency and accuracy of treatment. Accurate liver segmentation is fundamental for various computer-aided procedures including liver cancer diagnosis, hepatic disease interventions, and treatment planning.
In previous studies, many image segmentation methods based on convolutional neural networks, such as U-Net and FCN, have been proposed, which only can be thought as a kind of rough segmentation. Their results suffered from the loss of disconnected liver areas and inaccuracies around boundary information of small organs and blood vessels. With the limited number of data, it is very difficult to build models that are general enough to capture a large variability of the deformable organs, e.g., the liver. In this paper, we will propose a novel automatic segmentation method using a dense block and a feature selection block. This method turns the convolution process to a dense block with successive convolution layers based on U-Net. Dense connection combines underlying boundary features with upper semantic feature information, which extracts more information with fewer features as few as possible [16, 17]. We utilize a feature selection block [18] on the path of copy and concatenation in U-Net. We select the feature maps which are useful for the segmentation of objects and drop redundant feature information. By continuously enhancing the extraction of the fused features through the bottom-up strategy, it is possible to excavate more accurate segmentation processing of position, edge, and texture features with limited data. The new model is called dense feature selection U-Net (DFS U-Net).
2 Related work
Considering whether human interaction is necessary, we simply divide previous work into semi-automatic segmentation and automatic segmentation. Semi-automatic segmentation could better segment the target region with the guidance of humans [19]. But with stronger stability, the workload is greater. Liao et al. [20] proposed an efficient liver segmentation method based on graph cut and bottleneck detection using intensity, local context, and spatial correlation of adjacent slices. Yang et al. [21] came up with a hybrid semi-automatic segmentation method that consists of a fast-marching level-set and a threshold-based level-set.
The automatic segmentation method saves more work and time than the semi-automatic method does. The 3D deep supervised U-Net model [19, 22] obtained great achievements in medical image segmentation of CT volume, particularly in the accuracy of liver segmentation. The deep model ensembled the random feature subspace [23] performs effectively on the segmentation of liver tumors. Jin et al. [24] utilized an improved fully convolution network to segment livers. However, their results were not accurate and stable enough. In many cases, the segmented contours are not clear and smooth enough, and it is difficult to capture the feature information of small samples.
The H-DenseUNet [25] cascaded 2D dense U-Net and 3D residual dense U-Net, which has obtained more desired results in the segmentation of livers and lesions. This method aggregates the location information of 2D CT slices and the sequence spatial information of 3D CT volume. However, the shortcoming of this method is obvious that the huge network structure and a large number of parameters require high-end hardware for computation. Compared with the only 2D network, the computing of 3D convolution has an exponential time complexity. In terms of training time requirements, this cascaded hybrid network is also more demanding than other network models.
Recently, Krishna et al. [26] simplified the cascaded network by replacing the 3D structure with a 2D structure. However, the modified network is two-staged and still cascaded. Much information is redundant in a large number of feature maps generated by the successive dense convolution. In this paper, our work explores a novel end-to-end DFS method, which significantly improved the performance of liver segmentation.
3 Method
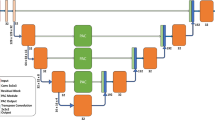
An overview of our proposed image segmentation with DFS U-Net and the entire pipeline are depicted in Fig. 2. The workflow consists of three major steps. The first step deals with data preprocessing and preparation for the neural network segmentation. In the second step, it feeds CT sequence slices into our DFS U-Net model and obtains the well-trained parameters for liver segmentation. In the final step, the neural network obtains the region of interest (ROI) by calculating the probability of each pixel. We use morphological optimization as a post-processing method to refine the final segmentation results.
3.1 Data processing
The data pre-processing generally includes data normalization and standardization, but due to the particular distribution of pixels in medical images, if the traditional normalization method is adopted, the image will lose a lot of important feature information. In order to adapt to the particularity of CT images, we keep the image values in an interval and use HU windowing [27] to pre-process the dataset.
First, we process the Pixel Padding Value in the acquired DICOM image. The abdominal interface of the human body is almost irregular. The DICOM image will automatically add a padding value to the image during the acquisition process. The padding value will increase the variation of the image range. Therefore, we first set the value of Pixel Padding in the dataset to 0, then remove the attribute from the DICOM image.
In the second stage, we need to use the following formula to convert the pixel values into HU values:
where the \(\mathrm{rescale\_slope}\) value and \(\mathrm{rescle\_intercept}\) value are set to 1 and −1024, respectively. The raw CT slices are windowed to a Hounsfield Unit in the range of −75 to 175 HU to neglect organs and tissues that are not interest [27]. Moreover, we enhance the contrast through histogram equalization. The intensities are better distributed by spreading out their frequencies so that the spacing out values are closely placed. Figure 3 shows the effect of our preprocessing to a raw medical slice.
3.2 DFS U-Net for liver segmentation
3.2.1 Architecture
The dense feature selection U-Net model proposed in this paper mainly includes convolution layer, pooling layer, dense connection block, transition layer, threshold deconvolution layer, and feature selection block. Figure 4 shows the architecture of the DFS U-Net. First, we utilize \(3\times 3\) filters to coarsely extract features of input CT images. After the first pool layer, we replace the convolution in the second layer with the dense connection block of different specifications between the coming adjacent pooling layers. We combine the convolution layer with \(1\times 1\) filters with a \(2\times 2\) max-pooling layer to reduce the resolution and dimension as the input of the next dense connection block, which is called the transition layer. The transition layer can also generate the number of feature maps we want for the skip connection.
In the deconvolution phase, we come up with a new method of restoring resolution, namely the threshold deconvolution layer. The size of the image is restored by the deconvolution operation. The feature maps from the transition layer and the deconvolution layer are concatenated so that the feature and location information will be reused in the down-sampling process. The improved resolution feature maps undergo subsequent continuous convolution operations and allow more accurate combination of information of skip connection and output of the deconvolution layer. In the process of deconvolution, the dense connection block is not used, because from f our experimental results, we found that the dense connection block could do well in the continuous exploration of new features, but not in reusing them. If the dense connection blocks are used blindly, they will make the network over parameterized, which will increase the training time but helpless with the accuracy of segmentation.
In the skip connection part, we add a module similar to residual connection in parallel to the skip connection, which is called feature selection block. With the deepening of network layers, the number of feature maps is increasing. Some of them are useless to our segmentation target and some are not informative.
All of these feature maps are redundant. Figure 5 shows feature maps that are not informative. The feature selection block can reflect the importance of each channel by a parameter. Thus, we could suppress redundant features and extract the feature maps we need.
3.2.2 Dense block
The basic idea of the dense block proposed is to continuously enhance the feature extraction of the target organ through successive convolutional layers. We have established a short path to make sure that the information transmission between the convolution layers is not affected. Briefly, the input to each layer comes from the output of all convolutional layers before in the same dense block. We define the output of the \(n^\mathrm{th}\) as follows:
where \(\left[ R_0,R_1,R_2, \ldots , R_{n-1}\right]\) refers to the feature map concatenations of \(0,\ldots , n-1\) layer, and \(F_n\) is a function that integrates BN, ReLU and convolution. In this way, each convolutional layer can access all previous feature maps in the dense block. To avoid the dimension being too large, we add a \(1\times 1\) convolution layer before each convolutional layer in the dense block, and made the output dimension of each convolution layer narrow (less than 100).
The short path between the convolution layers implies that each layer has access to the gradients from the upper layers and the input image directly, guiding to implicit in-depth supervision [15]. This structure can effectively reduce the number of parameters and control the complexity of the model. Through improving feature utilization and transmission rate, it can effectively alleviate the problem of vanishing gradient. To make the feature maps in one and the same block with coincident size, we set three blocks between each two pooling layers. We have set 6, 12, and 18 sets of convolution layers in the three dense blocks. Figure 6 shows the detailed architecture of six sets of dense blocks.
3.2.3 Feature selection block
The neural networks mainly consider semantic information but not consider the relationship between channels. The feature selection block is proposed based on this point. Our motivation is to explicitly establish the interdependence between feature channels. In addition, we do not intend to introduce a new dimension for feature channel fusion, but to adopt a new “feature re-calibration” strategy. Specifically, it acquires the importance of each feature channel by learning automatically and then according to this importance, it enhances useful features and suppress features that are not very helpful for the task.
Figure 7 shows the specific structure of the feature selection block. First, we compress the feature map with dimension \(w\times h\times c\) along the spatial dimension. This is achieved by using global average pooling to generate channel-wise statistics. We can define it as:
The \(1\times 1\times c\) feature maps have a global sensing field. They represent the global distribution of responses on feature channels and the dimension of output matches the number of feature channels of input. By fitting the correlation between channels through the full connection layer, the parameter W of \(F_\mathrm{c}\) layer generates a weight for each channel, and outputs the importance of each channel through the non-linear function:
where \(\delta\) refers to the ReLU function and \(\sigma\) refers to the sigmoid function. Finally, the importance of each channel after feature selection is weighted to the input feature maps by channel-wise multiplication to complete the re-selection of the input feature in the channel dimension. The final output of the block is defined as follows:
3.3 Optimization through DFS U-Net
3.3.1 Threshold deconvolution layer
To make the final output consistent with the original image size, we take advantage of the deconvolution layer to remap the feature with lower resolution to the denser space of higher resolution of input images. The zero values of the upsampled fill will make difference become large between max activation and other activation within each pooling region, which would increase inter-subject variance in local segmentation regions [28]. To deal with this issue, our new method fills values after comparing the mean value with the maximum activation value of the whole map instead of just filling zero value at the empty pixels. The mean value of the whole map is calculated when yielding pooled maps and filled in vacancy during upsampling. In this process, we need to select the value to be filled in the pooling area by using the following formula:
where f(x) is the filling method with two options; v,u denote maximum activation value after pooling and mean value of the entire map, respectively.
The specific process of the filling method is demonstrated in Fig. 8, where we have \(a,c< u\le b,d\). The output of the map is first enlarged to Map1 after filling maximum activation values a, b, c, and d. Then, for each of the maximum activation values, if they are larger than u other activation within corresponding pooling region is filled with u; otherwise, it is filled with value 0. After getting Map2, it still exists vacancy sparsely when \(v<u\). So we need to utilize deconvolution operation to further densify the outputs.
3.3.2 Improvement of loss function
Because of the peculiarities of enhanced CT images, two problems lead to the difficulty of our segmentation. (1) The target organ is divided into many disconnected parts in one CT slice. (2) Due to the possibility of being tangent to the boundary of the target organ in CT scanning, there are only a small part on some CT slices, even an area with tens or hundreds of pixels, which has a negative impact on accuracy. Figure 9 shows examples of these problems.
To solve the problem, we plan to change the distribution of back-propagation signals by improving the calculation of loss function, thus enhancing our network capture capability of the foreground during the training [29]. Because of the high imbalance between background and foreground voxels (organs, vessels, etc.) the network will concentrate on differentiating the foreground from the background voxels in order to minimize the loss function used for training. In order to avoid over-fitting and enhance the semaphore of loss function during the backpropagation, we add the second l_2 regularization term. By enhancing semaphores, the loss function can be more demanding on segmentation foreground so that the problem of the disconnected parts can be solved. The improved balanced cross-entropy loss function formula is as follows:
From the Eq. (7), our major idea is to balance the weight proportion and sample loss of foreground and background in the back propagation by introducing a new weight parameter \(\beta\), where \(1-\beta =|Y_+|/|Y|\) and \(\beta =|Y_-|/|Y|\). \(|Y_+|\) and \(|Y_-|\) denote the positive samples (foreground) and negative samples (background) of label sets (ground truth), respectively. In this way, even if the proportion of foreground is very small, the loss is more inclined to feature extraction of the foreground part.
3.3.3 Other optimization
We found that although the \(5\times 5\) convolution kernel has a large receptive field if we used two successive \(3\times 3\) convolution kernels, we could achieve the same effect as using a \(5\times 5\) convolution kernel, but the number of parameters would be less. Thus, we all used \(3\times 3\) kernels in our network. The initialization of kernels is “Xaiver” [30], which initializes the weights in the network by plotting them from the distribution of zero mean and specific variance. The weight distribution of the method is evenly distributed, which enables the network to converge fastest and the weights to reach superior values. The initial learning rate is \(10^{-4}\) and reduces at every 10 epochs. For the choice of pooling layer, we chose the max pooling. In general, the mean pooling can retain more background information of the image, and the max-pooling could retain more texture information. The detailed architecture of the final network is shown in Fig. 10.
3.4 Post processing
The binary mask achieved by softmax with loss prediction layer is processed by morphological operations for boundary enhancement and filling the internal holes.
4 Experiment
Training of the model for the weight-optimization process and segmentation based on the trained model, see Fig. 11. Given a training set of images and labels \(s=\{(i_n,l_n),n = 1,2,\ldots ,N\}\) , \(i_n\) denotes the raw CT images and denotes the ground truth label images. The image to be segmented is input into the trained model. During the training process, the input image is continuously extracted feature. Finally, the outputs are two probability maps, which represent the probability that each pixel belongs to a category. After thousands of iterations, we continuously reduce the loss value through feedback to get the model with the well-trained weights. We use this well-trained model for the final test phase segmentation.
4.1 Dataset preparation
In this experiment, to test the robustness and precision of our proposed novel network, we use two datasets to prevent occasions and coincidence. LiTS [25] is the dataset of liver and liver tumors for enhanced CT image segmentation challenge competition. Many researchers in the field of liver segmentation are using this dataset to show the superiority of their methods. The second dataset is DatasetA, which is composed of enhanced CT images of 180 patients from Affiliated Hospital of Jiangsu University through data preprocessing based on Housefield. The image of each patient is marked by a professional doctor as its ground truth.
For the LiTS dataset, we use 110 groups for training and the remaining 20 groups for testing. Similar to the setting of LiTS dataset, we divide datasetA into 150 cases and 30 cases for training and testing, respectively.
4.2 Experiment and evaluation
Based on the datasets introduced previously, the experimental results of contour show that three superiorities of DFS U-Net model: (I) the effectiveness of small region segmentation; (II) the robustness of disconnected region segmentation; (III) With limited training data, our deep network can still achieve accurate segmentation and avoid over-fitting. Figure 12 shows our segmentation of the liver.
We evaluate the proposed segmentation framework in the context of the automated segmentation of the liver. To evaluate the effectiveness of the DFS U-Net model on improving the accuracy of liver segmentation, we consider the following three evaluation measures.
The Dice coefficient is the function to calculate the coincidence degree of two sets in segmentation application, which is widely used to evaluate segmentation models. The Dice coefficient is calculated as follows:
where the set A and set B represent a result mask and its corresponding label, respectively. The higher Dice coefficient, the higher the segmentation accuracy is.
The Precision and Recall are also two commonly used evaluation coefficients in segmentation, which could calculate the relationship between positive and negative samples and prediction results. The formula defined is as follows:
The Precision reflects the relation between the correct prediction and the wrong prediction of background as a target. And Recall reflects the association between the correct prediction and the wrong prediction of target organ as background. In some ways, the two evaluation coefficients will not change in the same direction. But they could represent the stability, robustness and applicability of the segmentation model. The performance of DFS U-Net and various other algorithms on the LiTS dataset are showed in Table 1.
It can be seen from the table that the network model we proposed is obviously superior to other existing methods in all indicators. The proposed network is improved on the foundation of the traditional U-Net model, aiming at the problem that the previous semantic segmentation model can hardly solve. Thus, from Table 1, we can see that the segmentation effectiveness of our model is much better than that of FCN-8s and U-Net with the same data and preprocessing. Comparing with Krishna et al. [26] with both 2D dense U-Net structure, the Dice coefficients of our model are obviously better than those of them, which proves that our changes in other areas have indeed optimized our model.
In addition, we also present the comparison results of different algorithms on the Dataset A, which is shown in Table 2. By comparing the indicators of different algorithms, we can see that our algorithm has a better performance.
To show the advantages of our model in the segmentation of small samples and disconnected regions, we chose 60 CT slices from 20 groups of the testing part in the LiTS dataset, including small and disconnected liver regions. Besides, we used the same training and test dataset to compare with traditional U-Net and FCN-8s. In the process of training, we set the same values for the parameters of preprocessing, learning rate decay ratio, the total number of iterations, etc. Figure 13 shows the comparison of small section organs and Table 3 shows quantitative results.
The results prove the outstanding performance and capability of DFS U-Net for liver segmentation. From the results of the segmentation, it can be seen that the proposed DFS U-Net can effectively capture the feature information such as the position and boundary of a target organ. For small section organs, our model can still achieve good accuracy. Of course, the model also performs well for the segmentation of disconnected regions. Figure 14 shows the comparison results of the disconnected regions.
Without extracting 3D context information and features, it is worth noting that, the accuracy of our model is higher than the 3D DSN model of Dou et al. [19]. Therefore, this experiment verifies the DFS U-Net is powerful and robust in the liver segmentation problem.
5 Conclusion
This paper proposed a DFS U-Net segmentation model that combines traditional U-Net with densely connected blocks and feature selection block with an attention mechanism. The method first preprocesses the slice data of the enhanced CT. The preprocessing method uses the characteristics of the window width of the CT image to limit the variation range of the HU value in an image. The pre-processed datasets are more conducive to the training and convergence of our DFS model. Our experimental results showed that the insertion of dense connection blocks during the down-sampling process can significantly improve the accuracy of image segmentation. Moreover, the feature selection blocks does choose the useful feature maps for our task. The automatic segmentation results for the liver is not much different from the results of manual segmentation by the doctor. Our DFS U-Net model can still achieve more accurate segmentation in the case of insufficient data and can effectively avoid over-fitting.
References
Yu, Q., Xie, L., Wang, Y., Zhou, Y., Fishman, E.K., Yuille, A.L.: Recurrent saliency transformation network: Incorporating multi-stage visual cues for small organ segmentation, In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 8280–8289 (2018)
Yang, L., Zhang, Y., Chen, J., Zhang, S., Chen, D.Z.: Suggestive annotation: a deep active learning framework for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention, pp. 399–407 (2017)
Yang, X., Liu, C., Wang, Z., Yang, J., Le Min, H., Wang, L., Cheng, K.T.T.: Co-trained convolutional neural networks for automated detection of prostate cancer in multi-parametric MRI. In: Medical image analysis, pp. 212–227(2017)
Shelhamer, E., Long, J., Darrell, T.: Fully convolutional networks for semantic segmentation. In: IEEE transactions on pattern analysis and machine intelligence, pp. 640–651 (2017)
Chen, L.C., Papandreou, G., Kokkinos, I., Murphy, K., Yuille, A.L.: Semantic image segmentation with deep convolutional nets and fully connected crfs. arXiv:1412.7062 (2014)
Ronneberger, O., Fischer, P., Brox, T.: U-net: Convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention, pp. 234–241 (2015)
Wang, Z., Liu, C., Cheng, D., Wang, L., Yang, X., Cheng, K.T.: Automated detection of clinically significant prostate cancer in mp-MRI images based on an end-to-end deep neural network. In: IEEE transactions on medical imaging, pp. 1127–1139 (2018)
Nie, D., Gao, Y., Wang, L., Shen, D.: ASDNet: attention based semi-supervised deep networks for medical image segmentation. In: International conference on medical image computing and computer-assisted intervention, pp. 370–378 (2018)
Zhang, Y., Yang, L., Chen, J., Fredericksen, M., Hughes, D.P., Chen, D.Z.: Deep adversarial networks for biomedical image segmentation utilizing unannotated images. In: International conference on medical image computing and computer-assisted intervention, pp. 408–416 (2017)
Yang, D., Xu, D., Zhou, S.K., Georgescu, B., Chen, M., Grbic, S., Comaniciu, D.: Automatic liver segmentation using an adversarial image-to-image network, In: International conference on medical image computing and computer-assisted intervention, pp. 507–515 (2017)
Zheng, S., Jayasumana, S., Romera-Paredes, B., Vineet, V., Su, Z., Du, D., Torr, P.H.: Conditional random fields as recurrent neural networks. In: Proceedings of the IEEE international conference on computer vision, pp. 1529–1537 (2015)
Yan, Z., Yang, X., Cheng, K.T.: A skeletal similarity metric for quality evaluation of retinal vessel segmentation. In: IEEE transactions on medical imaging, pp. 1045–1057 (2017)
Xie, S., Tu, Z.: Holistically-nested edge detection. In: Proceedings of the IEEE international conference on computer vision, pp. 1395–1403 (2015)
Roth, H.R., Lu, L., Lay, N., Harrison, A.P., Farag, A., Sohn, A., Summers, R.M.: Spatial aggregation of holistically-nested convolutional neural networks for automated pancreas localization and segmentation. In: Medical image analysis, pp. 94–107 (2018)
Brügger, R., Baumgartner, C.F., Konukoglu, E.: A partially reversible U-Net for memory-efficient volumetric image segmentation. In: International conference on medical image computing and computer-assisted intervention, pp. 429–437 (2019)
Huang, G., Liu, Z., Van Der Maaten, L., Weinberger, K.Q.: Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 4700–4708 (2017)
Zhang, H., Patel, V.M.: Densely connected pyramid dehazing network. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 3194–3203 (2018)
Hu, J., Shen, L., Sun, G.: Squeeze-and-excitation networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 7132–7141 (2018)
Dou, Q., Chen, H., Jin, Y., Yu, L., Qin, J., Heng, P.A.: 3D deeply supervised network for automatic liver segmentation from CT volumes. In: International conference on medical image computing and computer-assisted intervention, pp. 149–157 (2016)
Liao, M., Zhao, Y.Q., Wang, W., Zeng, Y.Z., Yang, Q., Shih, F.Y., Zou, B.J.: Efficient liver segmentation in CT images based on graph cuts and bottleneck detection. In: Physica Medica, pp. 1383–1396 (2016)
Yang, X., Yu, H.C., Choi, Y., Lee, W., Wang, B., Yang, J., You, H.: A hybrid semi-automatic method for liver segmentation based on level-set methods using multiple seed points. In: Computer methods and programs in biomedicine, pp. 69–79 (2014)
Dou, Q., Yu, L., Chen, H., Jin, Y., Yang, X., Qin, J., Heng, P.A.: 3D deeply supervised network for automated segmentation of volumetric medical images. Med. Image Anal. 2017, 40–54 (2017)
Huang, W., Yang, Y., Lin, Z., Huang, G.B., Zhou, J., Duan, Y., Xiong, W.: Random feature subspace ensemble based extreme learning machine for liver tumor detection and segmentation. In: 2014 36th annual international conference of the IEEE engineering in medicine and biology society, pp. 4675–4678 (2014)
Jin, X., Ye, H., Li, L., Xia, Q.: Image segmentation of liver CT based on fully convolutional network. In: 2017 10th international symposium on computational intelligence and design (ISCID), pp. 210–213 (2017)
Li, X., Chen, H., Qi, X., Dou, Q., Fu, C.W., Heng, P.A.: H-DenseUNet: hybrid densely connected UNet for liver and tumor segmentation from CT volumes. In: IEEE transactions on medical imaging, pp. 2663–2674 (2018)
Kaluva, K.C., Khened, M., Kori, A., Krishnamurthi, G.: 2d-densely connected convolution neural networks for automatic liver and tumor segmentation. arXiv:1802.02182 (2018)
Christ, P.F., Ettlinger, F., Grün, F., Elshaera, M.E.A., Lipkova, J., Schlecht, S., Rempfler, M.: Automatic liver and tumor segmentation of CT and MRI volumes using cascaded fully convolutional neural networks. arXiv:1702.05970 (2017)
Zeiler, M.D., Krishnan, D., Taylor, G.W., Fergus, R.: Deconvolutional networks. In: 2010 IEEE Computer Society Conference on computer vision and pattern recognition, pp. 2528–2535 (2010)
Xie, S., Tu, Z.: Holistically-nested edge detection. In: Proceedings of the IEEE international conference on computer vision, pp. 1395–1403 (2015)
Glorot, X., Bengio, Y.: Understanding the difficulty of training deep feedforward neural networks. In: Proceedings of the thirteenth international conference on artificial intelligence and statistics, pp. 249–256 (2010)
Soler, L., Hostettler, A., Agnus, V., Charnoz, A., Fasquel, J., Moreau, J., Marescaux, J.: 3D image reconstruction for comparison of algorithm database: a patient specific anatomical and medical image database. In: IRCAD, Strasbourg, France, Tech. Rep. (2010)
Gauriau, R., Cuingnet, R., Lesage, D., Bloch, I.: Multi-organ localization with cascaded global-to-local regression and shape prior. In: Medical image analysis, pp. 70–83 (2015)
Wolz, R., Chu, C., Misawa, K., Fujiwara, M., Mori, K., Rueckert, D.: Automated abdominal multi-organ segmentation with subject-specific atlas generation. In: IEEE transactions on medical imaging, pp. 1723–1730 (2013)
He, B., Huang, C., Jia, F.: Fully automatic multi-organ segmentation based on multi-boost learning and statistical shape model search. In: VISCERAL Challenge@ ISBI, pp. 18–21 (2015)
Ben-Cohen, A., Diamant, I., Klang, E., Amitai, M., Greenspan, H.: Fully convolutional network for liver segmentation and lesions detection. In: Deep learning and data labeling for medical applications, pp. 77–85 (2016)
Ahmad, M., Yang, J., Ai, D., Qadri, S.F., Wang, Y.: Deep-stacked auto encoder for liver segmentation. In: Chinese conference on image and graphics technologies, pp. 243–251 (2017)
Rafiei, S., Karimi, N., Mirmahboub, B., Soroushmehr, S.M., Felfelian, B., Samavi, S., Najarian, K.: Liver segmentation in abdominal CT images by adaptive 3D region growing. arXiv:1802.07794 (2018)
Acknowledgements
The authors would like to thank Radiologists of the Medical Imaging Department of Affiliated Hospital of Jiangsu University. This work was supported by the National Natural Science Foundation of China (61772242, 62076130, 61572239, 61976106, 91846104), the China Postdoctoral Science Foundation (2017M611737), the Six Talent Peaks Project in Jiangsu Province (DZXX-122) and the Key Special Project of Health and Family Planning Science and Technology in Zhenjiang City (SHW2017019).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by X. Yang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, Z., Han, K., Wang, Z. et al. Automatic liver segmentation from abdominal CT volumes using improved convolution neural networks. Multimedia Systems 27, 111–124 (2021). https://doi.org/10.1007/s00530-020-00709-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00530-020-00709-x