Abstract
Chronic kidney disease (CKD) is a significant concern for individuals with type 1 diabetes (T1D), impacting their quality of life and healthcare costs. Identifying T1D patients at greater risk of developing CKD is crucial for preventive measures. However, it is challenging due to the asymptomatic progression of CKD and limited nephrologist availability in many countries. This study explores machine learning algorithms to predict CKD risk in T1D patients using ten years of retrospective data from the Epidemiology of Diabetes Interventions and Complications clinical trial. Eleven machine learning algorithms were applied to twenty-two readily available features from T1D patients’ routine check-ups and self-assessments to develop 10-year CKD risk prediction models. In addition, we also proposed a heterogeneous ensemble model (STK) using a stacking generalization approach. The models’ performance was evaluated using different evaluation metrics and repeated stratified k-fold cross-validation. Several predictive models showed reliable performance in CKD risk prediction, with the proposed ensemble model being the best performing with an average accuracy of 0.97, specificity of 0.98, sensitivity/recall of 0.96, precision of 0.98, F1 score of 0.97, Kappa and MCC score of 0.94, AUROC of 0.99, and Precision-Recall curve of 0.99. The proposed machine learning approach could be applicable for CKD risk prediction in T1D patients to ensure the necessary precautions to overcome the risk.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Chronic kidney disease (CKD) is a progressive condition in which the kidneys gradually lose function over time. It is characterized by a decline in kidney function, which can lead to a buildup of waste products and fluids in the body. The most accurate measure of total kidney function is the glomerular filtration rate (GFR), which indicates the volume of fluid our kidney filters in a unit of time. Kidney Disease Improving Global Outcomes (KDIGO) guidelines 2012 and current international guidelines define a person as having CKD if their estimated glomerular filtration rate (eGFR) has been less than \( 60\;{\text{mL}}/{\text{min}}/1.73\;{\text{m}}^{2} \) for over three months [1]. CKD is a major challenge to public health worldwide, as 10% of the total population is predicted to have CKD [2]. The yearly medical costs per patient with CKD can reach as high as $65,000 [3]. In addition, there is a higher chance of additional adverse health hazards, such as an increased risk of mortality, end-stage renal disease (ESRD) progression, and heart and artery problems. CKD is one of the major causes of death in the USA, and in 2019, it was the 12th leading cause of death globally [4, 5].
Diabetes is the leading cause of CKD, and 1 in 3 adults with diabetes may have CKD. For type 1 diabetes (T1D) patients, this ratio is even higher. More than 50% of patients with T1D have a chance of developing CKD [6]. The most common cause of end-stage renal disease in the West is diabetes CKD, which is also linked to a higher risk of cardiovascular events [7, 8]. In addition, when a diabetes patient is affected with CKD, their health-related quality of life decreases, and healthcare costs increase significantly [9, 10].
However, the positive side is that CKD is a non-communicable disease. The risk of CKD in T1D patients can be prevented or delayed through appropriate dietary and lifestyle adjustments and chronic kidney disease-targeted interventions [11,12,13]. Identifying T1D patients with CKD risk is crucial for this purpose. Unfortunately, this task can be difficult because CKD progression is asymptomatic in most cases [4]. In addition, in many countries, the nephrologists’ density is very low. According to the International Society of Nephrology Global Kidney Health Atlas (ISN-GKHA), in 2016 the nephrologist density in underprivileged countries was only 0.318 per million population [14]. Hence, an automated CKD prognosis model for T1D patients to identify the patients with a greater risk of developing CKD can be helpful in ensuring more intensive management to avoid CKD.
Recently, disease prediction and prognosis using machine learning (ML) techniques have shown considerable promise [15, 16]. We can find several ML-based CKD prognostic models in the literature. However, only a few of them were focused on diabetes patients, and even fewer were focused on T1D patients. Chan et al. (2021) utilized a combination of electronic health records and biomarkers of 1146 diabetes patients and random forest (RF) to develop a prognostic model to predict sustained eGFR decline or kidney failure within five years [17]. Though they achieved an area under the receiver operating characteristic curve (AUROC) of 0.77, their use of biomarkers made it challenging to implement this model in many places. Allen et al. (2022) used extreme gradient boosting (XGB) and RF machine learning algorithms to predict diabetic kidney diseases within five years upon diagnosis of type 2 diabetes and achieved an AUROC of over 0.75 [18]. In another study by Kanda et al. (2022), a large retrospective cohort from a Japanese insurance company was used to develop ML models to predict the risk of developing CKD and heart failure in type 2 diabetes patients [19]. Using the XGB algorithm, they achieved an AUROC of 0.718 for five years of CKD risk prediction. However, none of these two models considered type 1 diabetes patients.
Type 1 diabetes is distinct from type 2 diabetes [20]. Type 2 diabetes is closely related to lifestyle, food habits, and ethnicity, and its risk can be reduced by following a healthy diet and lifestyle. On the other hand, type 1 diabetes is a genetic disorder where patients’ immune system attacks and destroys insulin-producing cells in the pancreas. As a result, patients need to take insulin injections to control their blood glucose levels. Unlike type 2 diabetes, lifestyle changes cannot reduce the risk of type 1 diabetes. Research conducted by Kristófi et al. (2021) shows that type 1 diabetes patients have a 1.4–3.0-fold higher risk of CKD than type 2 diabetes patients [21]. As a result, a prognosis model dedicated to type 1 diabetes patients should be a more viable option. Unfortunately, very limited work has been done in this field.
In one study, Niewczas et al. (2017) studied the risk factors and mechanisms of end-stage renal disease (ESRD) in patients with type 1 diabetes (T1D) and chronic kidney disease [22]. The study analyzed serum metabolomic profiles in a prospective cohort of 158 T1D patients with proteinuria and impaired renal function. Over a median follow-up of 11 years, the study identified seven modified metabolites (C-glycosyltryptophan, pseudouridine, O-sulfotyrosine, N-acetylthreonine, N-acetylserine, N6-carbamoylthreonyladenosine, and N6-acetyllysine) in the patient’s serum that were strongly associated with renal function decline and the onset of ESRD, independent of clinical factors. This study also calculated estimated glomerular filtration rate slopes from serial serum creatinine measurements and established the time to start ESRD. In another study, Pilemann-Lyberg et al. (2019) considered two biomarkers (PRO-C6 and C3M) from 663 T1D patients with normoalbuminuric and macroalbuminuric. They estimated the relation of these biomarkers with adverse outcomes in patients with T1D, including a decline in eGFR and ESRD, using Cox proportional hazards models [23]. This research reported that sPRO-C6 was linked to a higher risk of renal function decline and the development of end-stage renal disease (ESRD). However, these models considered type 1 diabetes patients who already had CKD or other kidney complications. In addition, they used complex features like biomarkers or metabolites and tried to find their association with ESRD. None of these models used machine learning and were not suitable for predicting the risk of CKD in T1D patients.
Recently, Sripada et al. (2023) utilized data from the T1D exchange registry in the USA to develop a machine learning model to predict diabetic nephropathy in T1D patients [24]. This research achieved the best performance with an F1-score of 0.67 and AUC of 0.78 using the random forest model. Colombo et al. (2020) aimed to provide contemporary data on the rates and predictors of renal decline in individuals with type 1 diabetes [25]. The study also employed ridge regression to create a model for predicting renal disease progression in T1DM patients and achieved a mean squared correlation (Pearson \( r^2 \)) of 0.745. In one of our previous studies, we developed a nomogram-based CKD prediction model for T1D patients using multivariate logistic regression with 90.04% accuracy [26]. In another study, we evaluated the performance of traditional machine learning algorithms for predicting CKD in T1D patients [27]. However, these models are applicable to identifying existing CKD and are not suitable for predicting the risk of developing CKD in the future. In addition, the accuracy of the first two studies was relatively low.
Vistisen et al. (2021) focused on developing a robust prediction model for end-stage kidney disease (ESKD) in individuals with type 1 diabetes [28]. Their research utilized ridge regression for model development and a population-based cohort of over 5000 Danish adults with type 1 diabetes, spanning from 2001 to 2016. The prediction model, which accounted for the risk of death as a competing factor, incorporated various clinical parameters, including age, sex, diabetes duration, kidney function, albuminuria, blood pressure, HbA1c levels, smoking, and cardiovascular disease history. The model demonstrated excellent discrimination, particularly for the 5-year risk of ESKD with a C-statistic of 0.888. However, the model was designed to identify the risk of ESKD and was unsuitable for predicting general CKD risk in T1D patients. Additionally, the derivation cohort was imbalanced, with only 5.5% of the participants developing ESKD, and no steps were taken to address this imbalance. The C-statistic alone does not address the class imbalance, and the reported result may lead to biased estimations of the model’s performance. To our knowledge, no other prediction models have been developed to assess the risk of CKD progression in the type 1 diabetic population.
In this study, we sought to develop and validate a machine learning-based prognosis model that could predict the risk of developing CKD among type 1 diabetes patients without a history of kidney disease. The primary research question was: is it possible to identify the risk of developing CKD in T1D patients using readily available routine data? We hypothesize that applying various machine learning algorithms to the longitudinal data of T1D patients will enable accurate prediction of CKD risk. We applied eleven supervised machine learning classification algorithms, including linear, nonlinear, ensemble, bagging, artificial neural, and deep learning neural networks, to develop 10-year CKD risk prediction models for T1D patients. After analyzing the performance of these models, we proposed a robust heterogeneous ensemble model using a stacking generalization technique for CKD risk prediction in T1D patients through an innovative combination of the best-performing models from each category. To train our model, we consider the features easily available from T1D patients’ regular check-ups and self-assessments. Our main challenge was to develop a reliable risk prediction model using a simple dataset that would enable the identification of T1D patients at high risk of developing CKD within a 10-year time frame. Other challenges were identifying the most important features for CKD risk prediction from T1D patients’ routine check-up data and determining the optimal number of features for achieving the best machine learning model performance. We introduced a strategic feature ranking and optimization approach with combinations of different data pre-processing techniques to overcome these challenges.
Our research introduces a novel approach to predicting the risk of CKD in T1D patients. To the best of our knowledge, this would be the first machine learning-based 10-year CKD risk prediction model for type 1 diabetes patients. Unlike previous related models that primarily focus on ESRD outcomes, our study pioneers the prediction of general CKD risk in T1D patients over a 10-year horizon. Notably, our model relies solely on readily available features from patients’ regular check-ups and self-assessments, facilitating early interventions. Contrasting existing models that may depend on complex variables, our approach simplifies the process, making it accessible to a broader range of healthcare settings. The innovation extends to the development of an advanced heterogeneous ensemble model, combining diverse machine learning techniques to achieve superior performance even with straightforward features. Furthermore, this study introduces a strategic feature ranking and optimization approach to enhance model efficiency and accuracy. Another major contribution of our research is the provision of essential features from routine check-ups of T1D patients for CKD risk prediction.
By utilizing our proposed prognosis model, healthcare providers can identify T1D patients at high risk of developing CKD within a 10-year timeframe. This proactive approach empowers patients to take necessary precautions and interventions to address this potential threat. Furthermore, our model holds particular promise for T1D patients in developing nations, where access to nephrologists is limited. This research will serve as a valuable resource to bridge the healthcare gap and improve early CKD risk detection.
2 Methods
Our study followed a systematic process encompassing data collection, sample selection, data pre-processing, feature ranking, machine learning model training, and performance evaluation. A schematic diagram illustrating this process is provided in Fig. 1. Each step is comprehensively explained in the subsequent subsections.
2.1 Data source and study population
We reviewed 1375 T1D patients’ 10-year retrospective longitudinal data from the Epidemiology of Diabetes Interventions and Complications (EDIC) clinical trial. This trial was carried out by the National Institute of Diabetic, Digestive, and Kidney Diseases (NIDDK), USA, to examine how rigorous diabetes therapy affected the T1DM population [29, 30]. The EDIC trial started in 1994 at 28 sites in the USA and Canada and is still ongoing. In this trial, clinical parameters were measured following standard methodologies in the EDIC central biochemistry laboratory, and long-term quality control procedures were established to prevent measurement drift [30, 31]. Patients’ demographic and behavioral data were collected through self-assessments. The EDIC study measured patients’ body mass index, glycated hemoglobin level, blood pressure, serum creatinine level, and estimated GFR annually. In contrast, albumin excretion rate and fasting lipid levels were measured every two years [31]. The chronic kidney disease epidemiology collaboration (CKD-EPI) method was used to calculate the estimated GFR [31, 32]. More details of this dataset can be found in our previous two articles [26, 27].
To develop our model, we considered 10-year retrospective longitudinal data from the EDIC trial between the period of the year 1999 and the year 2008. Patients younger than 18 years old were excluded from our study. In addition, we excluded T1D patients with CKD or other kidney diseases at the baseline. We also excluded the patients who discontinued the EDIC trial or died for non-CKD reasons. All samples with missing values in the output class were also excluded. Ultimately, we selected 1309 samples, of which 110 (8.40%) developed CKD during the specified time frame, as depicted in Fig. 1.
2.2 Outcomes and variables
Our study aimed to solve a binary classification problem with two possible outcomes: CKD and non-CKD. CKD was defined as having an eGFR of less than \( 60\;{\text{mL}}/{\text{min}}/1.73\;{\text{m}}^{2} \). If a sample developed CKD during the 10-year follow-up period, it had the CKD class. We represented the CKD class with 1 and the non-CKD class with 0. We considered 22 variables to train our models. Among these variables, 2 were demographic characteristics: age and sex (Female); 5 were medical history: duration of insulin-dependent diabetes (IDDM_DUR), hypertension (HT), hyperlipidemia (HLIP), current smoking (SMOKE), current drinking (DRINK); 3 were medical treatment information: multiple daily insulin injections (MDI), on anti-hypertensive medication (ANTIHYP), on angiotensin-converting enzyme inhibitors or angiotensin receptor blockers medication (ACEARB); 4 were physical examination data: body mass index (BMI), systolic blood pressure (SBP), diastolic blood pressure (DBP), mean blood pressure (MBP); and 7 were laboratory values: glycated hemoglobin (HBA1C), albumin excretion rate (AER), serum creatinine (eSCR), total cholesterol (CHL), high-density lipoprotein (HDL), low-density lipoprotein (LDL), triglycerides (TRIG). Fourteen features (AGE, IDDM_DUR, BMI, SBP, DBP, MBP, HBA1C, CHOL, HDL, LDL, TRIG, eSCR, AER, eGFR) had numerical values, and other features had binary (yes/no) values. We used 1 to represent yes and 0 to represent no. Figure 2 represents the population distribution of all attributes.
2.3 Data pre-processing
Data cleaning and data pre-processing are vital to clinical model development. Different machine learning algorithms’ performance can vary significantly based on data pre-processing. We applied data augmentation, feature scaling, and outlier detection techniques in this study to process our data.
Our primary data had only 12 missing values in 5 samples. As we had longitudinal data, we replaced the missing values with patients’ next year’s data. Our dataset was imbalanced; among the 1309 samples, 110 samples were CKD-positive. Imbalance data can produce biased results. We applied the SMOTE-Tomek data augmentation technique [33] to balance the dataset. The SMOTE-Tomek approach combines the Synthetic Minority Oversampling Technique (SMOTE) [34] and the Tomek Links [35] under-sampling technique. Here, SMOTE produces artificial data for the minority class, and Tomek Links removes majority class samples most closely related to the minority group. In contrast to random oversampling, which only copies a few random examples from the minority class, SMOTE creates instances based on the distance between each data point and the minority class’s closest neighbors, resulting in new examples that are unique from the minority class’s original data [33]. We used self-written Python code and the imbalanced-learn open-source Python library [36] for data augmentation.
The numerical attributes of our dataset had a vast difference in range and magnitude, and feature scaling could help to increase our machine learning models’ accuracy and convergence speed. We explored three feature scaling techniques: min–max normalization (MinMax), standardization or z-score normalization (StdScal), and robust scaling (RobScal). The min–max normalization scales the numerical values of a feature to a range (usually 0 to 1) based on that feature’s maximum and minimum values. On the other hand, standardization transfers the feature’s value so that the mean becomes zero and the standard deviation becomes one. However, both techniques are sensitive to outliers, as outliers can often influence the sample mean/variance and min–max values negatively. The robust scaling technique changes the median and scales the data according to the quantile range; thus, this technique is less sensitive to outliers than the other two techniques. We used the open-source Python library Scikit-learn [37] to implement all feature scaling techniques.
Our data had outliers in several features. We applied the interquartile range (IQR) method and isolation forest (IF) algorithm [38] for outlier detection and removal. In the IQR method, we kept instances that are in the range of \(1.5 \times \left( {Q3 - Q1} \right)\), where Q1 and Q3 are the first quartiles and second quartiles, respectively. The IF algorithm is a random forest-based outlier detection technique that returns the anomaly score of each sample used in the algorithm [38]. We used the Scikit-learn library [37] for implementing outlier detection algorithms.
However, there is no general guideline for optimal data pre-processing procedures in machine learning-based applications. In this study, we have applied ten different combinations of these data pre-processing techniques to create ten separate datasets (DS-1 to DS-10). All machine learning models were applied to each dataset to determine the best-performing combination for each model.
2.4 Machine learning models development
2.4.1 Machine learning models
Twelve supervised machine learning classification algorithms were applied to develop 10-year CKD risk prediction models for T1D patients. We chose these machine learning algorithms from four categories: linear, nonlinear, ensemble method, and artificial neural network. We used three linear algorithms: logistic regression (LR) [39], linear discriminant analysis (LDA) [40], and Naïve Bayes (NB) [41]. These are classic machine learning algorithms widely used in classification problems. We also used three popular nonlinear algorithms: support vector classifier (SVC) [42], decision tree (DT) [43], and k-nearest neighbors (KNN) [44]. We used the open-sourced Python library Scikit-learn [37] to implement these algorithms.
Ensemble methods combine the predictions of a group of individually trained classifiers (such as decision trees) to classify new data points [45]. This relatively more complex approach usually provides better classification results than a single model [46]. In this study, we applied two bagging ensemble methods: random forest (RF) [47] and extremely randomized tree (ET) [48], and a boosting ensemble method: extreme gradient boosting (XGB) [49]. Scikit-learn open-source Python library [37] was used to implement all ensemble models.
In addition, we explored two artificial neural network models, multi-layer perceptron (MLP) [50] and TabNet [51] to build our prediction models. MLP is a classical neural network model widely used with many applications. TabNet is a relatively new approach that follows deep neural network (DNN) architecture and was developed by the Google AI team in 2019 [51]. TabNet is specially designed to work with tabular data. Although DNNs have shown significant success with audio, video, and image data, their performance was relatively poor with tabular data compared to different decision tree-based ensemble methods [51]. TabNet has a special sequential attention-based architecture, which enables it to outperform tree-based ensemble methods in many applications [51]. We used the PyTorch [52] implementation of the TabNet model, and for the MLP model, we used Scikit-learn [37].
In addition to the individual machine learning models, we proposed a powerful heterogeneous ensemble model (STK) using a stacking generalization approach [53]. The motivation behind adopting an ensemble approach lies in its ability to enhance prediction accuracy by leveraging the diverse strengths of multiple base models. The architecture of a stacking generalization model consists of two or more base models, also referred to as level-0 models, and a meta-model, designated as the level-1 model. The key concept here is that the meta-model learns how to best combine the predictions from the base models to produce an improved final output. This approach follows four algorithm steps.
-
1.
Base model selection: The first step is to choose a set of diverse base models (also known as level-0 models) that will form the foundation of the ensemble.
-
2.
Meta-model selection: Next, choose a meta-model (level-1 model) that will learn how to combine the predictions from the base models to optimize the final prediction.
-
3.
Training the meta-model: During the training phase, utilize the predictions generated by the base models, along with the original outputs (ground truth labels), as meta-data to train the meta-model. The meta-model learns to assign weights to each base model’s prediction to achieve the best combination.
-
4.
Weighted combination: Once the meta-model is trained, it assigns weights to the predictions of the base models. These weights reflect the importance or reliability of each base model’s output. When making predictions for new, unseen samples, the final prediction is determined by combining the outputs of the base models using the learned weights.
For our heterogeneous ensemble model (STK), we strategically selected the best-performing models from various categories, including linear (LDA), nonlinear (KNN), bagging ensemble method (RF), boosting ensemble method (XGB), and artificial neural networks (MLP), as our base models. We employed a logistic regression algorithm as the meta-model to harmonize base models’ predictions and derive an optimized ensemble output. During the training phase, we utilized the predicted outputs from the five base models, alongside the original outputs, as meta-data to train the meta-model. It learned to assign weights to each base model’s prediction, effectively capturing the unique strengths of each model. The final prediction for an unseen sample was then determined based on these learned weights using the following equation:
where
-
\(P\left( {Y = 1{|}X} \right)\) is the probability of the positive class (CKD).
-
\(X\) represents the input features, in our case, outputs of five base models.
-
\(x_{1}\), \(x_{2}\), …, \(x_{5}\) are the output of base model 1, base model 2, …, and base model 5.
-
\(w_{0}\), \(w_{1}\), \(w_{2}\), …, \(w_{5}\) are the learned weights assigned to each base model’s prediction.
-
\(e\) is the base of the natural logarithm.
The overall architecture of our STK model is depicted in Fig. 3. To implement this ensemble approach, we utilized the Scikit-learn open-source machine learning library for Python [37].
2.4.2 Cross-fold validation
We applied repeated stratified k-fold cross-validation from Scikit-learn [37] to train and test our ML models. A single train-test split or even a single run of the k-fold cross-validation procedure may produce a biased estimation of model performance. The repeated stratified k-fold cross-validation yields a more generalized result by doing the stratified cross-validation process [54] more than once and presenting the mean result across all folds from all runs. In our study, we used fivefold cross-validation with five times repetitions. Among the fivefold-split data, fourfolds (80% of the total sample) were used to train all ML models, and the remaining fifthfold (20% of the total sample) was used to evaluate the models. We used stratified k-fold, so the CKD and non-CKD class ratios were similar in every fold.
2.4.3 Hyperparameter optimization
Machine learning models have several parameters that must be learned from the data. We can fit the model parameters by training a model using existing data. However, machine learning models also have a special set of parameters known as hypermeters, which cannot be fit this way. Hyperparameters are used to customize a model and need to be set before training the model. As a result, hyperparameters can greatly influence model performance, and finding appropriate values for hyperparameters is essential. In this study, we applied a grid search approach [55] using the Scikit-learn [37] Python machine learning library to optimize hyperparameters. The list of hyperparameters from different models we selected to optimize is given in Table 1.
Here, the ‘solver’ hyperparameter in the LR algorithm is the optimization algorithm used to find the coefficients of the logistic regression model. Common choices include ‘liblinear,’ ‘lbfgs,’ ‘newton-cg,’ and ‘sag.’ The choice of solver impacts the convergence speed and is often selected based on the size and characteristics of the dataset. For example, ‘liblinear’ uses a coordinate descent algorithm suitable for small datasets, while ‘lbfgs’ uses a quasi-Newton method suitable for larger datasets. The ‘solver’ hyperparameter in LDA, NB has a similar role. The hyperparameter ‘learning rate’ in XGB, MLP, and TabNet controls the step size during the optimization process. It influences how quickly or slowly the model adapts to the training data. A lower learning rate makes the model converge more slowly but can result in better generalization, while a larger learning rate can speed up convergence but may lead to overfitting. The ‘n_estimators’ hyperparameter in RF, XGB, and ET determines the number of decision trees that will be used in the ensemble. Increasing the number of trees can lead to a more powerful model but can also make it more computationally intensive. It is crucial to strike a balance between model performance and computational resources. The ‘Max_depth’ hyperparameter specifies the maximum depth or levels of each decision tree in tree-based ensemble methods. It controls the complexity of individual trees. A shallow tree (low max_depth) is less complex but may underfit the data, while a deep tree (high max_depth) is more complex and may overfit. Setting an appropriate max_depth is crucial for balancing bias and variance. Similarly, other hyperparameters influence model performance in some way and need to be selected appropriately.
2.4.4 Feature selection
Our dataset had 22 features. We tried to optimize the number of features for each machine-learning model using a feature ranking approach. First, we used all features to train an ML model. Then, we ranked the features based on their importance in predicting CKD and created a ranked dataset. After that, we trained the same model using the top-1 features, top-2 features, top-3 features, and so on up to the top 22 features and reported the best-performing model with the minimum number of features. This process was repeated for each model across all datasets, from DS-1 to DS-10, ensuring a comprehensive evaluation. Our objective was to ascertain the most effective combination of essential feature sets, data pre-processing techniques, and machine learning algorithms for accurate CKD risk prediction in T1D patients. Five of our ML models (RF, XGB, ET, DT, and TabNet) had feature-importance methods, which we used for feature ranking while training these models. Other ML models (KNN, SVC, CNB, LDA, LR, MLP, STK) did not have the feature-importance method, and we used the XGB feature ranking algorithm to crate the ranked dataset before training these models. We chose XGB because, in our previous study [27], it provided the best feature ranking result on similar data.
2.5 Statistical analysis and performance metrics
We applied the Shapiro–Wilk test [56] on the dataset to identify numerical features that followed the Gaussian distribution. The homogeneity of variance for both the CKD and non-CKD groups was examined using Levene’s test [53]. We used the open-source Python package SciPy [57] and Pingouin [58] for the Shapiro–Wilk and Levene’s tests, respectively. In both tests, we used a p-value of 0.05. For baseline characteristics of the patients, quantitative features are displayed as means and standard deviation (Sd), while qualitative factors are shown as frequency and percentage (%). We compared these values with CKD and non-CKD groups using the two-sample T-test (quantitative attributes) and Chi-squared test (qualitative attributes) with a p-value 0f 0.05.
We applied several metrics to evaluate the developed ML models’ performance, including specificity (Sp), sensitivity (Sn), precision (Pr), recall (Re), accuracy (Acc), and F1 score. We also applied Cohen’s Kappa (Kappa) [59] and Matthews Correlation Coefficient (MCC) [60] to verify the models’ performance and reliability further. In addition, the area under the receiver operating characteristic (AUROC) curve [61] and the precision-recall (PR) curve [62] of the best-performing model from each algorithm were plotted to compare their performance. The Scikit-learn library [37] was used to calculate all metrics. We used the Python open-source libraries Matplotlib [63] and Seaborn [64] for our graphical representation and plotting. Our data were imbalanced, so we considered the F1 score as the primary evaluation metric.
3 Results
3.1 Baseline characteristics
A total of 1309 patients were included in this study; 620 were females (47.4%), and 689 were males (52.6%). During the ten-year time period, 110 patients developed CKD. Table 2 represents the baseline characteristics of the participants. The average age was 39.8 (+/6.9) years, the average diabetes duration was 18.3 (+/4.9) years, and the average eGFR was \( 108\;{\text{mL}}/{\text{min}}/1.73\;{\text{m}}^{2} \). According to the Shapiro–Wilk test result, only two features (DBP, MBP) had the normal distribution, and the other features had skewness. The population distribution of the features (see Fig. 1b) also represents similar findings. Ten features (AGE, DRINK, ACEARB, HLIP, BMI, SBP, DBP, CHOL, HDL, LDL) exhibit homogeneous variance for the two groups, according to Levene’s test result. HT, DRINK, ACEARB, ANTIHYP, MBP, HBA1C, CHOL, TRIG, eSCR, AER, and eGFR attributes’ values showed a significant difference in CKD and non-CKD groups.
3.2 Result of data pre-processing
We created ten separate datasets using different data pre-processing techniques and used all these datasets to train and test our ML models. The details of each dataset are presented in Table 3. Our primary dataset was imbalanced. Among 1309 samples, only 8.40% (110 samples) had CKD class. After applying the SMOTE-Tomek data augmentation technique, we got a balanced dataset (DS-2) of 2394 samples with 50.08% (1199 samples) of CKD samples. We used outlier removal techniques on the augmented dataset (DS-2). The IQR outlier detection technique removed samples more aggressively than the Isolation Forest (IF) algorithm. The sample size became 1705 and 2154 after applying IQR and IF outlier removal techniques, respectively. Figure 4 shows the impact of outlier removal on numerical attributes.
3.3 Performance of machine learning models
In this study, we applied 12 machine learning algorithms to develop a 10-year CKD risk prediction model for type 1 diabetes patients. The hyperparameters of each model were optimized using grid search (optimized values are given in Supplementary Table 1). All 12 models were applied to all ten datasets, DS-1 to DS-10, created by different pre-processing combinations. Detailed results for these models across the ten datasets are provided in Supplementary Tables 3 to 12. Notably, most models’ performance on the primary dataset, without any pre-processing (DS-1), was suboptimal. Despite achieving over 90% accuracy, this outcome was skewed due to dataset imbalance, rendering the results biased and misleading. None of the models attained an F1 score or Kappa value exceeding 50%, affirming their inadequate performance.
However, employing various data augmentation, outlier detection, and feature scaling methods significantly improved model performance, albeit with varying impact across models. Tree-based models demonstrated robustness against outliers and feature range differences, yielding consistent results across DS-2 to DS-10. Conversely, artificial neural network models (MLP, TabNet) proved sensitive to feature range differences, with improved performance observed with different feature scaling techniques (DS-8, DS-9, DS-10). Linear and nonlinear models also benefited from processed data, displaying enhanced performance. Table 4 outlines the performance of the models that achieved the best results in these ten datasets. Our proposed heterogeneous stacking ensemble model (STK) showed superior results in nearly all datasets, boasting F1 scores ranging from 0.94 to 0.97.
In Table 5, we summarize the performance of all models across all datasets, presenting the best-performing model for each algorithm, the pre-processed dataset, and the number of features (N) used to achieve optimal performance. We consider the F1 score, kappa values, and the number of features to be the primary evaluation metrics for selecting the best model. According to the evaluation metrics, our customized heterogeneous stacking ensemble model (STK) achieved the best performance with an average classification accuracy of 0.97, specificity of 0.98, sensitivity/recall of 0.96, precision of 0.98, F1 score of 0.97, Kappa and MCC score of 0.94, AUROC of 0.99, and Precision-Recall curve of 0.99. MLP and TabNet models came in second and third place with an accuracy and F1 score of 0.95 and 0.94, respectively. LDA, KNN, and RF were the best linear, nonlinear, and ensemble models with an accuracy and F1 score greater than 0.90. In contrast, the performance of NB and DT models was relatively poor compared to other models.
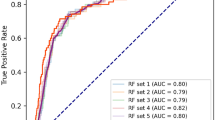
We also generated the Area Under the Receiver Operating Characteristic (AUROC) curve and precision-recall (PR) curve plots for all models across the ten datasets (DS-1 to DS-10), as depicted in Supplementary Figs. 1 to 10. The AUROC evaluates the trade-off between true positive and false positive rates, and the PR curves assess the precision-recall trade-off. AUROC is effective for balanced datasets, while PR curves are more suitable for imbalanced datasets, especially when precision is critical. Initially, models performed poorly on DS-1 based on PR curves but showed improvement on subsequent datasets (DS-2 to DS-10). In Fig. 5, we plot the AUROC and PR curves of models that achieved the best result for each of the ten datasets, and in Fig. 6, we compare the AUROC and PR curves of the best-performing models and the dataset to achieve the result. Both figures follow similar trends, as in Tables 4 and 5. STK achieved the highest AUROC and PR curve values for all datasets except DS-1. Notably, the STK model demonstrated perfect AUROC and PR curve values of 1 in DS-6, DS-7, DS-9, and DS-10. As shown in Fig. 6, other models also exhibited high AUROC and PR curve values (above 90%) using different datasets, except for NB and DT models. Like other performance metrics, these two models also achieved the lowest results here.
The feature ranking and the number of features varied significantly with different data pre-processing techniques and machine learning algorithms. Our heterogeneous stacking ensemble model (STK) achieved the best performance using SMOTE-Tomek data augmentation and Isolation Forest (IF) outlier removal technique (DS-7). This model used the top 20 features ranked by the XGB feature ranking algorithm, see Fig. 7. LR, LDA, SVC, and ET models also achieved their best results using the same dataset.
All tree-based models (DT, RF, ET, XGB) achieved their best performance with DS-2, which was prepared using only the SMOTE-Tomek data augmentation technique. RT, ET, and XGB models used 17 variables, while DT used only 12. However, the DT model’s performance was poor compared to other tree-based models. The KNN model had the best result using DS-5 (pre-processed using the SOMET-Tomek and RobSacl). The ANN models used DS-9, and the NB model used DS-6 to achieve their best performance. A complete list of ranked features used in each best-performing model is given in Supplementary Table 2.
4 Discussion
Chronic kidney disease (CKD) is a significant threat to global public health and is anticipated to affect 10% of the world’s population [2]. The medical treatment for CKD patients can be very expensive [3], and there is always a greater risk of adverse health complications. CKD was the 12th leading cause of global death in 2019 [5]. Type 1 diabetes (T1D) patients are most vulnerable to CKD, and more than 50% of T1D patients run the risk of developing CKD [6]. Diabetes CKD is the most common cause of end-stage renal disease in the West and is also linked to higher cardiovascular risk [7]. In addition, CKD significantly impacts type 1 diabetes patients’ health-related quality of life and healthcare costs [9, 10]. However, CKD is a non-communicable disease, and the risk of CKD can be reduced or prevented through proper medication, diet, and lifestyle [11,12,13]. For this purpose, the identification of T1D patients with a greater risk of developing CKD is vital to ensure proper treatment to avoid the risk.
However, CKD progression can be asymptomatic in most cases [4]. In addition, the nephrologist density is inferior in many countries. In 2016, there were only 0.318 nephrologists per million people in underprivileged countries, according to the International Society of Nephrology Global Kidney Health Atlas (ISN-GKHA) [14]. As a result, identifying CKD risk in T1D is challenging. To overcome these problems, a computer-aided CKD risk prediction model for T1D patients can be a valuable option. Unfortunately, limited research has been conducted in this sector. There are some machine learning (ML)-based CKD risk prediction models for type 2 diabetes (T2D) patients. However, a CKD risk prediction model dedicated to T1D patients would be more appropriate. T2D is mainly a lifestyle disease, whereas T1D is a genetic disorder where patients’ immune system attacks and destroys insulin-producing cells in the pancreas. T1D patients need to take insulin injections to control their blood glucose levels. Unlike type 2 diabetes, lifestyle changes cannot reduce the risk of type 1 diabetes. Moreover, T1D patients have a 1.4–3.0-fold higher risk of CKD than type 2 diabetes patients [21]. Unfortunately, we found limited studies on T1D patients for CKD risk prediction.
In this study, we employed a diverse set of ML models, including linear, nonlinear, bagging, boosting, and deep learning, to predict CKD risk in T1D patients over a 10-year period. The selection of these ML models was driven by their inherent strengths, each offering unique advantages. Logistic regression (LR), Naïve Bayes (NB), and linear discriminant analysis (LDA) provided interpretability, enabling us to understand the influence of individual features on CKD risk. Decision tree (DT) and k-nearest neighbors (KNN) excelled in capturing nonlinear relationships in the data, while the support vector classifier (SVC) offered robustness against noise. Random forest (RF), extremely randomized tree (ET), and extreme gradient boosting (XGB) models leveraged ensemble learning to enhance predictive performance. Multi-layer perceptron (MLP) and TabNet, our deep learning models, demonstrated the ability to handle complex patterns in the data. We evaluated the performance of these models on our dataset to identify the top-performing models. Finally, we proposed a strategic combination of the best-performing models from each category into a customized heterogeneous stacking ensemble model (STK) to leverage the strengths of every category. This ensemble approach was motivated by the desire to harness the complementary strengths of diverse models, ultimately improving prediction accuracy. The grid search method was applied to optimize hyperparameters for all ML models (see Table 3 and Supplementary Table 1).
We used 10-year retrospective longitudinal data of 1375 patients from the Epidemiology of Diabetes Interventions and Complications (EDIC) clinical trial [29, 30]. After applying excluding criteria (see Fig. 1), we selected 1309 samples where 8.40% of samples developed CKD within the 10-year timeframe. We considered 22 features readily available from T1d patients’ routine check-ups and self-assessment (see Table 2) in our study. We tried to solve a binary classification problem, where the outcomes were CKD and non-CKD classes. If a sample developed CKD within ten years, it had the CKD class; otherwise, it had the non-CKD class. For pre-processing the data, we applied ten different combinations of data augmentation, feature scaling, and outlier detection techniques and created ten separate datasets, DS-1 to DS-10 (see Table 3).
We employed a feature ranking approach to determine the optimal number of features for optimal model performance. We began by training a machine learning model using all available features and then ranked the features based on their significance in predicting CKD. Next, we trained and tested the same model using various subsets of the top-ranked features, starting with the top-1 feature and progressing up to the top-22 features. By doing so, we aimed to identify the model with the best performance using the least number of features. The RF, XGB, ET, DT, and TabNet models utilized their own feature importance methods for feature ranking. For models KNN, SVC, CNB, LDA, LR, MLP, and STK, we used the XGB algorithm for feature ranking before training. We observed that the feature ranking and optimal feature number varied depending on the combination of data pre-processing techniques and machine learning algorithms used. However, in most cases, the highest-ranked features were albumin excretion rate (AER), serum creatinine (eSCR), estimated glomerular filtration rate (eGFR), glycated hemoglobin (HBA1C), duration of insulin-dependent diabetes (IDDM_DUR), AGE, and current drinking (DRINK) (refer to Supplementary Table 2 for more details).
We used repeated stratified k-fold cross-validation with fivefold and 5-time repetitions to train and test all ML models. We used specificity, sensitivity/recall, precision, accuracy, F1 score, AUROC curve, and precision-recall curve to evaluate each model’s performance. Cohen’s Kappa (Kappa) and Matthews Correlation Coefficient (MCC) were also used to verify the models’ reliability. We iteratively applied each model across all datasets, from DS-1 to DS-10, to find the appropriate combination. Initially, model performance on the primary dataset (DS-1) was suboptimal but improved significantly by applying different data augmentation, outlier detection, and feature scaling techniques. However, the models’ performance varied on different datasets (refer to Supplementary Tables 3 to 12 for more details). Tree-based models showed robustness against different feature ranges and outliers, while neural network models were sensitive to feature scaling.
Overall, our proposed heterogeneous stacking ensemble model (STK) consistently demonstrated superior performance across nearly all datasets (see Table 4) and achieved the highest result using DS-7 and the top 20 features ranked by the XGB algorithm (see Table 5). Employing SMOTE-Tomek data augmentation and Isolation Forest (IF) outlier removal techniques during data pre-processing contributed to the STK model’s remarkable results. It achieved an average accuracy of 0.97, specificity of 0.98, sensitivity/recall of 0.96, precision of 0.98, F1 score of 0.97, Kappa and MCC score of 0.94, AUROC of 1.00, and Precision-Recall curve of 1.00. This model was closely followed by MLP and TabNet, with an average F1 score of 0.95. LDA and KNN were the best-performing linear and nonlinear models, with an average F1 score of 0.93 and 0.91, respectively. LDA had the best precision value of 1.0, and KNN had the best recall value of 1.0. RF was the best ensemble method with similar results. Five models (STK, LR, LDA, SVC, and ET) achieved their best performance using the SOMET-Tomek data augmentation and Isolation Forest outlier detection technique (DS-7). In comparison, tree-based models showed the most robustness against outliers and achieved the best performance without using feature scaling and outlier detection techniques (DS-2).
In the context of the current body of literature, our research fills a significant gap in the lack of predictive models for the risk of CKD progression in type 1 diabetes patients without previous experience of CKD or other kidney diseases. Prior studies have primarily focused on end-stage renal disease (ESRD) or existing CKD. In addition, these studies used different complex features, making their findings unsuitable for practical use in most cases. For example, studies conducted by Niewczas et al. [22] and Pilemann-Lyberg et al. [23] mainly focused on finding associations between ESRD in T1D patients and different biomarkers or metabolites. They also considered diabetes patients who already had CKD or other kidney disease. In opposition, our study targeted explicitly predicting general CKD risk in T1D patients without any previous kidney disease and included readily available features from T1D patients’ routine check-ups and self-assessments.
Sripada et al. [24] used the random forest algorithm to develop a prediction model for diabetic nephropathy in T1D patients and achieved the best performance with an F1-score of 0.67 and AUC of 0.78. Colombo et al. [25] employed ridge regression to create a model for predicting renal disease progression, achieving a mean squared correlation (Pearson \( r^2 \)) of 0.745. In one of our previous investigations, we created a 90.04 percent accurate nomogram-based CKD prediction model for T1D patients using multivariate logistic regression. However, these models were designed to identify existing CKD rather than predicting future risks. In contrast, our research was designed to predict the risk of developing CKD within a 10-year timeframe. This forward-looking approach addresses the critical need to identify patients at risk before the disease progresses to a severe stage.
Research conducted by Vistisen et al. [28] aimed to develop a robust prediction model for 5-year risk of ESKD in individuals with T1D. The model used ridge regression to demonstrate a C-statistic of 0.888 for end-stage kidney disease (ESKD) risk prediction. However, our model represents a substantial improvement over this approach. While they concentrated on predicting ESKD in T1D patients, our model forecasts the risk of developing CKD within a higher timeframe, allowing for early intervention. Vistisen et al.’s study did not address class imbalance adequately. In contrast, we meticulously addressed class imbalance through data pre-processing strategies. We also employed a wide range of evaluation metrics to ensure a comprehensive understanding of model efficacy. This comprehensive evaluation guarantees a thorough assessment of model performance under various conditions.
Our research introduces a pioneering approach to predicting the risk of CKD in patients with T1D. To our knowledge, this is the first machine learning-based model capable of forecasting CKD risk in T1D patients over a 10-year period, moving beyond the conventional focus on ESRD. Our model stands out by relying exclusively on readily available data from routine check-ups and patient self-assessments, streamlining the predictive process and enabling early interventions. In contrast to previous models that might incorporate complex variables, our approach prioritizes simplicity, widening its applicability across diverse healthcare settings. The innovation extends to the development of an advanced heterogeneous ensemble model, harnessing the strengths of various machine learning techniques to achieve superior predictive performance even with straightforward features. Furthermore, our systematic feature ranking and optimization approach enhanced model efficiency and provided a list of essential features for CKD risk prediction in T1D patients, making our research a valuable contribution to this field. Another major advantage of this study is that we used a dataset from the EDIC trial, which gathers data at 28 EDIC clinic locations throughout the USA and Canada, ensuring a variety of patient types.
However, certain limitations of our study should be acknowledged. The proposed research is solely dedicated to predicting CKD risk type 1 diabetes patients, though type 2 diabetes is more prevalent than type 1 diabetes. In future work, we plan to extend our research to encompass type 2 diabetes patients, leveraging our established methodology to develop tailored CKD risk prediction models. Secondly, we did not have an external validation dataset, which necessitates future testing on different cohorts to establish the model’s generalizability. We aim to collaborate with healthcare institutions to validate and implement our predictive models in real-world clinical settings, fostering their practical utility and impact. In addition, our approach employed a subset of available hyperparameters for optimization, suggesting that further exploration of hyperparameter space could yield even more refined models.
5 Conclusion
In this study, we applied twelve machine learning algorithms to develop 10-year CKD risk prediction models for type 1 diabetes patients. We used data from 1375 type 1 diabetes patients from the Epidemiology of Diabetes Interventions and Complications (EDIC) clinical trial to train our models. The dataset consisted of 22 readily available features, and we applied various data pre-processing techniques, including data augmentation, outlier detection, and feature scaling, to improve the data quality. We evaluated the performance of our machine learning models using repeated stratified k-fold cross-validation with fivefold and five-time repetitions. Specificity, sensitivity, precision, recall, accuracy, F1 score, Cohen’s Kappa (Kappa) value, and Matthews Correlation Coefficient (MCC) were used as evaluation metrics. After performing an extensive evaluation of all models, we found our customized heterogeneous stacking ensemble model (STK) as the best-performing CKD risk prediction model with an average accuracy of 0.97, specificity of 0.98, sensitivity/recall of 0.96, precision of 0.98, F1 score of 0.97, Kappa and MCC score of 0.94, AUROC of 0.99, and Precision-Recall curve of 0.99. The proposed model can be a valuable resource for identifying the risk of developing CKD in T1D patients, particularly those in developing nations with limited access to nephrologists.
Data availability statement
Restrictions apply to the availability of these data. Data were obtained from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (Bethesda, MD, USA) and are available (https://repository.niddk.nih.gov/studies/edic/, accessed on April 16, 2023) with the permission of NIDDK.
References
Khwaja A (2012) KDIGO clinical practice guidelines for acute kidney injury. Nephron Clin Pract 120(4):c179–c184. https://doi.org/10.1159/000339789
Bikbov B et al (2020) Global, regional, and national burden of chronic kidney disease, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. The Lancet 395(10225):709–733. https://doi.org/10.1016/S0140-6736(20)30045-3
Wang V, Vilme H, Maciejewski ML, Boulware LE (2016) The economic burden of chronic kidney disease and end-stage renal disease. Semin Nephrol 36(4):319–330. https://doi.org/10.1016/j.semnephrol.2016.05.008
Centers for Disease Control and Prevention. Chronic Kidney Disease in the United States, 2021. Atlanta, GA: US Department of Health and Human Services, Centers for Disease Control and Prevention; 2021
Cockwell P, Fisher L-A (2020) The global burden of chronic kidney disease. The Lancet 395(10225):662–664. https://doi.org/10.1016/S0140-6736(19)32977-0
Costacou T, Orchard TJ (2018) Cumulative kidney complication risk by 50 years of type 1 diabetes: the effects of sex, age, and calendar year at onset. Diabetes Care. https://doi.org/10.2337/dc17-1118
Saran R et al (2016) US renal data system 2016 annual data report: epidemiology of kidney disease in the United States. Am J Kidney Dis 69(3):2017. https://doi.org/10.1053/j.ajkd.2016.12.004
Valmadrid CT, Klein R, Moss SE, Klein BEK (2000) The risk of cardiovascular disease mortality associated with microalbuminuria and gross proteinuria in persons with older-onset diabetes mellitus. Arch Intern Med 160(8):1093. https://doi.org/10.1001/archinte.160.8.1093
Azmi S, Goh A, Muhammad NA, Tohid H, Rashid MRA (2018) The cost and quality of life of Malaysian type 2 diabetes mellitus patients with chronic kidney disease and anemia. Value Health Reg Issues 15:42–49. https://doi.org/10.1016/j.vhri.2017.06.002
Verberne WR et al (2019) Development of an international standard set of value-based outcome measures for patients with chronic kidney disease: a report of the international consortium for health outcomes measurement (ICHOM) CKD working group. Am J Kidney Dis 73(3):372–384. https://doi.org/10.1053/j.ajkd.2018.10.007
Evangelidis N, Craig J, Bauman A, Manera K, Saglimbene V, Tong A (2019) Lifestyle behaviour change for preventing the progression of chronic kidney disease: a systematic review. BMJ Open 9(10):e031625. https://doi.org/10.1136/bmjopen-2019-031625
Kalantar-Zadeh K, Jafar TH, Nitsch D, Neuen BL, Perkovic V (2021) Chronic kidney disease. The Lancet 398(10302):786–802. https://doi.org/10.1016/S0140-6736(21)00519-5
Kelly JT et al (2021) Modifiable lifestyle factors for primary prevention of CKD: a systematic review and meta-analysis. J Am Soc Nephrol 32(1):239–253. https://doi.org/10.1681/ASN.2020030384
Bello AK et al (2017) Assessment of global kidney health care status. JAMA 317(18):1864. https://doi.org/10.1001/jama.2017.4046
Haque F, Reaz MBI, Chowdhury MEH, Hashim FH, Arsad N, Ali SHM (2021) Diabetic sensorimotor polyneuropathy severity classification using adaptive neuro fuzzy inference system. IEEE Access 9:7618–7631. https://doi.org/10.1109/ACCESS.2020.3048742
Chowdhury MEH et al (2020) Can AI help in screening viral and COVID-19 pneumonia? IEEE Access 8:132665–132676. https://doi.org/10.1109/ACCESS.2020.3010287
Chan L et al (2021) Derivation and validation of a machine learning risk score using biomarker and electronic patient data to predict progression of diabetic kidney disease. Diabetologia 64(7):1504–1515. https://doi.org/10.1007/s00125-021-05444-0
Allen A et al (2022) Prediction of diabetic kidney disease with machine learning algorithms, upon the initial diagnosis of type 2 diabetes mellitus. BMJ Open Diabetes Res Care 10(1):e002560. https://doi.org/10.1136/bmjdrc-2021-002560
Kanda E et al (2022) Machine learning models for prediction of HF and CKD development in early-stage type 2 diabetes patients. Sci Rep 12(1):20012. https://doi.org/10.1038/s41598-022-24562-2
Aspriello SD et al (2011) Diabetes mellitus-associated periodontitis: differences between type 1 and type 2 diabetes mellitus. J Periodontal Res 46(2):164–169. https://doi.org/10.1111/j.1600-0765.2010.01324.x
Kristófi R et al (2021) Cardiovascular and renal disease burden in type 1 compared with type 2 diabetes: a two-country nationwide observational study. Diabetes Care 44(5):1211–1218. https://doi.org/10.2337/dc20-2839
Niewczas MA et al (2017) Circulating modified metabolites and a risk of ESRD in patients with type 1 diabetes and chronic kidney disease. Diabetes Care 40(3):383–390. https://doi.org/10.2337/dc16-0173
Pilemann-Lyberg S et al (2019) Markers of collagen formation and degradation reflect renal function and predict adverse outcomes in patients with type 1 diabetes. Diabetes Care 42(9):1760–1768. https://doi.org/10.2337/dc18-2599
Sripada S, Sripada S, Belapurkar S (2023) 17-LB: diabetic nephropathy prediction with machine-learning models for patients with type 1 diabetes. Diabetes. https://doi.org/10.2337/db23-17-LB
Colombo M et al (2020) Predicting renal disease progression in a large contemporary cohort with type 1 diabetes mellitus. Diabetologia 63(3):636–647. https://doi.org/10.1007/s00125-019-05052-z
Chowdhury NH et al (2022) Nomogram-based chronic kidney disease prediction model for type 1 diabetes mellitus patients using routine pathological data. J Pers Med 12(9):1507. https://doi.org/10.3390/jpm12091507
Chowdhury NH et al (2021) Performance analysis of conventional machine learning algorithms for identification of chronic kidney disease in type 1 diabetes mellitus patients. Diagnostics 11(12):2267. https://doi.org/10.3390/diagnostics11122267
Vistisen D et al (2021) A validated prediction model for end-stage kidney disease in type 1 diabetes. Diabetes Care 44(4):901–907. https://doi.org/10.2337/dc20-2586
The DCCT/EDIC Research Group (2011) Intensive diabetes therapy and glomerular filtration rate in type 1 diabetes. N Engl J Med 365(25):2366–2376. https://doi.org/10.1056/NEJMoa1111732
American Diabetes Association (1999) Epidemiology of Diabetes Interventions and Complications (EDIC), “Long-term renal outcomes of patients with type 1 diabetes mellitus and microalbuminuria: an analysis of the Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications cohort.” Diabetes Care 22(1):99–111. https://doi.org/10.2337/diacare.22.1.99
Perkins BA et al (2019) Risk factors for kidney disease in type 1 diabetes. Diabetes Care 42(5):883–890. https://doi.org/10.2337/dc18-2062
Silveiro SP, Araújo GN, Ferreira MN, Souza FDS, Yamaguchi HM, Camargo EG (2011) Chronic kidney disease epidemiology collaboration (CKD-EPI) equation pronouncedly underestimates glomerular filtration rate in type 2 diabetes: figure 1. Diabetes Care 34(11):2353–2355. https://doi.org/10.2337/dc11-1282
Zeng M, Zou B, Wei F, Liu X, Wang L (2016) Effective prediction of three common diseases by combining SMOTE with Tomek links technique for imbalanced medical data. In: 2016 IEEE international conference of online analysis and computing science (ICOACS). IEEE, pp 225–228. https://doi.org/10.1109/ICOACS.2016.7563084
Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP (2002) SMOTE: synthetic minority over-sampling technique. J Artif Intell Res 16:321–357. https://doi.org/10.1613/jair.953
Tomek I (1976) Two modifications of CNN. IEEE Trans Syst Man Cybern SMC-6(11):769–772. https://doi.org/10.1109/TSMC.1976.4309452
Lema G, Nogueira F, Aridas CK (2017) Imbalanced-learn: a python toolbox to tackle the curse of imbalanced datasets in machine learning. J Mach Learn Res 18(17):1–5. https://doi.org/10.48550/arXiv.1609.06570
Pedregosa F et al (2011) Scikit-learn: machine learning in python. J Mach Learn Res 12(85):2825–2830
Liu FT, Ting KM, Zhou Z-H (2008) Isolation forest. In: 2008 eighth IEEE international conference on data mining. IEEE, pp 413–422. https://doi.org/10.1109/ICDM.2008.17
LaValley MP (2008) Logistic regression. Circulation 117(18):2395–2399. https://doi.org/10.1161/CIRCULATIONAHA.106.682658
Izenman AJ (2013) Linear discriminant analysis. pp 237–280. https://doi.org/10.1007/978-0-387-78189-1_8
Huang Y, Li L (2011) Naive Bayes classification algorithm based on small sample set. In: 2011 IEEE international conference on cloud computing and intelligence systems. IEEE, pp 34–39. https://doi.org/10.1109/CCIS.2011.6045027
Noble WS (2006) What is a support vector machine? Nat Biotechnol 24(12):1565–1567. https://doi.org/10.1038/nbt1206-1565
Safavian SR, Landgrebe D (1991) A survey of decision tree classifier methodology. IEEE Trans Syst Man Cybern 21(3):660–674. https://doi.org/10.1109/21.97458
Peterson L (2009) K-nearest neighbor. Scholarpedia 4(2):1883. https://doi.org/10.4249/scholarpedia.1883
Opitz D, Maclin R (1999) Popular ensemble methods: an empirical study. J Artif Intell Res 11:169–198. https://doi.org/10.1613/jair.614
Ardabili S, Mosavi A, Várkonyi-Kóczy AR (2020) Advances in machine learning modeling reviewing hybrid and ensemble methods, pp 215–227. https://doi.org/10.1007/978-3-030-36841-8_21
Breiman L (2001) Random forests. Mach Learn 45(1):5–32. https://doi.org/10.1023/A:1010933404324
Geurts P, Ernst D, Wehenkel L (2006) Extremely randomized trees. Mach Learn 63(1):3–42. https://doi.org/10.1007/s10994-006-6226-1
Chen T, Guestrin C (2016) Xgboost: a scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining, New York, NY, USA: ACM, pp 785–794. https://doi.org/10.1145/2939672.2939785
Hush DR (1989) Classification with neural networks: a performance analysis. In: IEEE international conference on systems engineering. IEEE, pp 277–280. https://doi.org/10.1109/ICSYSE.1989.48672
Arik SO, Pfister T (2019) TabNet: attentive interpretable tabular learning
Imambi S, Prakash KB, Kanagachidambaresan GR (2021) PyTorch, pp 87–104. https://doi.org/10.1007/978-3-030-57077-4_10
Schultz BB (1985) Levene’s test for relative variation. Syst Biol 34(4):449–456. https://doi.org/10.1093/sysbio/34.4.449
Berrar D (2019) Cross-validation. In: Encyclopedia of bioinformatics and computational biology. Elsevier, pp 542–545. https://doi.org/10.1016/B978-0-12-809633-8.20349-X
Liashchynskyi P, Liashchynskyi P (2019) Grid search, random search, genetic algorithm: a big comparison for NAS
Mudholkar GS, Srivastava DK, Thomas Lin C (1995) Some p-variate adaptations of the shapiro-wilk test of normality. Commun Stat Theory Methods 24(4):953–985. https://doi.org/10.1080/03610929508831533
Virtanen P et al (2020) SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods 17(3):261–272. https://doi.org/10.1038/s41592-019-0686-2
Vallat R (2018) Pingouin: statistics in Python. J Open Source Softw 3(31):1026. https://doi.org/10.21105/joss.01026
Guggenmoos-Holzmann I (1996) The meaning of kappa: Probabilistic concepts of reliability and validity revisited. J Clin Epidemiol 49(7):775–782. https://doi.org/10.1016/0895-4356(96)00011-X
Chicco D, Jurman G (2020) The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genomics 21(1):6. https://doi.org/10.1186/s12864-019-6413-7
Hanley JA, McNeil BJ (1982) The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143(1):29–36. https://doi.org/10.1148/radiology.143.1.7063747
Keilwagen J, Grosse I, Grau J (2014) Area under precision-recall curves for weighted and unweighted data. PLoS ONE 9(3):e92209. https://doi.org/10.1371/journal.pone.0092209
Hunter JD (2007) Matplotlib: A 2D Graphics Environment. Comput Sci Eng 9(3):90–95. https://doi.org/10.1109/MCSE.2007.55
Seaborn Swarm Plot. https://seaborn.pydata.org/generated/seaborn.swarmplot.html. Accessed 26 Dec 2021
Acknowledgements
We would like to thank the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (Bethesda, MD, USA) for providing the Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications (DCCT/EDIC) database. The database is available on request from the NIDDK website. (https://repository.niddk.nih.gov/studies/edic/, accessed on April 16, 2023). This work is partially supported by the ICT Division, Ministry of Posts, Telecommunications and Information Technology, Government of Bangladesh, through the ICT Division fellowship Program (Grant No 56.00.0000.052.33.004.22-0S) and the International Centre for Theoretical Physics (ICTP), Italy, through the Affiliated Centers Program and STEP Program.
Funding
This research is supported by a grant from the Ministry of Higher Education (KPT), Malaysia, Grant No. FRGS/1/2021/TK0/UKM/01/4 and Universiti Kebangsaan Malaysia under Grant No. DIP-2020-004.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chowdhury, M.N.H., Reaz, M.B.I., Ali, S.H.M. et al. Machine learning algorithms for predicting the risk of chronic kidney disease in type 1 diabetes patients: a retrospective longitudinal study. Neural Comput & Applic 36, 16545–16565 (2024). https://doi.org/10.1007/s00521-024-09959-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-024-09959-6