Abstract
Recently, numerous research works in retinal-structure analysis have been performed to analyze retinal images for diagnosing and preventing ocular diseases such as diabetic retinopathy, which is the first most common causes of vision loss in the world. In this paper, an algorithm for vessel detection in fundus images is employed. First, a denoising process using the noise-estimation-based anisotropic diffusion technique is applied to restore connected vessel lines in a retinal image and eliminate noisy lines. Next, a multi-scale line-tracking algorithm is implemented to detect all the blood vessels having similar dimensions at a selected scale. An openly available dataset, called “the STARE Project’s dataset,” has been firstly utilized to evaluate the accuracy of the proposed method. Accordingly, our experimental results, performed on the STARE dataset, depict a maximum average accuracy of around 93.88%. Then, an experimental evaluation on another dataset, named DRIVE database, demonstrates a satisfactory performance of the proposed technique, where the maximum average accuracy rate of 93.89% is achieved.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Recently, numerous research works in the retinal-structure analysis has been performed to analyze retinal images for diagnosing and preventing ocular diseases, such as diabetic retinopathy (DR), which is the most common cause of vision loss in this world.
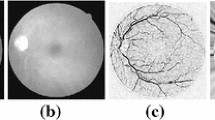
These approaches are also employed in the diagnosis of certain systematic disorders such as several cardiovascular diseases, hypertensive retinopathy and risk of stroke. Actually, the retinal segmentation techniques assist the ophthalmologists and the eye-pathology experts by minimizing the time required in eye screening, reducing the costs and providing the most efficient disease treatment and management systems [1–4]. Earlier, eye-structure detection methods were defined in terms of structures that they would segment [5–7]. For example, the blood vessel extraction was classified into two sets, such as pixel-processing-based methods and vessel–tracking-based methods [8], and the optic disk segmentation techniques included the deformable-based and shape-based template matching techniques [9]. However, the use of these techniques in retinal disease diagnosis has been enhanced greatly in the last few decades. Quantitative analysis of the retinal structures (vessels, optic disk, fovea, macula and optical nerve fiber layer) may be complex and needs to have attention and time. Figure 1 illustrates these main structures. Figure 1a shows the optic disk and the fovea (orange circles). The bright area in which many vessels converge is the optic disk, the connection between the brain and the eye. The dark area in the image is the fovea which is in charge of creating an image from the incident light. Figure 1b presents a manual segmentation of a retinal vessel network that is in charge of the blood and the oxygen supply to the retina. The retinal blood vessel network can inform us abnormal changes induced by eye disorders and first symptoms of some systemic diseases like the DR (Fig. 2). In other words, the information about the vessel morphologic changes, the branching pattern, the length, the width and the tortuosity gives us data on both the abnormal changes and the degrees of disease severity [10–13]. Thus, the automatic detection of a retinal vessel network becomes the most important issue in the detection of the DR. A lot of works have been put forward to detect the 2D complex vessel network [14–16]. The retinal vessel detection is a specific line detection problem, so several works of vessel segmentation are based on the line detection operator.
An accurate vessel detection algorithm in retinal images is generally a complex task because of several reasons. The difficulties include the low contrast of images, the anatomical variability of vessels and the presence of noise due to the complex acquisition system. Aiming to solve those pre-mentioned problems, generally there exist two stages in the retinal vessel detection process: preprocessing and vessel extraction. Some approaches are only based on the second step [17, 18], which is not desirable because the obtained experimental results contain false-positive vessel pixels caused by the existing noise in the original image. Therefore, the preprocessing stage is often necessary before extracting any relevant information from an image. The aim of this stage is to improve the visual appearance of images by reducing noise without affecting small vessels and highlighting edges within the image. The main technique used in preprocessing is filtering. The accuracy of the vessel segmentation process increases by the filtering capacity. The captured images are often corrupted by various noise, which is a random variation in intensity values. The most common kinds of noise found in medical imaging are the additive noise, called also the Gaussian noise, the Speckle (or multiplicative) noise and the Shot or Poisson noise. In the literature, there exist numerous techniques that mainly aim to filter out the noise from images. Those filtering techniques can be divided into two categories, linear and nonlinear. The linear filters, such as the Gaussian and Winner filters, are especially good for removing the Gaussian noise and in several cases for the other kinds of noise as well. The linear filter updates the value of a pixel by weighted the sum of its neighborhood in successive windows. The main problem with the linear filters is that they tend to degrade the image details and to blur the sharp discontinuities in an image. Instead of linear filters, the nonlinear filters are very effective in removing noise while retaining image details. They remove especially certain types of noise that are not additive such as the Speckle noise. The anisotropic diffusion of Perona and Malik (PMAD) is the most powerful nonlinear filter, which is a multi-scale smoothing and edge detection scheme [19]. Several versions of this filter have been developed to denoise monochrome images corrupted especially by the Speckle noise such as the speckle reducing anisotropic diffusion (SRAD) [20], the Flux-based anisotropic diffusion (FBAD) [21] and the detail preserving anisotropic diffusion (DPAD) [22]. The problem of such methods is that the noise type and its characteristics are assumed to be known in advance and constant over the whole image, which is not actually valid in practical circumstances. Hence, these approaches cannot give an optimal image quality for images corrupted by other unknown noise models. To improve the effectiveness of these filtering algorithms, it is required to adjust their parameters according to an accurate noise model.
In this work, a retinal vessel extraction process is implemented, which is based on two stages. The first stage is to denoise a retinal image using a powerful version of the anisotropic diffusion technique, named the Adaptive noise-reducing anisotropic diffusion filter (ANRAD). This filter is able to determine the accurate model of noise present in the retinal image, through which its parameters are adjusted to improve its efficacy. The second stage presents a multi-scale line-tracking algorithm to detect all the blood vessels having similar dimensions at a selected scale.
The present paper is structured as follows. Section 2 introduces an overview of the existing literature reviews on retinal vessel detection. In Sect. 3, the segmentation method is elaborated in detail, where it is divided in two basic steps. The first one presents the preprocessing task, and the second one describes the vessel extraction process. Some of our experiment results are addressed in Sect. 4. In Sect. 5, a general conclusion is given about the proposed method.
2 Related works
Several methodologies have been developed to segment retinal vessels and evaluated in the literature [5–7, 10, 13, 23–26]. These techniques are grouped into two different categories. The first one consists of the pixel-processing based approaches, and the second one concerns tracking-based approaches. The pixel-processing-based approaches extract the vessels in two steps. First, the enhancement of the vessel structures is effected using detection processes such as morphological preprocessing approaches and adaptive filters. The second stage is the vessel shape recognition by a thinning operation or by branch point detection to identify each pixel as either a vessel point or a background one. All of these methods treat the image pixel-by-pixel applying a series of processing operations on each pixel. Some of them use neutral networks and time frequency analysis for prediction of individual pixels in the image if they are vessel or background. Various specific processing pixel operations are used by several works in the literature. In [27], the extraction of retinal vessels was carried out using the Matched filter, which was a simple operator that combined optical and spatial properties of objects to recognize. In this work, the gray level profile of retinal vessel cross section was assumed as a Gaussian shaped curve. Then, the authors constructed 12 different kernels and employed them to search vessel segments along all possible directions. After that, at each pixel, only the maximum of their responses was retained. Finally, a thresholding algorithm was applied on the resulting image response in order to eliminate false detections and to obtain a binary representation of the retinal network. In [28], another version of matched filter was presented, which used local and global thresholding properties to segment the retinal vasculature. Some algorithms based on image ridges using differential filters have been utilized to detect vessels from retina images. In [29], the authors employed a vessel centerline detection approach and multi-directional morphological bit plane slicing to extract the blood vessel network from the background of the retinal image. First, the blood vessel centerlines were detected, in four orientations, using the first order derivative of the Gaussian filter. The final response of the vessel centerlines was given by combining the obtained four responses. Then, two maps were obtained for the shape and orientation of vessels using a multi-directional morphological top-hat operator combined with directional structuring elements followed by bit plane slicing of the vessel’s enhanced gray level image. The resulting centerline images were combined with these maps to provide the vasculature tree. The second category required locating, manually or automatically, the pixels belonging to a vessel for training the vessels using measures of some local image properties. Most of the approaches of this category have been based on the Gaussian functions as a model for the vessel profile. In [30], the authors developed an algorithm that introduced an effective approach based on a priori knowledge of the blood vessel morphology and local neighborhood informations of the vessel’s segment position, orientation and width. In [31], the authors proposed an algorithm that used a model of twin Gaussian functions for the quantification of vessel diameters in retinal images, and then they described the variation of the vessel diameter in the direction of the axe vessel using a tracking technique based on the parameters of modeled intensity profiles through a cross section of every vessel. In [32], a tracking process of the retinal vasculature network was suggested. In this work, a second-order derivative Gaussian matched filter was employed to determine the location of the center line and width of a vessel. Beside this, the Kalman filter was used to accurately estimate the next possible vessel location. In [33], an accurate tracing process of retinal vasculature was suggested. This method automatically detected the seed-tracking points; then the vessel boundaries were explored by a set of 2D correlation kernels. These points were used to trace the vasculature recursively.
3 Proposed system
Figure 3 shows the block diagram of the proposed methodology for the blood vessels’ segmentation in RGB color fundus images. There are two main steps: the preprocessing process followed by the extraction vessel process. The preprocessing process includes the removal of image noise using the ANRAD filter, and then the RGB image is converted to grayscale image. The ANRAD introduces an automatic RGB noise model estimator in a partial differential equation system similar to the SRAD diffusion. The estimated noise model, also called the noise level function (NLF), is considered as a function of standard deviation depending on the RGB pixel intensities in the image. The extraction vessel process using a multi-scale line-tracking algorithm is employed to detect the vessels in the image, firstly the vessels having similar dimensions at a selected scale r. The extraction of the vessels having a ray r is achieved by computing the similarity measure to a vessel following the steps mentioned below (see Fig. 3):
-
Determine the local orientations, at each pixel x in the image, using the hessian matrix.
-
Compute the gradient image at each pixel x.
-
Compute the two gradient images using a bilinear interpolation at a distance ±r from x in the orthogonal direction of the vessel axis.
-
Retain the maximum of the two computed gradient quantities.
Thus, several responses of a vessels’ tree for several scales are obtained. Then, the final vessel tree is achieved by retaining the maximum of all obtained vessel response maps, called the multi-scale vesselness response. A detailed description of the proposed methodology is presented below.
4 Materials and proposed segmentation methodology
4.1 Image enhancement
The quality of the retinal images is not always good enough to use if one wishes to detect automatically the 2D complex retinal vessel network [34, 35]. These images present a good amount of noise that makes the vessel detection a difficult task. To reduce noise in digital images, most algorithms in the literature have used the noise model with some assumptions such as additive, identically distributed throughout the image and independent of the RGB color data. These approaches can not effectively extract the “true” data (or its best approximation) from the noisy images since an accurate noise model estimator from a single retinal image is required to enhance the denoising task.
4.1.1 Noise model in retinal images
In the Charge-Coupled Device (CCD)-based fundus camera, the light filaments passed through the lens are translated to electrical charge in the CCD sensor. After that, this amount of charge is treated and digitally enhanced to become a binary image. Generally, the images acquired by the CCD digital camera systems are characterized by good quality. However, these images are not completely free from some kind of distortions/artifacts. In [36, 37], the authors mentioned that there existed mainly four noise types such as the dark current noise, the shot noise and the amplifier and quantization noise, noted, respectively, as Nc, Ns and Nq. Nq was the minimal noise connected to the imaging system and is neglected in this work.
Taking into account the previously presented noise sources (Nc and Ns), the real CCD sensor output IN can be modeled as:
where L is the image irradiance, Ns defines a multiplicative noise modeled by a standard Gaussian distribution having a mean equal to unity and a variance L·σ 2s , and Nc represents an additive Gaussian distribution noise that has a zero mean and a relative variance σ 2c [38]. Thus, the appropriate noise model indicates a multiplicative noise component that is defined as a function of image irradiance (or brightness).
In the ideal imaging system, the image radiance is recorded at the image sensor of a camera as an image irradiance L. This irradiance image is then converted according to the radiometric response functions of a camera f into an image intensity I which is the output signal of the camera. In general, it is a nonlinear mathematical function of the image irradiance. Hence, the ideal imaging system is expressed by a one-variable mathematical function of the image irradiance that may be expressed as follows:
In a reverse process of (Eq. 2), the measured intensities are transformed into irradiance measurements. As a consequence, (1) can be written as:
Till now, the noise introduced in the output CCD camera is white (uncolored). Indeed, the raw data of the image sensor are treated by various image processing steps, including demosaicing, gamma and color corrections, JPEG compression etc., which implies that the noise properties in the final output deviate significantly from the most used noise model, which is uncolored (or white), uniform and either additive or multiplicative. Therefore, to completely know the accurate noise model, it is necessary to estimate color noise model instead the white noise. The appropriate noise model of Eq. (3) can be written as follows [39, 40]:
where IE[.] defines the expected value of a discrete random variable, and IN and I are, respectively, the noisy output image and the noise-free output image. The proposed Noise level function (NLF) (or noise variance model) describes a nonlinear function of the intensity depending on the imaging system parameters.
4.1.2 Noise model estimation process
In the suggested work, an iterative noise model estimation process is introduced to determine the accuracy NLF (or ∑2) in retinal images [41, 42]. The principal steps of this process are summarized and presented in Fig. 4. To compute the NLF, a one-channel image is considered in this paragraph (assuming that the similar described operations are performed for each RGB component of color image). The noise modeling is performed in two parts. In the first part, a database containing all the NLFs of the existing CCD cameras is created. Then, by applying the Principal component analysis (PCA) on the database, a general form of the approximation model of the NLF is given and expressed as follows:
where \(\overline{{\sum\nolimits^{2} }}\) and ωη are, respectively, the mean and the eigenvectors of the NLF obtained by the PCA. α1 ,…, αm are unknown parameters of the noise model with m being the retained number of principal components. In the second part (see Fig. 4), the original noisy image is denoised (Fig. 4a) by a low-pass filter, and then the smoothed image is segmented into homogeneous regions using the k-means clustering algorithm [43]. Next, the mean of noise-free signal and the noise variance for each region are computed and plotted on a graphe to form a scatter plot of samples of noise variances on the estimated noise-free signals (Fig. 4b, c). The x-axis of image intensity is discretized into κ uniform intervals, and at each one, the region having the minimum noise variance is taken (the blue stars in Fig. 4d). The lower envelope drawn below the sample points in the scatter plot is the estimated NLF curve (Fig. 4e). However, the estimated noise variance of each region in the image is an overestimate of the real noise level because it may contain a signal; thus the drawn curve is an upper bound of the real NLF.
To determine the real noise model, the goal is to infer the accurate NLF model from the lower envelope of the samples. In other words, it is sufficient to find the unknown αη in the expression of Eq. (5). To resolve that, an inference problem in a probabilistic framework was formulated. Using the maximum likelihood estimator (MLE), the best NLF approximation is given (Fig. 4f).
4.1.3 Noise-estimation-based anisotropic diffusion
To improve the retinal image quality, a method called the Adaptive Noise-Reducing Anisotropic Diffusion (ANRAD) [41] is employed in this work. The diagram of this method is summarized in Fig. 5.
It is an improved algorithm of the Speckle Reducing Anisotropic Diffusion (SRAD) method [20], where the described iterative accurate automatic RGB noise model estimator in the last paragraph is introduced in its partial differential equation (see Fig. 5). To compute the noise variance (or noise model), the SRAD needs to take a homogeneous region from the original image and takes the average of all the local variances as NLFs. Although it is not hard for a user to select the homogeneous region, it is non-trivial for the computer machine. Also, the noise level is uniform and not depends neither on the intensities and nor on the color in the image. To deal with these inconveniences, the ANRAD employs the explained automatic algorithm to estimate the accurate noise model. The NLF is a function of noise variance depending on the RGB pixel intensities in the image instead of the constant value of the noise variance used in the SRAD filter. Its general expression can be written as follows:
where ∇I and div are, respectively, the gradient and the divergence operators, Δt defines the step time, Ii,j;t and i, j are, respectively, the discrete image and the spatial coordinates of a pixel x in the image, wi,j defines the sliding window centered at the current pixel, and |wi,j| is its size. wi,j is used to compute the local statistics for each pixel, and it is equal to 5 × 5. ϕ(…) is the diffusion function expressed by:
with
and
where ci,j;t defines the image instantaneous coefficient of variation of the image which has the role of the identification of edges and homogeneous regions in an image, c 2n (i, j; t) controls the amount of smoothing applied to the image—called the instantaneous coefficient of variation of the noise, ∑2(I,i,j;t) is the noise level function, and Var(I,i,j;t) and \(\overline{I}^{2}_{{ {i,j;t} }}\) are, respectively, the local variance and the mean values in the image. This filter is adapted to denoise images, containing a mixed color noise produced by nowadays CCD digital camera, and deals with the data adaptive to the amount of color noise at each pixel in the image.
4.2 Vessel extraction process
4.2.1 Extraction of local orientations
In various algorithms, such as [44] and [45], the intensity profile of the cross section of a retinal vessel is assumed to be similar to a Gaussian distribution (Fig. 6b). Also, in [44, 46], the authors assumed that the intensity values along the retinal vessels do not change much. These common assumptions are employed in the present work.
Consider a point M(i, j) of a surface S, belonging to a retinal blood vessel, defined by the vessel pixel intensity k = I(i; j) in the global-coordinate system (i, j, k). Let this function k be at least two times continuously differentiable at i and j. We choose an orthonormal local landmark, an original M, containing the plane tangent to S at M (plane carried by the unit vector normal to the surface) (Fig. 7). With this construction of landmark, the derivatives of the first order are 0. There are only derivatives of the second order (and higher). We assume that they did not cancel the point M. In developing the Taylor series of u of the second order, we get:
where the derivatives \(\frac{{\partial^{2} I}}{{\partial i^{2} }}\), \(\frac{{\partial^{2} I}}{{\partial j^{2} }}\), \(\frac{{\partial^{2} I}}{\partial ij}\) are computed at the point M. The simplest possible masks for computing the second-order partial derivatives on a sampled image are shown in Fig. 8.
Equation (6) represents a conic whose center is M (or summit M in the parabola case). The conical coefficients are the elements of the Hessian matrix:
The Hessian matrix H is symmetric. It has thus two real eigenvalues, λ1 and λ2, corresponding to the two principal curvatures of the surface, and two eigenvectors, v1 and v2, corresponding to the two main directions. This matrix characterizes the form of the conical: hyperbolic paraboloid, elliptic paraboloid, paraboloid cylindrical …. Therefore, it defines the nature of the stationary point M, which can be a peak point, a ridge, a saddle ridge, a flat, minimal surface, a pit, a valley or a saddle valley. The quantity W = λ1 + λ2 is often used to identify the surface patches [47, 48]:
-
If W < 0, the stationary point is a peack, ridge or saddle ridge.
-
If W = 0, the stationary point belongs to a flat or a minimal surface.
-
If W > 0, the stationary point is a pit, a valley or a saddle valley.
Several works on blood vessel segmentation like [49] and [50] have used the eigenvalues of H to compute the “vesselness” measure (or the similarity to a line) of a pixel. Table 1 summarizes the possible relations between the eigenvalues of H to detect the structures of various geometrical forms. In particular, a pixel vessel has a small λ2 and a large positive λ1. Its corresponding eigenvectors indicate singular directions: v2 gives the direction along the vessel, in which the intensities change a little, and v1 is its orthogonal (see Fig. 9).
From left-to-right, top-to-bottom: a part from green channel retinal image; map of eigenvectors of Hessian matrix; zoom on map of eigenvectors: Green vectors describing vessel directions and blue vectors their orthogonals; map of greatest eigenvalue of Hessian matrix; map of weak eigenvalues; denoised image with proposed method
4.2.2 Multi-scaled analysis-based vesselness
-
Multi-scale analysis.
The original motivation for developing a notion of scale-space representation of a given data set comes from the fact that the existing objects in this world are composed of different shapes over certain ranges of scale and may so be identified in several different ways according to the chosen scale of observation. To better understand this, the concept of a branch of a tree is taken as a good example, where it makes sense only from a few centimeters to no more than a few meters. At the nanometer or kilometer scales, the tree concept is meaningless. At those scale levels, it is more pertinent to talk, respectively, about molecules that constitute the tree leaves and the forest in which the tree grows. In image analysis, to calculate any kind of representation from image data, it is necessary to detect information using some operators according to the data. It means that the information that can be extracted is widely determined using both the size of the structures of the image and the used size of the operators. Then, some basic questions will be proposed, such as: What is the type of operators to use? How large should they be? And where can we apply them? Firstly, the scale-space representation of 1D signals was introduced by Witkin [51], and then by Koenderink [52], for digital images. The idea behind the concept of scale space is intended to represent the input data/signal at multiple levels or scales, in such a way that fine scales of structures are removed. In addition, a continuous scale parameter is associated to all the scale levels in the multi-scale representation [51–56].
For any two-dimensional data I:IR 2 → IR, their scale-space representation T:IR 2 × IR → IR+ is given by:
and
For some family G:IR 2 × IR+ → IR of convolution kernels, t is the continuous scale parameter. Besides, the Gaussian kernel constitutes a good canonical choice for producing a scale-space representation. A demonstration of this unicity can be found in [52] or with more details in [51]. In addition, the scale-space family of any signal is defined as the solution of the heat equation. The Gaussian kernel is chosen as the unique scale-space kernel to change the scale. Based on this concept, the scale-space derivatives at any scale in a scale space may be obtained directly by differentiating the scale-space representation or simply by convolving the input data with the differentiation of the Gaussian operators:
where β = (β1, β2,…, βN) constitutes a multi-index notation for the derivative operator \(\partial_{{x^{\beta } }} = \partial_{{x_{1}^{{\beta_{1} }} }} , \ldots ,\partial_{{x_{N}^{{\beta^{N} }} }}\). More generally, these Gaussian operators are used as a basis to solve a large number of visual tasks, such as motion estimation, feature detection, stereo matching and feature classification.
-
Vessel tree detection.
The general idea of multi-scale analysis-based methods is to define a scale range which can be defined from tmin and tmax (corresponding to σmin and σmax). Then, it is discretized utilizing a log scale to have precision for the low scales and finally calculate a response map for all the scales from the original input image [57]. In the case of retinal images, retinal vessels appear in different scales from thin to large (Fig. 10). For this, the minimal and maximal vessel radii to detect are determined by the user. Then, computing the response map for one single scale needs different stages (see Fig. 11). First of all, vessel pixels should be preselected using the analysis by the Hessian matrix’s eigenvalues, as mentioned above. These pixels have to be close to the center axis of the vessel. Afterward, the vesselness response for every preselected pixel is computed at a chosen scale σ. This response needs to use the eigenvectors of the Hessian matrix in order to define for all the pixels of the image the orientation Θ(σ, x), which is orthogonal to the vessel center axis that goes through the current point (the point M in Fig. 11). From this current point M and in this direction Θ, both points are located at an equal distance r. These points are noted by M1 and M2 in Fig. 12. The response function Γσ(I) at the current point M is defined as the maximum of both absolute values of the first derivative of the image intensity in the direction Θ among these two points. Let
and
Then, the similarity measure to a vessel can be expressed as:
where \(\overrightarrow {d}\) presents the unitary vector in the direction of Θ, and \(\overrightarrow {d} = \overrightarrow {v}_{1}\) . Tx (.,σ) defines the image gradient at the chosen scale σ, which is obtained by convolving the original image intensity with the first Gaussian derivative function, with σ being its standard deviation. The gradient vector Tx at the point \((x + \sigma\cdot\overrightarrow {d} )\) is given by a bilinear interpolation.
If one winches to detect a retinal vessel having a radius r, a scale t is perceived for this latter. Therefore, the scales’ correspondents for each vessel radius are used for a multi-scale vesselness response (see Fig. 12). For a given scalet, a response vesselness image Γt(I) is computed from the initial image.
Accordingly, different responses for different scales are obtained, and the multi-scale vesselness response for the whole image Γmulti(I) is defined as the maximal response over all scales. For a given pixel x and a scale range [tmin, tmax]:
Γmulti(x) is interpreted as an indicator whose xψ belongs to a vessel and Γt(x) as an indicator whose ψx belongs to a vessel having a radius t.
4.2.3 Bilinear interpolation
Interpolation is a technique that estimates an approximate continuous value of a function. Many different interpolation techniques, including nearest neighbor, bicubic, bilinear, are available for application in several tools for image processing like Photoshop [58]. Among the interpolation applications, we can cite: image resampling, image zooming, image scaling, image resolution enhancement, sub-pixel image registration, and correcting spatial distortions, and a lot more [59, 60]. In this work, bilinear interpolation is used to compute the gradient vector Tx at the point \((x + \sigma\cdot\overrightarrow {d} )\), which is a resampling method that takes the distance-weighted average of the four neighborhood pixels values to estimate a new pixel one.
The principle is illustrated in Fig. 13, where it uses interpolation in both horizontal and vertical directions, which leads to give a better result than the nearest neighbor method and takes less computation time compared to the bicubic method. Let (x, y) be the point whose unknown intensity value I′ is to be found. It is assumed that the intensity values of its four nearest neighbors \(Q_{11} \, = \, I_{(i, \, j)}, \, Q_{12} \, = \, I_{(i \, + \, 1, \, j)}, \, Q_{21} \, = \, I_{(i, \, j \, + \, 1)}, \,\) and \(Q_{22} \, = \, I_{(i + 1, \, j + 1)}\,\) are known in advance. Also, it is supposed that the area of the square formed around (i',j') is 1.
The point in the position (i',j') is used to divide the square into four areas. Each area defines the weight of its nearest pixel. For instance, a.b defines the weight of the pixel I(i, j), and so forth. As a result, the new value of the pixel (i',j') is given by a weighted average of the four nearest pixel values and is written as follows:
5 Results
The proposed approach is evaluated using two publicly available database of real retinal images [61–63]. The input parameters \(r_{ \hbox{min} } ,r_{\hbox{max} }\), which are necessary to measure the performance of the method, are the smaller and larger vessel radii that want to detect from the original image with \(r_{\hbox{min} } = 1.25\) and \(r_{\hbox{max} } = 7\). The range between these two parameters is discretized by log scale on 4 scales. Also, \(i\,{\text{ter }}\) is used as the number of iterations of the ANRAD.
In the first experiment, the preprocessing task is applied to remove noise in the image. To investigate the efficacy of the filtering process, some experiments are done. Firstly, the ANRAD is tested on a synthetic image corrupted with additive noise, and then in case of real noise in retinal images. The numerical accuracy is evaluated using two parameters: the SNR rate [64], where the higher the SNR is the better the result is, and the MSSIM [65]. The latter is utilized to give the similarity between the noise-free image and the processed data, which belongs to the range [0, 1].
Figure 14 presents the filtering results using the ANRAD and SRAD filters. It shows that the recovered image by applying the ANRAD filter has a better visual quality in comparison with the SRAD filter. The ANRAD produces a smoother result, where the edges are well preserved and the contrast is improved better. The parameters of each filter are mentioned in Table 2. For the ANRAD, the smoothing step time is set to 0.2, and the denoising process runs adaptively with 100 iterations. According to the obtained results, the ANRAD shows better results for both the SNR and the MSSIM. It presents a good performance compared to the SRAD filter, since it has the greatest SNR value, which is equal to 76.7737, and the highest MSSIM score (close to 1), which is equal to 0.983.
The proposed filtering process with the ANRAD filter is developed for retinal images, which are corrupted with color signal-dependent noise. The ANRAD uses a general NLF as an input parameter instead a constant variance value like in the SRAD filter. The utilized noise model is three continuous functions describing the noise variance as a function of local intensity in the whole image for each color channel. Figures 15 and 16 present two color retinal images and their three color corresponding model noises (green, red and blue channels). Each curve of the noise model describes the relationship between the intensities’ values and their corresponding values of the noise level in the image. Furthermore, there are spatial correlations introduced by the effect of three color components of the image. Figure 16 indicates that the estimated NLFs are significantly modeled even though the color distribution does not span the full range of intensities, which explains the ability of the method to estimate the NLF beyond the observed image intensities. To show the efficacy of the proposed denoising process, Fig. 17 presents an example, where the green scale is selected to show the filtering results because it presents a higher contrast between the vessels and the retinal background. From the results shown in Fig. 17c, e, f, the thin small vessel at the bottom right hand side corner of the image in Fig. 17b is markedly altered or lost. On the other side, from Fig. 17d, it is noticeable that the proposed method is much more capable to enhance out the flat areas, keep the thin vessels and preserve the contours better than the other methods. Thus, the ANRAD approach is able to reduce the noise and at the same time to preserve very well the major region boundaries and the thin details.
a Green channel image of original retinal image in Fig. 5a; b part of original image (a); c result with PMAD method (Thres = 15; iter = 30; Δt = 0.05); d with ANRAD (iter = 30; Δt = 0.2); e with SRAD (iter = 30; Δt = 0.2); f and with DPAD (iter = 30; Δt = 0.2)
The second experiment is the vessel segmentation task and it is applied to detect all vessels in the retinal image. In the retinal blood vessel segmentation, the results are generally evaluated over a pixel-based classification. Each pixel in an image is classified into vessel or non-vessel. Four different classes of pixels should be identified to achieve a good classification: the True Positive (TP) and the True Negative (TN), when a pixel in the output image is correctly detected as a vessel or non-vessel, and the False Negative (FN) and the False Positive (FP), which are two misclassification quantities. The FN appears when a pixel in a vessel is detected in the non-vessel region and the FP when a non-vessel pixel is detected as a vessel pixel. From these classifications, there are two widely known measurements used to evaluate the performance of the proposed vessel segmentation process: the TP Rate (TPR) (or sensitivity) and the FP Rate (FPR) [66–70]. These performance measures are defined as follows:
Another measure is used, which is the Maximum average accuracy (MAA), where the maximal accuracy is determined by varying a rounding threshold from 0 to 1 to obtain a binary image that matches the vessel segmentation image to a high level. The accuracy term is defined as the ratio of the sum of the number of pixels correctly classified as a background and as a foreground divided by the number of all pixels in the image:
where P and N define the total number of vessel and non-vessel pixels in the segmented image.
5.1 STARE database
In this section, the suggested method is assessed firstly on a publicly available database of real retinal images, known as the STARE Project database [61]. It contains twenty fundus color images. Ten of them are from healthy eyes and the others from unhealthy ones. These images are captured by a special camera. They are digitized on 24 bits for a grayscale resolution and have a size of \(7 0 0 { } \times 6 0 5 { }\) pixels. This dataset provides two groups of hand-labeled segmentations that are segmented with hand by specialists. Each of these images is adapted as “a ground truth” to evaluate our approach. To demonstrate the efficiency of the segmentation process with the filtering task, Fig. 18 provides the segmentation results before and after the application of the ANRAD filter. This figure shows the improvements rendered by the ANRAD model, where it maintains efficiently the vessels while making the background more homogeneous. Therefore, the ANRAD filter is a principle step before the segmentation process since it preserves the needed information. Figure 19 depicts the segmented images and the manually labeled images for the STARE dataset.
ANRAD-filter effect on blood vessel segmentation process from left-to-right, top-to-bottom: Color retinal image; Sub-image of the original retinal image; Hand-labeled “truth” images of first and second eye specialists; Segmentation result without ANRAD filter; Segmentation result with ANRAD filter N = 10
To better evaluate the proposed method, the experiment results on 20 images from the STARE dataset are presented in Table 3. In Table 4, the current approach is compared versus the most recent approaches in terms of TPR, FPR and MAA. In Table 4, the performance measurements of some methods are reported from their papers, such as Staal et al. [66, 69], Hoover et al. [45], Zhang et al. [71], Chaudhuri et al. [27, 72], Martinez-Perez et al. [73] and Mendonca and Campilho [74], are presented. Moreover, these performance results are the average values for the whole set of 20 images, except the approach of Staal et al. [66, 69], which used 19 out of 20 images of the STARE images, among which ten were healthy and nine were unhealthy.
Table 2 shows our results obtained on all 20 images in the STARE database, estimated using the hand-labeled segmentation images. These results are the mean of the TPR = 0.6801 corresponding to an FPR of around 0.0289 and an MAA = 0.9388. The results demonstrate that our technique has a competitive maximum average accuracy value where it performs better than the approach of Hoover [45] and the Matched filter [27, 72]. In addition, it remains close to the others.
5.2 DRIVE database
The results of the proposed method are also compared with those on 20 images from the DRIVE database [62, 63]. Figure 20 shows the segmented images and the manually labeled images for the DRIVE dataset. The experiment results of the TPR, the FPR and the MAA are depicted in Table 5, where the images hand-labeled by a human expert are used as a ground truth.
The experimental results show an MAA around of 0.9389. Also, we compare the performance of the suggested technique with the sensitivities and specificities of the methods cited in Table 6. It is found that for the DRIVE database, the method provides a sensitivity of 0.6887 and a specificity of 0.9765. It is clear that the proposed method performs well with a low specificity even in the presence of lesions in some images.
6 Limitations and future work
The proposed segmentation methodology has achieved competitive results with the existing methods, but at the same time it has some disadvantages. However, the user defines by themselves and manually the scale range of the width of vessels, which cannot be accurate and can affect the ability or the efficiency to detect the whole vessel network in the image. In addition, the method responds not only to vessel pixels but also to non-vessel ones. For example, the border of the optic disk and the fovea appear clearly in the obtained results of Figs. 19 and 20. To overcome the sensitivity to non-vessel pixel detection, the method needs to be improved by employing a process of discrimination between vessel and non-vessel. The segmented image can provide pathological changes as vessel pixels (Fig. 21), which can be considered as another inconvenient. Also, it can extract very well the large vessels but not those very thin ones.
Our future work will involve on the optimization of the performance of the proposed vessel segmentation from retinal images having pathological changes and on investigating solutions that can more accurately segment thin vessels. Moreover, it is intended to work more closely with ophthalmologists, to evaluate the method and to improve it according to their feedback.
7 Conclusion
The goal of this paper is to segment blood vessels in real retinal images to help interpret the retinal vascular network. The general idea is to combine a new version of an anisotropic diffusion method to remove noise with a multi-scale vesselness response that is based on the Hessian matrix’s eigenvectors and the gradient information image to detect all vessels from retinal images. In fact, the main advantage of the present technique is its capability to extract large and small vessels at different image resolutions. In addition, the ANRAD filter has a vital role in denoising images and in decreasing the difficulty of vessel extraction especially for thin vessels. The first results demonstrate the robustness of our technique against noise and its capability of detecting blood vessels.
Change history
30 December 2021
A Correction to this paper has been published: https://doi.org/10.1007/s00521-021-06819-5
21 May 2024
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1007/s00521-024-10026-3
Abbreviations
- ANRAD:
-
Adaptive noise-reducing anisotropic diffusion filter
- DPAD:
-
Detail preserving anisotropic diffusion
- FBAD:
-
Flux-based anisotropic diffusion
- NLF:
-
Noise level function
- MLE:
-
Maximum likelihood estimator
- \({\text{FPR}}\) :
-
False-positive rate
- \({\text{MAA}}\) :
-
Maximum average accuracy
- \({\text{MSSIM}}\) :
-
Mean structural similarity index measure
- PMAD:
-
Anisotropic diffusion of perona and malik
- \({\text{SNR}}\) :
-
Signal-to-noise ratio
- SRAD:
-
Specle reducing anisotropic diffusion
- \({\text{TPR}}\) :
-
True positive rate
- x :
-
Image pixel
- f :
-
Response function of a camera
- L :
-
Irradiance image
- I :
-
Image intensity
- IN :
-
Noisy image
- Ns :
-
Multiplicative noise
- Nc :
-
Additive noise
- σ 2c :
-
Variance of additive noise
- σ 2s :
-
Variance of multiplicative noise
- Nq :
-
Quantization noise
- ∑2 :
-
Noise model
- IE(.):
-
Expectation of a random variable
- \(\overline{{\sum^{2} }}\) :
-
Mean of principal components
- ωη :
-
Eigenvectors of principal components
- m :
-
Number of retained eigenvectors
- αη :
-
Unknown parameters of noise model
- η :
-
Index of unknown parameters of noise model
- i, j :
-
Spatial coordinates of current pixel x
- wi,j :
-
Window centered at current pixel
- c :
-
Instantaneous coefficient of the variation of the image
- c 2n :
-
Instantaneous coefficient of the variation of the noise
- φ :
-
Diffusion function
- Var:
-
Local variance
- \(\overline{I}^{2}\) :
-
Square of local mean intensity
- Δt :
-
Step time
- iter:
-
Iteration number of ANRAD filter
- ∇:
-
Gradient operator
- div:
-
Divergence operator
- κ :
-
Discretization number
- t :
-
Continuous scale parameter
- G :
-
Convolution kernel
- ∂ :
-
Derivative operator
- σmin :
-
Minimal scale
- σmax :
-
Maximal scale
- Θ :
-
Image orientation
- Γσ :
-
Response function at scale σ
- Γmulti :
-
Multi-scale response
- \(\overrightarrow {d}\) :
-
Unitary vector of direction Θ
- \(\overrightarrow {v}_{1}\) :
-
First eigenvector of Hessian matrix
- \(\overrightarrow {v}_{2}\) :
-
Second eigenvector of Hessian matrix
- λ1 :
-
First eigenvalue of Hessian matrix
- λ2 :
-
Second eigenvalue of Hessian matrix
- r :
-
Radius vessel
- [tmin, tmax]:
-
Scale range
- \(I^{ '} \,\) :
-
Interpolated image
- \(Q_{11} \, , \, Q_{12} \, , \, Q_{21} \, , \, Q_{22}\) :
-
Four nearest pixel values of pixel \(x\)
- di :
-
Displacement along i-axis
- dj :
-
Displacement along j-axis
- Thres:
-
Threshold on norm gradient of image
- \(N\) :
-
Iteration number of PMAD method
References
Asad AH, Azar AT, Hassaanien AE (2012) Integrated features based on gray-level and hu moment-invariants with ant colony system for retinal blood vessels segmentation. Int J Syst Biol Biomed Technol (IJSBBT) 1(4):60–73
Pal NR, Pal SK (1993) A review on image segmentation techniques. Pattern Recogn 26(9):1277–1294
El-Baz AS, Acharya R, Mirmehdi M, Suri JS (2011) Multi modality state-of-the-art medical image segmentation and registration methodologies, vol 1. Springer, New York
Kauppi T et al (2010) Eye fundus image analysis for automatic detection of diabetic retinopathy. Lappeenranta University of Technology, Lappeenranta
Asad AH, Azar AT, Hassanien AE (2013) Ant colony-based system for retinal blood vessels segmentation. In: Proceedings of seventh international conference on bio-inspired computing: theories and applications (BICTA 2012) advances in intelligent systems and computing volume 201, 2013, pp 441–452. doi:10.1007/978-81-322-1038-237
Asad AH, Azar AT, Hassanien AE (2014) A new heuristic function of ant colony system for retinal vessel segmentation. Int J Rough Sets Data Anal 1(2):15–30
Asad AH, Azar AT, Hassanien AE (2014) A comparative study on feature selection for retinal vessel segmentation using ant colony system. Recent Adv Intell Inform Adv Intell Syst Comput 235(2014):1–11. doi:10.1007/978-3-319-01778-51
Fritzsche K, Can A., Shen H, Tsai C, Turner J, Stewart C, Roysam B (2003) Automated model based segmentation, tracing and analysis of retinal vasculature from digital fundus images. In: Suri JS, Laxminarayan S (eds) State-of-the-art angiography, applications and plaque imaging using MR, CT, ultrasound and X-rays. Academic Press, pp 225–298
Cheng J, Liu J, Yanwu X, Yin F, Wong DWK, Tan N-M, Tao D, Cheng Ching-Yu, Aung T, Wong TY (2013) Superpixel classification based optic disc and optic cup segmentation for glaucoma screening. IEEE Trans Med Imaging 32(6):1019–1032
Malek J, Azar AT (2016) 3D Surface Reconstruction of Retinal Vascular Structures. International Journal of Modelling, Identification and Control (IJMIC), Inderscience Publishers, Olney, UK. (in press)
Malek J, Azar AT, Tourki R (2015) Impact of retinal vascular tortuosity on retinal circulation. Neural Comput Appl 26(1):25–40. doi:10.1007/s00521-014-1657-2
Malek J, Azar AT, Nasralli B, Tekari M, Kamoun H, Tourki R (2015) Computational analysis of blood flow in the retinal arteries and veins using fundus image. Comput Math Appl 69(2):101–116
Malek J, Azar AT (2016) A computational flow model of oxygen transport in really retinal network. International Journal of Modelling, Identification and Control (IJMIC), Inderscience Publishers, Olney, UK. (in press)
Sofka M, Stewart CV (2006) Retinal vessel centerline extraction using multiscale matched filters, confidence and edge measures. IEEE Trans Med Imaging 25(12):1531–1546
Zhang B, Zhang L, Zhang L, Karray F (2010) Retinal vessel extraction by matched filter with first-order derivative of Gaussian. Comput Biol Med 40(4):438–445
Hutchinson A, McIntosh A, Peters J, O’keeffe C, Khunti K, Baker R, Booth A (2000) Effectiveness of screening and monitoring tests for diabetic retinopathy—a systematic review. Diabet Med 17(7):495–506
Hou Y (2014) Automatic segmentation of retinal blood vessels based on improved multiscale line detection. J Comput Sci Eng 8(2):119–128
Nguyen UT, Bhuiyan A, Park LA, Ramamohanarao K (2013) An effective retinal blood vessel segmentation method using multi-scale line detection. Pattern Recogn 46(3):703–715
Perona P, Malik J (1990) Scale-space and edge detection using anisotropic diffusion. IEEE Trans Pattern Anal Mach Intell 12(7):629–639
Yu Y, Acton ST (2002) Speckle reducing anisotropic di_usion. IEEE Trans Image Process 11(11):1260–1270. doi:10.1109/TIP.2002.804276
Krissian K (2002) Flux-based anisotropic diffusion applied to enhancement of 3-D angiogram. IEEE Trans Med Imaging 21(11):1440–1442
Aja-Fernández S, Vegas-Sánchez-Ferrero G, Martín-Fernández M, Alberola-López C (2009) Automatic noise estimation in images using local statistics. Additive and multiplicative cases. Image Vis Comput 27(6):756–770
Ben Abdallah M, Malek J, Azar AT, Montesinos P, Belmabrouk H, Monreal JE, Krissian K (2015) Automatic extraction of blood vessels in the retinal vascular tree using multiscale medialness. Int J Biomed Imaging 2015:519024-1–519024-16. doi:10.1155/2015/519024
Emary E, Zawbaa H, Hassanien AE, Schaefer G, Azar AT (2014b) Retinal vessel segmentation based on possibilistic fuzzy c-means clustering optimised with cuckoo search. In: IEEE 2014 international joint conference on neural networks (IJCNN 2014), July 6–11, Beijing International Convention Center, Beijing, China
Asad AH, Azar AT, Hassanien AE (2013) An improved ant colony system for retinal vessel segmentation. In: 2013 federated conference on computer science and information systems (FedCSIS), Krakow, Poland, September 8–11, 2013
Emary E, Zawbaa H, Hassanien AE, Schaefer G, Azar AT (2014a) Retinal blood vessel segmentation using bee colony optimization and pattern search. In: IEEE 2014 international joint conference on neural networks (IJCNN 2014), July 6–11, Beijing International Convention Center, Beijing, China
Chaudhuri S, Chateterjee S, Katz N, Nelson M, Goldbaum M (1989) Detection of blood vessels in retinal images using two-dimensional matched filters. IEEE Trans Med Imaging 8(3):263–269
Chanwimaluang T, Fan G (2003) An efficient algorithm for extraction of anatomical structures in retinal images. In: Proceedings of ICIP, pp 1193–1196
Fraz MM, Barman SA, Remagnino P et al (2012) An approach to localize the retinal blood vessels using bit planes and centerline detection. Comput Methods Programs Biomed 108(2):600616
Zhou L, Rzeszotarski MS, Singerman LJ, Chokreff JM (1994) The detection and quantification of retinopathy using digital angiograms. IEEE Trans Med Imaging 13(4):619–626
Goa X, Bharath A, Stanton A, Hughes A, Chapman N, Thom S (2001) A method of vessel tracking for vessel diameter measurement on retinal images. In: Proceedings ICIP, pp 881–884
Chutatape O, Zheng L, Krishnan SM (1998) Retinal blood vessel detection and tracking by matched Gaussian and Kalman filters. In Proceedings 20th annual international conference IEEE engineering in medicine and biology, pp 3144–3149
Can A, Shen H, Turner JN, Tanenbaum HL, Roysam B (1999) Rapid automated tracing and feature extraction from retinal fundus images using direct exploratory algorithms. IEEE Trans Inf Technol Biomed 3(2):125–138
Hani AFM, Soomro TA, Faye I, Kamel N, Yahya N (2014) Denoising methods for retinal fundus images. In: 2014 IEEE international conference on intelligent and advanced systems (ICIAS), Kuala Lumpur, 3–5 June, 2014, pp 1–6. doi:10.1109/ICIAS.2014.6869534
Malek J, Tourki R (2013) Inertia-based vessel centerline extraction in retinal image. In: IEEE 2013 international conference on control, decision and information technologies (CoDIT), pp 378–381
Healey GE, Kondepudy R (1994) Radiometric CCD camera calibration and noise estimation. IEEE Trans Pattern Anal Mach Intell 16(3):267–276
Irie K, McKinnon AE, Unsworth K, Woodhead IM (2008) A model for measurement of noise in CCD digital-video cameras. Meas Sci Technol 19(4):045207
Liu X, Tanaka M, Okutomi M (2013) Estimation of signal dependent noise parameters from a single image. In: ICIP, pp 79–82
Gravel P, Beaudoin G, De Guise JA (2004) A method for modeling noise in medical images. IEEE Trans Med Imaging 23(10):1221–1232
Liu C, Szeliski R, Kang SB, Lawrence Zitnick C, Freeman WT (2008) Automatic estimation and removal of noise from a single image. IEEE Trans Pattern Anal Mach Intell 30(2):299–314
Ben Abdallah M, Malek J, Azar AT, Belmabrouk H, Monreal JE, Krissian K (2016) Adaptive noise-reducing anisotropic diffusion filter. Neural Comput Appl 27(5):1273–1300
Ben Abdallah M, Malek J, Tourki R, Monreal JE, Krissian K (2013) Automatic estimation of the noise model in fundus images. In: IEEE 2013 10th international multi-conference on systems, signals & devices (SSD), pp 1–5
Arthur D, Vassilvitskii S (2007) k-means + + : the advantages of careful seeding. In: Proceedings of the eighteenth annual ACMSIAM symposium on discrete algorithms. New Orleans, pp 1027–1035. 7–9. doi:10.1145/1283383.1283494
Wu CH, Agam G, Stanchev P (2007) A general framework for vessel segmentation in retinal images. In: IEEE 2007 International symposium on computational intelligence in robotics and automation CIRA 2007, pp 37–42
Hoover A, Kouznetsova V, Goldbaum M (2000) Locating blood vessels in retinal images by piecewise threshold probing of a matched filter response. IEEE Trans Med Imaging 19(3):203–210
Qian X, Brennan MP, Dione DP, Dobrucki WL, Jackowski MP, Breuer CK, Sinusas AJ, Papademetris X (2009) A non-parametric vessel detection method for complex vascular structures. Med Image Anal 13(1):49–61
Hai TTT, Augustin L (2003) Extraction de Caract´eristiques locales: Crêtes et Pics. Book: RIVF
Ersoy I, Bunyak F, Mackey MA, Palaniappan K (2008) Cell segmentation using Hessian-based detection and contour evolution with directional derivatives. In: 15th IEEE international conference on image processing, 2008. ICIP 2008
Sato Y, Nakajima S, Atsumi H, Koller T, Gerig G, Yoshida S, Kikinis R (1997) 3D multi-scale line filter for segmentation and visualization of curvilinear structures in medical images, CVRMed-MRCAS’97
Frangi AF, Niessen WJ, Vincken KL, Viergever MA (1998) Multiscale vessel enhancement filtering, medical image computing and computer-assisted interventation-MICCAI’98. Springer, New York
Witkin AP (1984) Scale-space filtering: a new approach to multiscale description. In: Acoustics, speech, and signal processing, IEEE international conference on ICASSP’84, vol 9, pp 150–153
Koenderink JJ (1984) The structure of images. Biol Cybern 50(5):363–370
Lindeberg T (1994) Scale-space theory: a basic tool for analyzing structures at different scales. J Appl Stat 21(1–2):225–270
Sporring J, Florack L, Nielsen M, Johansen P (1997) Gaussian scale-space theory. Kluwer Academic Publishers, Dordrecht
Florack L (1997) Image structure. Kluwer Academic Publishers, Dordrecht
Haar Romeny BM (2003) Front-end vision and multi-scale image analysis: multi-scale computer vision theory and applications, written in mathematica, vol 27. Springer, New York
Abdallah MB, Malek J, Krissian K, Tourki R (2011) An automated vessel segmentation of retinal images using multiscale vesselness. In: 2011 8th international multi-conference on systems signals and devices (SSD). IEEE, pp 1–6
Prajapati A, Naik S, Mehta S (2012) Evaluation of different image interpolation algorithms. Int J Comput Appl 58(12):6–12
Zhang M, Wang J, Li Z, Li Y (2010) An adaptive image zooming method with edge enhancement. In: 3rd international conference on advanced computer theory and engineering (ICACTE), pp 608–611
Tam WS, Kok CW, Siu WC (2009) A modified edge directed interpolation for images. In: 17th European signal processing conference (ESPC)
Hoover A (1995) STARE database. http://www.ces.clemson.edu/ahoover/stare
Niemeijer M, Staal J, van Ginneken B, Loog M, Abramoff MD (2004) Comparative study of retinal vessel segmentation methods, on a new publicly available database Medical. Imaging 2004:648–656
Niemeijer M, van Ginneken B (2002) Drive database URL www.isi.uu.nl/Research/Databases/DRIVE/results.php
Martens JB, Meesters L (1998) Image dissimilarity. Signal Process 70(3):155–176. doi:10.1016/S0165-1684(98)00123-6
Aja-Fernandez S, Alberola-Lopez C (2006) On the estimation of the coefficient of variation for anisotropic diffusion speckle filtering. IEEE Trans Image Process 15(9):2694–2701. doi:10.1109/TIP.2006.877360
Staal J, Abr`amoff MD, Niemeijer M, Viergever MA, van Ginneken B (2004) Ridge-based vessel segmentation in color images of the retina. IEEE Trans Med Imaging 23(4):501–509
Soares JVB, Leandro JJG, Cesar RM Jr, Jelinek HF, Cree MJ (2006) Retinal vessel segmentation using the 2-D Gabor wavelet and supervised classification. IEEE Trans Med Imaging 25(9):1214–1222
Soares JVB, Leandro JJG, Cesar RM Jr, Jeline KHF, Cree MJ (2006) Retinal vessel segmen-tation using the 2D Morlet wavelet and supervised classification. IEEE Trans Med Imaging 25(9):1214–1222
Staal JJ, Abramoff MD, Niemeijer M, Viergever MA, van Ginneken B (2004) Ridge based vessel segmentation in color images of the retina. IEEE Trans Med Imaging 23(4):501–509
Sinthanayothin C, Boyce J, Williamson CT (1999) Automated localisation of the optic disk, fovea, and retinal blood vessels from digital colour fundus images. Br J Ophthalmol 83:902–910
Zhang B, Zhang L, Zhang L, Karray F (2010) Retinal vessel extraction by matched filter with first-order derivative of Gaussian. Comput Biol Med 4(40):438–445
Chaudhuri S, Chatterjee S, Katz N, Nelson M, Goldbaum M (1989) Detection of blood vessels in retinal images using two-dimensional matched filters. IEEE Trans Med Imaging 8(3):263–269
Martinez-Perez ME, Hughes AD, Thom SA, Bharath AA, Parker KH (2007) Segmentation of blood vessels from red-free and uorescein retinal images. Med Image Anal 11(1):47–61
Mendonca AM, Campilho A (2006) Segmentation of retinal blood vessels by combining the detection of centerlines and morphological reconstruction. IEEE Trans Med Imaging 25(9):1200–1213
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Ben Abdallah, M., Azar, A., Guedri, H. et al. RETRACTED ARTICLE: Noise-estimation-based anisotropic diffusion approach for retinal blood vessel segmentation. Neural Comput & Applic 29, 159–180 (2018). https://doi.org/10.1007/s00521-016-2811-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-016-2811-9