Abstract
The discovery of microRNAs (miRNAs) as novel regulators of gene expression has led to a marked change in how gene regulation is viewed, with important implications for development and disease. MiRNAs are endogenous, small, noncoding RNAs that largely repress their target mRNAs post-transcriptionally. The regulation of gene expression by miRNAs represents an evolutionarily conserved mechanism that is broadly applicable to most biological processes. Recent studies have begun to define the role of miRNAs in different cell lineages during kidney development, and to implicate specific miRNAs in developmental and pathophysiological processes in the kidney. This review will focus on novel insights into the role(s) of miRNAs in kidney development, and discuss the implications for pediatric renal disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The initial discovery that a small regulatory RNA could specifically silence the function of a gene in the nematode, Caenorhabditis elegans [1], has subsequently led to revolutionary changes in how we view gene regulation, advances in the ‘toolbox’ of scientists for gene silencing, and perhaps more importantly, potential novel therapeutic approaches for disease. microRNAs (miRNAs) comprise a class of endogenous, non-coding RNA molecules that generally cause their mRNA targets to undergo post-transcriptional repression. Over the past two decades, it has become clear that miRNA-mediated regulation of gene expression represents an evolutionarily conserved mechanism that is broadly applicable to most biological processes. There are over 21,000 miRNAs reported in 168 species to date, from plants to animals (miRBase version 18 [2]), and it has been estimated that up to ½ of all transcripts are regulated by miRNAs [3]. There are several recent, comprehensive reviews of miRNAs in the kidney [4–10]. This review will focus on novel insights into the role(s) of miRNAs in kidney development, and discuss the implications for pediatric renal disease.
miRNA biogenesis and function
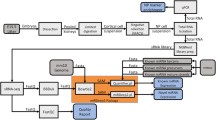
Much like other genes, miRNA genes are transcribed by RNA polymerase II into primary miRNA transcripts (Fig. 1) [11]. This primary transcript is processed by the Microprocessor complex in the nucleus to produce stem-loop precursor miRNAs, which are then exported into the cytoplasm via Exportin-5. These precursor miRNAs are subsequently cleaved by Dicer to form mature miRNAs. The mature miRNA recognizes and binds to its respective target mRNAs, recruiting the RNA-induced silencing complex (RISC) (Fig. 1) [11]. Following the recruitment of RISC, miRNAs decrease expression of their mRNA targets through translational repression, deadenylation and/or enhanced mRNA decay [12]. Thus, miRNAs generally function as negative regulators of gene expression.
Schematic diagram of microRNA biogenesis. miRNA genes are transcribed by RNA polymerase II into primary miRNA (pri-miRNA) transcripts. Pri-miRNAs are processed into precursor stem-loop miRNAs (pre-miRNAs) by the Microprocessor complex (DGCR8/Drosha), which are then exported into the cytoplasm by Exportin-5. Pre-miRNAs are cleaved by Dicer to produce mature miRNAs. Mature miRNAs recognize their respective target mRNAs, recruit the RNA-induced silencing complex (RISC) complex, and mediate post-transcriptional repression of their targets through translational repression, deadenylation and/or enhanced mRNA decay (reprinted from Fig. 1 in reference #11 with permission from the Journal of the American Society for Nephrology [11])
How do miRNAs recognize their targets? With a few exceptions, the key feature of miRNA target recognition is mRNA sequence complementarity to the 8-nucleotide (nt) ‘seed’ miRNA sequence, and most miRNA target sites occur in the 3’-untranslated region [13]. A number of bioinformatic algorithms have been developed based on this observation, in addition to incorporating other sequence features that are thought to confer increased specificity to miRNA target prediction [14]. Based on the experimental datasets from which these algorithms were derived, it has been suggested that mammalian miRNAs have on average several hundred mRNA targets per conserved miRNA family [15].
The production of miRNAs is regulated at multiple levels. For example, transcription of miRNA genes is largely transcription factor-dependent, and recent studies have systematically identified candidate miRNA promoters using chromatin immunoprecipitation (ChIP)-sequencing for chromatin marks specific to transcriptional initiation sites [16]. These experiments are revealing insights into how miRNAs and transcription factors are linked in global gene regulatory networks [17]. Some miRNAs are transcribed in clusters from a single primary miRNA transcript, in which the individual miRNAs are processed from a common precursor transcript, allowing for coordinated expression and the potential for cooperative function. A large number of miRNAs are also subject to post-transcriptional regulation, at the level of processing of primary miRNA transcripts and precursor miRNAs, as well as RNA editing [18–21]. Finally, although most miRNAs repress their respective target mRNAs, some miRNAs have been shown to activate targets depending on the cellular context [22].
There are distinct features associated with miRNA-mediated gene regulation [17]. miRNA knockdown studies show that the degree of miRNA repression is relatively modest for individual proteins, and that a miRNA can modulate the expression of hundreds to thousands of proteins [23, 24]. Although the effect of individual miRNA-mRNA target interactions may be modest, since many miRNAs target multiple members of a signaling pathway, the combinatorial effect is likely to be more robust [25]. Another distinguishing feature is the potential speed of miRNA repression, since miRNAs act following transcription [26]. Furthermore, miRNAs can distribute to different subcellular compartments, based on their association with the site of protein translation [27, 28]. Broadly speaking, miRNAs are thought to “fine-tune” existing transcriptional programs, and it may thus be more informative to describe miRNA activity in the context of modulating signaling pathways at multiple levels, as part of larger regulatory networks [25].
miRNAs in early kidney development
The concept that miRNAs are crucial regulators of developmental processes has its roots in the original description of the first miRNA, lin-4, in controlling developmental timing in C. elegans [1]. Since then, it has become clear that miRNAs are necessary for the development of multiple tissues, and are critical in the regulation of immunity, oncogenesis and cardiac disease. Interestingly, miRNAs are implicated as key regulators of embryonic stem cells, and specific miRNA families have recently been shown to have the ability to reprogram somatic cells into induced pluripotent cells [29, 30]. Given their importance in numerous developmental processes, it is not surprising that miRNAs play an important role during kidney development.
To briefly review, kidney development begins when a small group of mesodermal cells are induced to form nephron progenitors by the ureteric bud (reviewed in [31]). In response to signals from the ureteric bud and surrounding stromal cells, nephron progenitors are capable of undergoing both self-renewal and differentiation into the multiple cell types of the mature nephron. The ureteric bud branches in response to reciprocal signals from nephron progenitors, and goes on to form the collecting system of the kidneys. This process continues in an iterative fashion during kidney development, such that new nephrons are continually being induced in the nephrogenic zone just below the renal capsule. Thus, there is a corticomedullary gradient of differentiating nephrons, with the most immature cells present in the renal cortex.
Given the growing interest in miRNAs, large-scale efforts to profile and document gene expression during embryonic kidney development have expanded to include small RNA expression [32–34]. Using several different technological platforms (miRNA microarrays, small RNA cloning or deep RNA-sequencing), these studies have now made available a number of datasets that describe miRNA expression in the developing kidney. Recently, the observation that modified locked nucleic acid nucleotides can be used to detect miRNAs by in situ hybridization has led to further information regarding the cellular localization of specific miRNAs [33, 35, 36]. Together these studies provide an emerging, though still incomplete, picture of the spatial and temporal expression patterns of miRNAs during kidney development.
miRNAs in kidney progenitor cells
The first functional studies addressing roles for miRNAs in specific cell lineages in the kidney have used a conditional approach to knock down Dicer, which is required for the production of mature miRNAs (Fig. 1) [11]. Conditional Dicer models have been reported for nephron progenitors, ureteric epithelium, podocytes, proximal tubules and juxtaglomerular cells [35, 37–42]. The loss of miRNAs in nephron progenitors and their cellular descendants results in a premature depletion of progenitors notable by embryonic day 15 in the mouse, and a marked decrease in nephron number [35, 39]. This appears to be mediated by increased apoptosis in the progenitor population, and up-regulation of the pro-apoptotic protein Bim in the absence of miRNAs [35]. Although disruption of Dicer activity did not grossly affect nephron patterning, there is evidence to suggest that apoptosis is also elevated in proximal segments of the developing nephron [35, 39]. These findings raise the intriguing question of whether miRNAs serve to regulate congenital nephron endowment by modulating the balance between apoptosis and survival in nephron progenitors.
miRNAs in the ureteric bud lineage
Loss of miRNAs in the ureteric lineage results in hypoplastic, cystic kidneys with varying degrees of hydronephrosis, depending on the efficiency of conditional deletion of Dicer [39, 43]. The hypoplasia is thought to be secondary to early termination of branching morphogenesis, and is associated with decreased expression of Wnt11 and c-ret, two critical regulators of normal ureteric bud branching [39]. The appearance of cysts in the ureteric epithelium is evident by embryonic day 15, and is accompanied by ciliary changes, increased proliferation, and elevated apoptosis [39].
miRNA function in the mature nephron
Podocyte-specific loss of Dicer activity results in marked proteinuria by two weeks of age, followed by rapid progression to renal failure in mice [37, 38, 41]. The severe proteinuria is coupled with histological and ultrastructural abnormalities including crescent formation, glomerulosclerosis, foot process effacement, tubular simplification, and atrophy [37, 38, 41]. While the initial specification of podocytes occurs normally in these mice, the maintenance of podocyte structure requires miRNAs. The mutant podocytes demonstrate decreased expression of several cytoskeletal proteins, including synaptopodin, ezrin and podocalyxin, as well as the slit-diaphragm associated proteins, nephrin and podocin [37, 38, 41]. Unlike in nephron progenitors or the ureteric epithelium, there are minimal changes in apoptosis or proliferation in the mutant podocytes. In initial attempts to identify miRNAs that are responsible for the disruption of podocyte structure, bioinformatic analysis of upregulated mRNA transcripts in mutant glomeruli suggested the miR-30 family members as possible candidates [41]. Interestingly, the inducible deletion of another miRNA processing enzyme Drosha in podocytes in 2 to 3-month-old mice also results in a similar phenotype, demonstrating an ongoing need for miRNA function in the structure and function of mature podocytes [44].
Mice with a Dicer deletion in renin-secreting cells in the juxtaglomerular apparatus demonstrate loss of juxtaglomerular cells, decreased circulating renin, reduced blood pressure, striped interstitial fibrosis and vascular abnormalities [40]. Within the areas of fibrotic bands, there are vascular alterations ranging from near replacement of arterioles by interstitial cells to distorted arterioles with fibroplasia. In the absence of increased cell death, the decrease in juxtaglomerular cells is thought to result from alterations in the determination of the renin lineage. Recent work implicates two specific miRNAs, miR-330, and miR-125b-5p, in modulating the renin lineage [45].
Unlike the other conditional Dicer models in the kidney, there is no developmental or functional renal defect seen in mice with disruption of Dicer in the proximal tubule [42]. In fact, these mice possess a resistance to ischemia-reperfusion injury, with decreased histological evidence of tubular injury, smaller increases in serum creatinine, and decreased apoptosis. This report further described miRNAs that were up- or down-regulated in response to renal ischemia-perfusion injury, as a means to begin evaluating the mechanisms by which miRNAs might be mediating this response to injury.
miRNAs and kidney disease
Over the past decade, there has been a rapid expansion in the number of studies addressing potential role(s) for miRNAs in kidney disease. This work can broadly be divided into two main areas: the analysis of differential miRNA expression in renal disease, and the study of specific miRNAs that regulate pathologic processes (see recent reviews for a systematic description [4, 5, 8, 10]). These studies implicate transforming growth factor-β regulation of miRNA expression in diabetic nephropathy [46–49], p53 induction of miR-34a in ischemic acute kidney injury [50], miR-15a regulation of the cell cycle regulator Cdc25A in polycystic kidney disease [51], and the oncomir miR-17 ~ 92 cluster in renal cell carcinoma and Wilms’ tumor [52–54]. Differential miRNA profiles have been described in rodent disease models as well as from patient samples (urine, blood, and renal biopsies) for diseases including acute kidney injury [42, 55, 56], polycystic kidney disease [57, 58], acute rejection [59–61], lupus nephritis [62] and IgA nephropathy [63].
A recent illustrative example is that of miR-21 and renal fibrosis. miR-21 is up-regulated in response to transforming growth factor-β signaling, and has been functionally implicated in the fibrosis seen in mouse models of cardiac hypertrophy and idiopathic pulmonary fibrosis [64, 65]. Several recent studies have gone on to demonstrate decreased renal fibrosis following unilateral ureteral obstruction after miR-21 knockdown in wild-type mice or miR-21 knockout mice [66–68].
Novel therapeutic applications that take advantage of miRNA-mediated pathophysiological processes are becoming more feasible. One possibility is the use of miRNAs as novel biomarkers, particularly given their marked stability. One recent study suggests that their average half-life may be as long as ~5 days [69]. Furthermore, miRNAs are transported in the plasma [70] and can be isolated from urine [71, 72], increasing their potential utility.
There are several experimental approaches currently in development to modulate miRNA activity in vivo. Modified oligonucleotides, such as antagomirs or locked nucleic acid oligonucleotides, have successfully been used to target endogenous miRNAs in mammals, and can target miRNAs in the kidney [10, 66, 73]. Other approaches include the use of “miRNA sponges” to sequester endogenous miRNAs, or the introduction of oligonucleotide “target maskers” that protect miRNA targets against miRNA-mediated repression [10, 74].
Challenges in miRNA research
What are the challenges moving forward in understanding the roles of miRNAs in kidney development? While the Dicer models have provided crucial insights into functional requirements for miRNAs in the kidney, the challenge now is the identification of specific miRNAs that are responsible, at least in part, for the observed phenotypes. One example is the description of the role of the miR-30 family in regulating pronephric development in Xenopus [75]. Knockdown of miR-30a-5p was sufficient to phenocopy almost all of the pronephric defects that are caused by global loss of miRNAs using Dicer or Dgcr8 morpholino knockdown, suggesting that the miR-30 family is essential in normal pronephric development. Moreover, this study went on to demonstrate that a transcription factor known to be essential for kidney development in the mouse and frog, Xlim1/Lhx1, is a target of miR-30.
From a scientific standpoint, defining biologically relevant miRNA-mRNA target interactions continues to be a difficult task. The bioinformatic target prediction algorithms are hampered by a high false positive prediction rate, and can fail to predict the most biologically important miRNA targets [76]. Nevertheless, they remain an important tool, and continue to be the basis behind which most miRNA-mRNA target interactions have been verified experimentally. A more recent approach involves high-throughput RNA sequencing of RISC-bound RNA fragments, which allows for the identification of RISC-associated miRNAs and their target mRNAs [77–79]. These newer experimental approaches to miRNA target identification offer the possibility of more robust target predictions, and the ability to better refine bioinformatic algorithms that describe miRNA-mRNA target interactions.
Despite the rapid growth of information regarding miRNAs, there remains much to be learned about miRNA-mediated regulation of normal and abnormal kidney function. As we move towards defining the activity of specific miRNAs and their target(s) of interest, this offers the possibility of innovative approaches to the diagnosis and therapy of renal diseases. Many challenges remain, from the development of safe and effective drug delivery methods for small RNAs, to understanding the role of miRNAs within larger gene regulatory networks to minimizing off-target effects. However, it is encouraging to see how much progress has been made since these small RNA molecules were first described two decades ago.
References
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854
Kozomara A, Griffiths-Jones S (2011) miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39:D152–D157
Rajewsky N (2006) microRNA target predictions in animals. Nat Genet 38(Suppl):S8–S13
Bhatt K, Mi QS, Dong Z (2011) microRNAs in kidneys: biogenesis, regulation, and pathophysiological roles. Am J Physiol Renal Physiol 300:F602–F610
Li JY, Yong TY, Michael MZ, Gleadle JM (2010) Review: the role of microRNAs in kidney disease. Nephrology (Carlton) 15:599–608
Wessely O, Agrawal R, Tran U (2010) MicroRNAs in kidney development: lessons from the frog. RNA Biol 7:296–299
Karolina DS, Wintour EM, Bertram J, Jeyaseelan K (2010) Riboregulators in kidney development and function. Biochimie 92:217–225
Kato M, Arce L, Natarajan R (2009) MicroRNAs and their role in progressive kidney diseases. Clin J Am Soc Nephrol 4:1255–1266
Saal S, Harvey SJ (2009) MicroRNAs and the kidney: coming of age. Curr Opin Nephrol Hypertens 18:317–323
Lorenzen JM, Haller H, Thum T (2011) MicroRNAs as mediators and therapeutic targets in chronic kidney disease. Nat Rev Nephrol 7:286–294
Ho J, Kreidberg JA (2012) The long and short of MicroRNAs in the kidney. J Am Soc Nephrol 23:400–404
Djuranovic S, Nahvi A, Green R (2011) A parsimonious model for gene regulation by miRNAs. Science 331:550–553
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136:215–233
Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27:91–105
Friedman RC, Farh KK, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19:92–105
Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, Calabrese JM, Dennis LM, Volkert TL, Gupta S, Love J, Hannett N, Sharp PA, Bartel DP, Jaenisch R, Young RA (2008) Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134:521–533
Hobert O (2008) Gene regulation by transcription factors and microRNAs. Science 319:1785–1786
Lee EJ, Baek M, Gusev Y, Brackett DJ, Nuovo GJ, Schmittgen TD (2008) Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA 14:35–42
Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM (2006) Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev 20:2202–2207
Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K (2007) RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep 8:763–769
Kim YK, Heo I, Kim VN (2010) Modifications of small RNAs and their associated proteins. Cell 143:703–709
Vasudevan S, Tong Y, Steitz JA (2007) Switching from repression to activation: microRNAs can up-regulate translation. Science 318:1931–1934
Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP (2008) The impact of microRNAs on protein output. Nature 455:64–71
Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455:58–63
Shirdel EA, Xie W, Mak TW, Jurisica I (2011) NAViGaTing the micronome–using multiple microRNA prediction databases to identify signalling pathway-associated microRNAs. PLoS One 6:e17429
Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W (2006) Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125:1111–1124
Ashraf SI, McLoon AL, Sclarsic SM, Kunes S (2006) Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell 124:191–205
Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME (2006) A brain-specific microRNA regulates dendritic spine development. Nature 439:283–289
Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, Morrisey EE (2011) Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 8:376–388
Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, Tanaka F, Saito T, Nishimura J, Takemasa I, Mizushima T, Ikeda M, Yamamoto H, Sekimoto M, Doki Y, Mori M (2011) Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 8:633–638
Dressler GR (2006) The cellular basis of kidney development. Annu Rev Cell Dev Biol 22:509–529
Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, Magen A, Canidio E, Pagani M, Peluso I, Lin-Marq N, Koch M, Bilio M, Cantiello I, Verde R, De Masi C, Bianchi SA, Cicchini J, Perroud E, Mehmeti S, Dagand E, Schrinner S, Nurnberger A, Schmidt K, Metz K, Zwingmann C, Brieske N, Springer C, Hernandez AM, Herzog S, Grabbe F, Sieverding C, Fischer B, Schrader K, Brockmeyer M, Dettmer S, Helbig C, Alunni V, Battaini MA, Mura C, Henrichsen CN, Garcia-Lopez R, Echevarria D, Puelles E, Garcia-Calero E, Kruse S, Uhr M, Kauck C, Feng G, Milyaev N, Ong CK, Kumar L, Lam M, Semple CA, Gyenesei A, Mundlos S, Radelof U, Lehrach H, Sarmientos P, Reymond A, Davidson DR, Dolle P, Antonarakis SE, Yaspo ML, Martinez S, Baldock RA, Eichele G, Ballabio A (2011) A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol 9:e1000582
Thiagarajan RD, Cloonan N, Gardiner BB, Mercer TR, Kolle G, Nourbakhsh E, Wani S, Tang D, Krishnan K, Georgas KM, Rumballe BA, Chiu HS, Steen JA, Mattick JS, Little MH, Grimmond SM (2011) Refining transcriptional programs in kidney development by integration of deep RNA-sequencing and array-based spatial profiling. BMC Genomics 12:441
Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T (2007) A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129:1401–1414
Ho J, Pandey P, Schatton T, Sims-Lucas S, Khalid M, Frank MH, Hartwig S, Kreidberg JA (2011) The pro-apoptotic protein bim is a MicroRNA target in kidney progenitors. J Am Soc Nephrol 22:1053–1063
Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH (2005) MicroRNA expression in zebrafish embryonic development. Science 309:310–311
Harvey SJ, Jarad G, Cunningham J, Goldberg S, Schermer B, Harfe BD, McManus MT, Benzing T, Miner JH (2008) Podocyte-specific deletion of dicer alters cytoskeletal dynamics and causes glomerular disease. J Am Soc Nephrol 19:2150–2158
Ho J, Ng KH, Rosen S, Dostal A, Gregory RI, Kreidberg JA (2008) Podocyte-specific loss of functional microRNAs leads to rapid glomerular and tubular injury. J Am Soc Nephrol 19:2069–2075
Nagalakshmi VK, Ren Q, Pugh MM, Valerius MT, McMahon AP, Yu J (2011) Dicer regulates the development of nephrogenic and ureteric compartments in the mammalian kidney. Kidney Int 79:317–330
Sequeira-Lopez ML, Weatherford ET, Borges GR, Monteagudo MC, Pentz ES, Harfe BD, Carretero O, Sigmund CD, Gomez RA (2010) The microRNA-processing enzyme dicer maintains juxtaglomerular cells. J Am Soc Nephrol 21:460–467
Shi S, Yu L, Chiu C, Sun Y, Chen J, Khitrov G, Merkenschlager M, Holzman LB, Zhang W, Mundel P, Bottinger EP (2008) Podocyte-selective deletion of dicer induces proteinuria and glomerulosclerosis. J Am Soc Nephrol 19:2159–2169
Wei Q, Bhatt K, He HZ, Mi QS, Haase VH, Dong Z (2010) Targeted deletion of Dicer from proximal tubules protects against renal ischemia-reperfusion injury. J Am Soc Nephrol 21:756–761
Pastorelli LM, Wells S, Fray M, Smith A, Hough T, Harfe BD, McManus MT, Smith L, Woolf AS, Cheeseman M, Greenfield A (2009) Genetic analyses reveal a requirement for Dicer1 in the mouse urogenital tract. Mamm Genome 20:140–151
Zhdanova O, Srivastava S, Di L, Li Z, Tchelebi L, Dworkin S, Johnstone DB, Zavadil J, Chong MM, Littman DR, Holzman LB, Barisoni L, Skolnik EY (2011) The inducible deletion of Drosha and microRNAs in mature podocytes results in a collapsing glomerulopathy. Kidney Int 80:719–730
Medrano S, Monteagudo MC, Sequeira-Lopez ML, Pentz ES, Gomez RA (2011) Two microRNAs -miR-330 and miR-125b-5p- mark the juxtaglomerular cell and balance its smooth muscle phenotype. Am J Physiol Renal Physiol 302:F29–F37
Kato M, Arce L, Wang M, Putta S, Lanting L, Natarajan R (2011) A microRNA circuit mediates transforming growth factor-beta1 autoregulation in renal glomerular mesangial cells. Kidney Int 80:358–368
Kato M, Putta S, Wang M, Yuan H, Lanting L, Nair I, Gunn A, Nakagawa Y, Shimano H, Todorov I, Rossi JJ, Natarajan R (2009) TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol 11:881–889
Kato M, Wang L, Putta S, Wang M, Yuan H, Sun G, Lanting L, Todorov I, Rossi JJ, Natarajan R (2010) Post-transcriptional up-regulation of Tsc-22 by Ybx1, a target of miR-216a, mediates TGF-{beta}-induced collagen expression in kidney cells. J Biol Chem 285:34004–34015
Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, Natarajan R (2007) MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci USA 104:3432–3437
Bhatt K, Zhou L, Mi QS, Huang S, She JX, Dong Z (2010) MicroRNA-34a is induced via p53 during cisplatin nephrotoxicity and contributes to cell survival. Mol Med 16:409–416
Lee SO, Masyuk T, Splinter P, Banales JM, Masyuk A, Stroope A, Larusso N (2008) MicroRNA15a modulates expression of the cell-cycle regulator Cdc25A and affects hepatic cystogenesis in a rat model of polycystic kidney disease. J Clin Invest 118:3714–3724
Chow TF, Mankaruos M, Scorilas A, Youssef Y, Girgis A, Mossad S, Metias S, Rofael Y, Honey RJ, Stewart R, Pace KT, Yousef GM (2010) The miR-17-92 cluster is over expressed in and has an oncogenic effect on renal cell carcinoma. J Urol 183:743–751
Chow TF, Youssef YM, Lianidou E, Romaschin AD, Honey RJ, Stewart R, Pace KT, Yousef GM (2010) Differential expression profiling of microRNAs and their potential involvement in renal cell carcinoma pathogenesis. Clin Biochem 43:150–158
Kort EJ, Farber L, Tretiakova M, Petillo D, Furge KA, Yang XJ, Cornelius A, Teh BT (2008) The E2F3-Oncomir-1 axis is activated in Wilms' tumor. Cancer Res 68:4034–4038
Shapiro MD, Bagley J, Latz J, Godwin JG, Ge X, Tullius SG, Iacomini J (2011) MicroRNA expression data reveals a signature of kidney damage following ischemia reperfusion injury. PLoS One 6:e23011
Lorenzen JM, Kielstein JT, Hafer C, Gupta SK, Kumpers P, Faulhaber-Walter R, Haller H, Fliser D, Thum T (2011) Circulating miR-210 predicts survival in critically ill patients with acute kidney injury. Clin J Am Soc Nephrol 6:1540–1546
Pandey P, Brors B, Srivastava PK, Bott A, Boehn SN, Groene HJ, Gretz N (2008) Microarray-based approach identifies microRNAs and their target functional patterns in polycystic kidney disease. BMC Genomics 9:624
Pandey P, Qin S, Ho J, Zhou J, Kreidberg JA (2011) Systems biology approach to identify transcriptome reprogramming and microRNA targets during the progression of Polycystic Kidney Disease. BMC Syst Biol 5:56
Anglicheau D, Sharma VK, Ding R, Hummel A, Snopkowski C, Dadhania D, Seshan SV, Suthanthiran M (2009) MicroRNA expression profiles predictive of human renal allograft status. Proc Natl Acad Sci USA 106:5330–5335
Sui W, Dai Y, Huang Y, Lan H, Yan Q, Huang H (2008) Microarray analysis of MicroRNA expression in acute rejection after renal transplantation. Transpl Immunol 19:81–85
Lorenzen JM, Volkmann I, Fiedler J, Schmidt M, Scheffner I, Haller H, Gwinner W, Thum T (2011) Urinary miR-210 as a mediator of acute T-cell mediated rejection in renal allograft recipients. Am J Transplant 11:2221–2227
Dai Y, Sui W, Lan H, Yan Q, Huang H, Huang Y (2009) Comprehensive analysis of microRNA expression patterns in renal biopsies of lupus nephritis patients. Rheumatol Int 29:749–754
Wang G, Kwan BC, Lai FM, Choi PC, Chow KM, Li PK, Szeto CC (2010) Intrarenal expression of microRNAs in patients with IgA nephropathy. Lab Invest 90:98–103
Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, Kaminski N, Abraham E (2010) miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med 207:1589–1597
Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S (2008) MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 456:980–984
Zarjou A, Yang S, Abraham E, Agarwal A, Liu G (2011) Identification of a microRNA signature in renal fibrosis: role of miR-21. Am J Physiol Renal Physiol 301:F793–F801
Zhong X, Chung AC, Chen HY, Meng XM, Lan HY (2011) Smad3-mediated upregulation of miR-21 promotes renal fibrosis. J Am Soc Nephrol 22:1668–1681
Chau BN, Xin C, Hartner J, Ren S, Castano AP, Linn G, Li J, Tran PT, Kaimal V, Huang X, Chang AN, Li S, Kalra A, Grafals M, Portilla D, Mackenna DA, Orkin SH, Duffield JS (2012) MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci Transl Med 4:121ra118
Gantier MP, McCoy CE, Rusinova I, Saulep D, Wang D, Xu D, Irving AT, Behlke MA, Hertzog PJ, Mackay F, Williams BR (2011) Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res 39:5692–5703
Vickers KC, Remaley AT (2010) MicroRNAs in atherosclerosis and lipoprotein metabolism. Curr Opin Endocrinol Diabetes Obes 17:150–155
Wang G, Kwan BC, Lai FM, Chow KM, Kam-Tao Li P, Szeto CC (2010) Expression of microRNAs in the urinary sediment of patients with IgA nephropathy. Dis Markers 28:79–86
Yamada Y, Enokida H, Kojima S, Kawakami K, Chiyomaru T, Tatarano S, Yoshino H, Kawahara K, Nishiyama K, Seki N, Nakagawa M (2011) MiR-96 and miR-183 detection in urine serve as potential tumor markers of urothelial carcinoma: correlation with stage and grade, and comparison with urinary cytology. Cancer Sci 102:522–529
Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M (2005) Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438:685–689
Davidson BL, McCray PB Jr (2011) Current prospects for RNA interference-based therapies. Nat Rev Genet 12:329–340
Agrawal R, Tran U, Wessely O (2009) The miR-30 miRNA family regulates Xenopus pronephros development and targets the transcription factor Xlim1/Lhx1. Development 136:3927–3936
Thomas M, Lieberman J, Lal A (2010) Desperately seeking microRNA targets. Nat Struct Mol Biol 17:1169–1174
Chi SW, Zang JB, Mele A, Darnell RB (2009) Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460:479–486
Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T (2010) Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141:129–141
Zisoulis DG, Lovci MT, Wilbert ML, Hutt KR, Liang TY, Pasquinelli AE, Yeo GW (2010) Comprehensive discovery of endogenous argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol 17:173–179
Acknowledgments
Dr. Ho’s laboratory is supported by the National Institutes for Diabetes, Digestive and Kidney Diseases (NIDDK) grant 1K99DK087922, the Pennsylvania Department of Health, Children’s Hospital of Pittsburgh of UPMC Research Advisory Committee and a Norman S. Coplon Extramural Grant. Dr. Kreidberg’s laboratory is supported by NIDDK grants R01DK087794 and R01DK091295, and the Harvard Stem Cell Institute.
Disclosures
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ho, J., Kreidberg, J.A. MicroRNAs in renal development. Pediatr Nephrol 28, 219–225 (2013). https://doi.org/10.1007/s00467-012-2204-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00467-012-2204-y