Abstract
Hydrolyzed polyacrylamide (HPAM) biotransformation in an up-flow anaerobic sludge blanket reactor including biodegradation performances, biodegradation mechanisms, key enzymes, and functional microorganisms was explored. Response surface methodology was applied to further improve HPAM degradation. The predicted degradation ratios of HPAM and CODCr were 46.2% and 83.4% under the optimal conditions. HPAM biodegradation ratio and total organic carbon removal ratio reached 40.5% and 38.9%. Total nitrogen concentration was dramatically decreased with the increasing fermentation time during the fermentation, while low ammonia nitrogen (NH4+–N) and nitrite nitrogen (NO2−–N) were generated. NH4+–N and NO2−–N increased slightly on the whole. Enzyme activity change was correlated with HPAM biodegradation. Dehydrogenase activity had a decline of 21.3–41.0%, and the minimum value occurred at 300 mg/L of HPAM. Urease activity was varied from 28.7 to 78.7% and the maximal inhibition ratio occurred at 200 mg/L of HPAM. Mechanisms for the biodegradation of HPAM were also explored by FT-IR, HPLC, and SEM. The results indicated that long-chain HPAM was broken into micromolecule compounds and the amide groups of HPAM were transformed into carboxyl groups. Based on the sequencing results on an Illumina MiSeq platform, Proteobacterias, Bacteroidetes, and Chloroflexi were turned out to be the critical microorganisms involved in HPAM degradation. This work lays a basis for HPAM-containing wastewater treatment and offers a support for water saving and emission reduction. It is of great significance to the sustainable development of oilfield.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Hydrolyzed polyacrylamide (HPAM) made of either acrylamide or a combination of acrylamide and acrylic acid [1] is widely applied in various industries, especially as an oil displacement agent in the petroleum industry [2, 3]. The application of HPAM is now raising a new environmental issue. In general, HPAM is considered as not toxic [4], but the intermediates of HPAM degradation, especially the acrylamide monomer (AMD) due to incomplete polymerization reaction [5], are hazardous to the peripheral nerves [6]. Produced wastewater that enters the surface water and the groundwater system which can exist for a long time poses potential risks to humans and other organisms [7]. Thus, it is urgent to convert HPAM into harmless matter quickly.
The possibility of biodegradation of HPAM by microorganisms was of great concern and the researchers found that some microorganism could use HPAM as nutrient. The results of HPAM biodegradation performance seem contradictory. Haveroen et al. [8] illustrated that microbial activities and methanogenesis could be stimulated by PAM in anaerobic environments which lacked nitrogen sources. However, it was shown that carbon backbone of PAM could be broken and the amide could be used as nitrogen source in anaerobic or aerobic process environments [8]. Grula et al. [9] demonstrated that PAM could be transformed into polyacrylates through inducible amidase, but it cannot be used by microorganisms as a carbon source.
Recent studies have mainly paid attention to the various operation parameters and performance during the bio-treatment process. Several reports can be found focusing on the toxicity of HPAM and the intermediate products on the performance of bio-treatment which is regarded as critical factors. Enzyme and functional microorganism also could sensitively characterize the performance of activated sludge [10]. In general, microbial activity of activated sludge was represented by enzyme, for example, urease, phosphatase, and amidase [11] which tended to be restrained by toxic substance [12]. When microbial community was exposed to the noxious substance, its structure was often considerably changed in activated sludge [13]. Due to the different microbial species, microorganisms had varied affects on pollutants’ biodegradation [14, 15]. However, the study about improving the removal of HPAM by optimizing the structure of microbial community in anaerobic activated sludge fermentation systems has not been reported.
The objectives of this paper are to: (1) determine how HPAM could be utilized by microorganisms; (2) reveal the mechanisms of amide hydrolyzation and carbon chain breakdown of HPAM under the optimal operating conditions obtained from RSM; (3) clarify the effects of HPAM concentration on enzymatic activity; and (4) explore the effect of HPAM on the microbial community by the Illumina MiSeq platform.
Materials and methods
Activated sludge sample, HPAM and wastewater
Activated sludge was collected from Qingdao Shazikou Wastewater Treatment Plant. The main characteristic of activated sludge was as follows: 1–2 mm of average particle size, pH 6.9 ± 0.2, 31.2 of Sludge Volume Index (SVI), 12.8 g L− 1 of volatile suspended solid (VSS), and 23.5 g L− 1 of total suspended solid (TSS). The constitution of the laboratory wastewater was as follows: 1500 mg L− 1–1600 mg L− 1 of chemical oxygen demand (CODCr), 500 mg L− 1 of HPAM, pH 6.5–7.5. The addition of glucose was to regulate CODCr and the addition of NaHCO3 was to control pH value. With NH4Cl, KH2PO4, and trace-element addition (Fe, Mn, Cu, Co, and Ni), the growth of microorganisms could be positively improved [16].
Experimental setup and start-up
The anaerobic biodegradation experiment of HPAM was conducted in an UASB reactor. The working volume of UASB whose height is 80 cm with an internal diameter of 8 cm was 7.5 L. The MLSS concentration in the UASB reactor was maintained about 5000 mg L−1. Continuous operation was carried out during the whole experiment. In the start-up period, the reactor was fed with the laboratory sewage as described above. In addition, the organic loading ratio was gradually promoted by increasing a set of concentration of HPAM from 50 to 500 mg L−1. During the whole acclimation period, HPAM degradation ratio and CODCr removal ratio were examined. The UASB reactor reached stable state after 49 days. When HPAM removal was inclined to steady, sludge and effluent samples were collected to assess HPAM biodegradation, enzyme activity, and microbial community.
Determination of HPAM removal
In this paper, HPAM was examined via the method of starch–cadmium iodine [17]. CODCr, TN, NH4+–N, and NO2−–N were measured by the standard methods. A TOC analyzer (TOC-Vcpn, Shimadzu, Japan) was used for determining TOC. The preparation of HPAM samples for SEM, HPLC, and FT–IR analysis was provided by Pi et al. [18]. The prepared samples were analyzed by SEM (Model TM-1000, Hitachi, Japan), HPLC (Waters 2998, Agilent, America), and FT-IR (Model Tensor 27, BRUKER, Germany), respectively. The content and kind of VFAs from the fermentation period were also detected.
Response surface methodology for optimizing HPAM degradation
Traditional multi-factor optimization experiment ignored the joint effect among influencing factors [19]. During the operating process of UASB, response surface methodology (RSM) was applied to evaluate interactive influence of operating factors. The factors investigated were initial hydraulic retention time (HRT) (A), pH (B), and temperature (C). Three-level and three-variable box-behnken designs (BBD) were conducted to achieve the highest biodegradation of HPAM and CODCr in UASB reactor. The levels of independent variables determined by single-factor experiments are illustrated in Table 1. To fit the experimental data, a predictive quadratic model with multiple regressions was selected. To graphically analyze the statistical significance of the models, analysis of variance (ANOVA) was employed. The model terms were estimated by the p value, which was recommended that low p value (< 0.05) of the quadratic model was appropriate for both responses [20].
Enzyme activity analysis
Dehydrogenase widely existed in the tissue of animals and plant and the microorganisms’ cells. It can initiate enzymatic reactions which catalyze the oxidation of a wide range of substrates. Oxidization capability of activated sludge and microorganism activity [21] can be explored by means of measuring dehydrogenase activity. It was determined using 2,3,5-triphenyltetrazolium chloride (TTC) [22]. Urease catalyzes urea hydrolysis to ammonia and CO2 [23]. Urease found in large-scale microorganisms and plants plays a significant role in urea conversion. The products of enzymatic reactions can provide nitrogen source. Urease activity could describe the soil N supplying capacity. It was tested by the amount of the production of ammonia. Urease activity was determined by the method of Yao et al. [15]. Due to widely existence of phosphatase in soil organisms, it has also acted as an ordinary biochemical indicator in characterization activity of activated sludge. Phosphatase activity was measured using the method of Joner and Johansen [24]. At the same time, activities of dehydrogenase, urease, and phosphatase were monitored. Each treatment was sampled for three times of each enzyme activity, and average figures would be available for the three enzymes.
Determination of the critical microorganisms of HPAM degradation
The samples of activated sludge from different operating’ stages of UASB (feed sludge, acclimated activated sludge, and accumulated sludge after long-term UASB operation) were collected to determine critical microorganisms for degrading HPAM. The microorganisms were sequenced on an Illumina MiSeq platform.
Results
Optimized biodegradation condition of HPAM by response surface methodology
To optimize the operating parameter of UASB, RSM was carried out with three variables at three levels and 17 runs. The optimal combination of the initial HRT (A), pH (B), and temperature (C) was explored by a Box–Behnken design (BBD). The BBD design achieved the highest biodegradation of HPAM that was further validated. Two best fit model equations for the biodegradation of HPAM (Eq. 1 and Eq. 2) were generated as follows:
where A, B, and C are independent factors described above; RHPAM degradation means the predicted response of the biodegradation ratio of HPAM; RCOD degradation means the predicted response of the degradation ratio of CODCr. Two models of F reflected a fairly low p value (< 0.05), indicating that obtained models were significant and displayed a good prediction for experiment data [25]. R2 value was checked to evaluate the adequacy of the models. R2 value was generally recommended higher than 0.9 to make sure the model to adjust the models to the experimental data [26]. The values of the predicted R2 (R2 = 0.995 for RHPAM degradation, and 0.962 for RCOD degradation) and the adjusted R2 (R2adj = 0.988 for RHPAM degradation, and 0.914 for RCOD degradation) indicated that the models were sufficiently reliable. Adequacy precision ratio was evaluated to characterize signal-to-noise (S/N), which was considered sufficiently satisfactory for the model if the ratio was above four (adequacy precision ratio = 31.8 for RHPAM degradation and adequacy precision ratio = 11.1 for RHPAM degradation) [27]. If a model’s CV is no more than 10%, it can be considered reasonably reproducible [28]. Consequently, the relatively low coefficients of variation (CV = 3.5% for RHPAM degradation, and 2.3% for RCOD degradation) implied high accuracy of fitting and the reliable prediction.
To explore the relationship between three variables, 3D surface graphs of RSM are displayed in Fig. 1. HPAM removal clearly illustrated the significantly influence by the combinations of factors. The optimal conditions (i.e., 7.5 of pH, 31 h of HRT and 33 °C of temperature) were reached maximum biodegradation ratio of HPAM and CODCr simultaneously. With this condition, the predicted degradation ratio of HPAM and CODCr was 46.2% and 83.4%. However, the optimum operating parameters are: 7.5 of pH, 31 h of HRT and 33 °C of temperature considering the actual situation of production.
HPAM removal efficiency
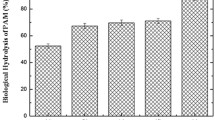
To explore HPAM removal efficiency, ultraviolet spectrophotometer and TOC analyzer were employed to test the wastewater samples without other carbon source before and after UASB treatment were analyzed. The HPAM biodegradation ratio based on ultraviolet spectrophotometer under the conditions of the optimum operating parameters can reach about 40.5%, while the results of HPAM removal efficiency based on TOC are shown in Fig. 2a. The HPAM biodegradation ratio was about 38.9% based on TOC analyzer. In addition, the effluent from the first two outlets of UASB which accumulated most of the activated sludge contributed more to the HPAM biodegradation ratio based on TOC (Fig. 2a).
HPAM removal based on nitrogen transformation
The HPAM wastewater samples before and after biodegradation without other nitrogen source were analyzed to explore the nitrogen biotransformation of HPAM. The TN was approximately 5000 µmol L−1 and was found in the influent, while no NO2−–N, NO3—N, and NH4+–N was observed (Fig. 2b). Regarding the effluent, TN concentration was dramatically decreased with the increasing fermentation time during the fermentation, while low NH4+–N and NO2−–N were generated. NH4+–N and NO2−–N increased slightly on the whole.
HPAM biodegradation based on SEM
For purpose of comparing the surface morphology change of HPAM during the anaerobic fermentation processes, HPAM of different stages, including solid, solution, after UASB treatment, were observed via SEM (Fig. 3). SEM micrographs revealed that the surface of HPAM solid was not smooth, which had sharp corner (Fig. 3a), while the HPAM solution (Fig. 3b) became flat; Fig. 3c shows that the HPAM molecular was cleaved into short rod-like after anaerobic fermentation, which can be be taken as an indirect evidence that high-molecular HPAM could be furthered biodegraded into small molecular fragments by microorganisms.
HPLC analysis before and after HPAM biodegradation
Before and after the anaerobic fermentation processes without other carbon resource, the HPAM wastewater samples and a standard acrylamide monomer sample were characterized via HPLC to explore the structure change of HPAM and whether the degradation product contained acrylamide monomer. The HPLC profiles of HPAM before and after UASB treatment are shown in Fig. 4a. In the liquid chromatogram of the wastewater sample, the appearances of two peaks were at 4.459 and 5.034 min of retention times, respectively. After biodegradation, retention times of the two peaks transferred to 4.346 and 4.943 min (Fig. 4a). In addition, the areas of these peaks were reduced as well. As shown in Fig. 4a, the peak of the acrylamide monomer (retention time was 6.897 min) had not been found in chromatogram of wastewater after biodegradation. In addition, during fermentation time, acetic acid was detected.
FT-IR analysis before and after HPAM biodegradation
The samples incubated under the optimum operating conditions were analyzed by infrared spectroscopy (Fig. 4b). The changes of structure were determined through comparing absorption peaks of samples. The peaks at 1475, 1564, 1120, 1666, and 3440 cm− 1 represented absorption peaks of N–H, C–O, C–N, and C=O bonds and amide group [29]. The peak at 3440 cm− 1 which represented amidogen became wider after biodegradation. It indicated the amidogen level had decreased. However, the peaks of C–N stretching vibration (1120 cm− 1) were still found in the samples after biodegradation, which indicated the present of amide groups and the appearance of partly hydrolyzed HPAM in the samples. Simultaneously, the peak at 1666 cm− 1 disappeared and became a weak peak at 1564 cm− 1, indicating that amide had been transformed into the carboxyl. Besides, peaks of C–N (1120 cm− 1) and C=O (1666 cm− 1) became weaker after biodegradation than that before biodegradation.
Enzyme activity analysis in the process of anaerobic HPAM degradation
The activities of phosphatase urease and dehydrogenase varied with HPAM concentrations (Fig. 5a) and different fermentation times (Fig. 5b). The enzyme activities were also inhibited by different concentrations of HPAM and fermentation times. In Fig. 5a, dehydrogenase activity had a decline of 21.3–41.0%, and the minimum value occurred at 300 mg L−1 of HPAM. Urease activity was varied from 28.7 to 78.7% and the maximal inhibition ratio occurred at 200 mg L−1 of HPAM. In addition, phosphatase activity declined slightly with HPAM concentration on the whole.
The enzyme activity with different fermentation times and corresponding 500 mg L−1 HPAM degradation ratio is shown in Fig. 5b. In the first 7 days, the treatment of 500 mg L−1 HPAM had slight inhibitory effect on dehydrogenase activity compared with control group. During the rest time, dehydrogenase activity increased with time up to the 28th day and then showed slightly decrement. Compared with the control, dehydrogenase activity was characterized by significant inhibitory effect. In general, HPAM can inhibit the activation of dehydrogenase in both the short term and the long term, indicating that HPAM is harmful to indigenous microorganisms. Urease activity was significantly activated by HPAM present in the 14th–35th day. In the 42th day, urease activity tended to the level of control. In general, the activation of urease can be lasted for a long time which shows the stimulation effect on the indigenous microorganisms. Except for the 7th day, phosphatase activity tended to the level of control during the fermentation time of 42 days. In addition, phosphatase activity was slightly activated in the 7th day, which revealed that HPAM treatment had some certain effects of activation on phosphatase.
Enzyme activity change was correlated with HPAM biodegradation during fermentation. As shown in Fig. 5b, the tendency of HPAM removal showed certain correlation with the content of dehydrogenase, which indicated that dehydrogenase activity change was well coincident with HPAM removal. The tendency of HPAM biodegradation was consistent with the change of urease which worked on linear amide bond. Compared with the tendency of HPAM removal, the trend of phosphatase showed in opposite direction, as shown in Fig. 5b.
Key microorganisms involved in HPAM degradation
To explore the relationship between microbial distribution characteristics and the biodegradation of HPAM, a long-period running UASB of HPAM wastewater treatment was carried. The samples were gathered under different cultivation stages (the seed sludge, acclimated activated sludge in UASB, accumulated activated sludge in UASB after 10 months’ cultivation). In the UASB after fermentation stages, the microbial community structures were studied on an Illumina MiSeq platform to evaluate bacteria biodiversity of sludge from different fermentation stages. Table 2 shows that the sequences of bacteria from different stages in the UASB were identified to contain 2269, 2049, and 1522 OTUs, respectively.
The Shannon diversity index was applied to represent the samples collected from the UASB under different cultivation stages (Table 2). Obvious differences were found in the diversities of the three microbial populations. Seed sludge had the highest diversity (6.0) followed by the accumulated activated sludge (5.0) and the acclimated activated sludge (4.1). This probably reflected part of microorganisms in the seed sludge which could not adapt to the wastewater containing HPAM were eliminated by complicated and changing wastewater environment, while part of microorganisms in the seed sludge which adapted to high concentrations of wastewater containing HPAM survived and then reproduced.
From Fig. 6, significant changes were observed in the microbial communities of sludge to adapt to the operation conditions and the HPAM environments. Proteobacteria, Bacteroidetes, Planctomycetes, and Chloroflexi accounted for 28.8%, 20.3%, 10.6%, and 7.4% in seed sludge, respectively, whereas those in acclimated activated sludge were determined to be 36.5%, 33.4%, 2.9%, and 4.0%, respectively. Besides, Fusobacteria was also one of the predominant phyla in the acclimated activated sludge. Proteobacteria, Bacteroidetes, Planctomycetes, and Chloroflexi were present in the accumulated sludge after long-term UASB operation and accounted for 23.3%, 39.1%, 3.8%, and 4.3%, respectively.
Discussion
HPAM biotransformation
Based on starch–cadmium iodine method, the removal of HPAM exceeded that reported in the previous researches [26, 30, 31]. HPAM removal under the same conditions conducted by anaerobic baffled reactor (ABR) [32] can reach 48.1% higher than by UASB reactor, while the removal efficiency based on TOC conducted by ABR [32] can merely reach 32.9% lower than by UASB. Results of two methods seem different. It can be explain by determination method of HPAM biodegradation based on starch–cadmium iodide method which only measures the amount of amide–N. Results of TOC indicated C–C bond break was associated with HPAM degradation, which is also reported in the literature [33]. TOC of HPAM sample after degradation decreased dramatically manifested long-chain HPAM was partly broken into micromolecule compounds and inorganic substance. Similar to TOC analysis, the results of SEM (Fig. 3) suggested that high-molecular-weight HPAM could be bio-transformed into small molecular organics by microorganisms. As shown in Fig. 2b, the generation of NH4+–N and NO2−–N revealed that anaerobic biodegradation involved in partial nitrification, which indicated that the amidogen of the HPAM could be partly utilized by microorganism. According to HPLC analysis (Fig. 4a), it can be implied that HPAM was biodegraded into smaller fragments after fermentation. Furthermore, HPAM was the unique carbon source in this system to biotransform acetic acid. The appearance of acetic acid indicated that long carbon chains could be broken down into short-chain micromolecule, which could be taken as the evidence of how microorganisms utilize part of HPAM as energy source for metabolism [34]. Meanwhile, no existence of acrylamide implied HPAM biodegradation did not result in the appearance and accumulation of acrylamide monomers. During the biodegradation of HPAM, amide of HPAM was transformed to carboxyl groups and is shown in Fig. 4b, which was coincident with the previous studies [18, 35]. In addition, the level of C–N and C=O after biodegradation became lower than that before biodegradation. It is verified that the utilization of nitrogen and carbon source by microorganisms contributed to HPAM biotransformation in anaerobic environment. Such a phenomenon was also observed in other biological systems with PAM as the substrate [1]. According to the TOC, SEM, HPLC, FT-IR, and nitrogen analyses, the results indicated that long-chain HPAM was partly broken into micromolecule compounds and amide of HPAM was transformed into carboxyl groups. Besides, HPAM biotransformation did not accumulate acrylamide monomers.
Change of enzyme activity
Dehydrogenase is related to the oxidation–reduction reactions, which is an intracellular enzyme in the cell. Its function is to transfer H+ between metabolic intermediates using various mediators in cells. The change of dehydrogenase with different concentrations of HPAM and different fermentation times (Fig. 5b) indicated that HPAM was involved in metabolic pathways. The previous studies have reported that dehydrogenase activity was the most sensitive microbial parameter closely related to pollution [36]. During the biodegradation, dehydrogenase involves in the reducing reaction and improves the potential of H+ and e− release for energy [37]. Alcohol dehydrogenase also can convert the polyacrylic acid to acetyl-CoA during intermediate reactions of HPAM biodegradation [38].
Urease is a type of amidase, which could transform organic nitrogen to ammonium nitrogen by catalyzing the hydrolyzation of the amido bond in an organic substance, and thus, the content of i nitrogen in soil could be improved [39]. The analysis of urease activity above (Fig. 5b) showed urease content exhibited a highly significant correlation with HPAM degradation and was increased by HPAM. Previous reports showed that it was a similar situation for the aerobic environments [40] and urease activity was improved by the use of both organic and inorganic fertilizers [41]. Therefore, it is suggested that urease is a best indicator of soil quality to evaluate existing conditions and potential changes in activated sludge.
Phosphatase enzyme is associated with the transformation of organic and inorganic phosphorus compounds, which is reported that its function is to inhibit substrate by eliminating the phosphate group, where the substrate could not be further degraded [37]. Lower phosphatase activity observed with higher removal (Fig. 5b) supports high-efficiency utilization of substrate promoting the potential of H+ and e− release for energy. Thus, when substrate was highly utilized, the content of phosphatase could be at low level. The decrease of phosphatase activity with time was a good indicator for utilizing substrate [42].
Preliminary hypotheses of HPAM degradation mechanism
Based on these studies mentioned above, the metabolic mechanism for the biodegradation of HPAM was proposed. According to the proposed metabolic pathways (Fig. 7), the C–N of HPAM was cut, which was catalyzed by amidase such as urease analyzed above. Urease plays a role in soil N supplying capacity. It is reported that amidase was identified in numerous genera of bacteria, such as Pseudomonas and Bacillus [31] which was also observed in three samples of activated sludge. The generation of NH4–N and NO2–N (Fig. 2b) revealed that anaerobic biodegradation involved in partial nitrification. The change in the concentration of generated NO2−–N illustrated the consumption of NO2−–N, which presumed denitrification or anaerobic ammonium oxidation (ANAMMOX) [43]. –OH replaced –NH2, and then, HPAM was hydrolyzed into acetyl-CoA and pyruvic acid by dehydrogenase mentioned above. Besides, the carbon chain of HPAM experienced the catalysis by monooxygenases oxidation [29]. During HPAM biodegradation, the carbon chain was broken and converted to smaller substances, which could act as energy source for microorganisms. At last, the degradation products of HPAM included volatile fatty acid (acetic acid, propionic acid, etc.) and excluded acrylamide monomer. The metabolites of VFSs resulted from the utilization of long-chain fatty acids, and finally, all of the acids were transformed to CH4 and H2O [44].
Key microorganisms involved in HPAM degradation
After the operation of UASB for a long time, the relative abundance of Bacteroidetes increased, and Proteobacteria was the most frequently encountered microbial group from beginning to end (Fig. 6). Protebacteria and Bacteroidete were dominant microorganism in the microbial communities of UASB, indicating that they might play a key role during fermentation of HPAM, which was also demonstrated by Yan et al. [40] and Dai et al. [38]. High amounts of Chloroflexi in the reactor made filamentous bacteria become another key microorganism besides Bacteroidetes and Proteobacteria. The filamentous bacteria played the bridge roles among aggregates to make the granules remain steady. The filamentous granules were filled with bacteria which resulted in the formation of compact granules [45]. Planctomycetes also accounted for a certain proportion in the bacterial community structure, but its function is still unknown [46], so further studies would be required to determine the function of Planctomycetes. Additionally, denitrification and higher utilization of nitrogen source were primarily caused by the existence of Nitrospirae. Methanogens such as Methanobacteria and Methanomicrobia were also found in the samples, which could increase the utilization of VFAs and convert acetate to methane [47]. In a word, the phyla Proteobacterias, Bacteroidetes, and Chloroflexi were turned out to be the significant microorganisms involved in HPAM biodegradation.
Conclusions
The predicted degradation ratios of HPAM and CODCr were 46.2% and 83.4% under the optimal conditions (7.5 of pH, 31 h of HRT and 33 °C of temperature). Biodegradation performance of HPAM in UASB was examined with the parameters such as TN, NH4+–N, and TOC. HPAM biodegradation ratio and TOC removal ratio reached 40.5% and 38.9%. TN concentration was dramatically decreased with the increasing fermentation time during the fermentation, while low NH4+–N and NO2−–N were generated. NH4+–N and NO2−–N increased slightly on the whole. Dehydrogenase activity had a decline of 21.3–41.0%, and the minimum value occurred at 300 mg L−1 of HPAM. Urease activity was varied from 28.7 to 78.7% and the maximal inhibition ratio occurred at 200 mg L−1 of HPAM. In addition, phosphatase activity declined slightly with HPAM concentration on the whole. Long-chain HPAM was broken into micromolecule compounds and the amide groups of HPAM were transformed into carboxyl groups. Proteobacterias, Bacteroidetes, and Chloroflexi were turned out to be the critical microorganisms involved in HPAM degradation.
References
Cui YZ, Zhang J, Sun M, Zhai LF (2015) Bioelectricity-assisted partial degradation of linear polyacrylamide in a bioelectrochemical system. Appl Microbiol Biotechnol 99(2):947–956
Zhao X, Liu L, Wang Y, Dai H, Wang D, Cai H (2008) Influences of partially hydrolyzed polyacrylamide (HPAM) residue on the flocculation behavior of oily wastewater produced from polymer flooding. Sep Purif Technol 62(1):199–204
Zhang H, Zhong Z, Xing W (2013) Application of ceramic membranes in the treatment of oilfield-produced water: effects of polyacrylamide and inorganic salts. Desalination 309:84–90
Rudén C (2004) Acrylamide and cancer risk-expert risk assessments and the public debate. Food Chem Toxicol 42(3):335–349
Luo YL, Yang ZH, Xu ZY, Zhou LJ, Zeng GM, Huang J, Xiao Y, Wang LK (2011) Effect of trace amounts of polyacrylamide (PAM) on long-term performance of activated sludge. J Hazard Mater 189(1):69–75
Woodrow JE, Seiber JN, Miller GC (2008) Acrylamide release resulting from sunlight irradiation of aqueous polyacrylamide/iron mixtures. J Agric Food Chem 56(8):2773–2779
Guezennec AG, Michel C, Bru K, Touzé S, Desroche N, Mnif I, Motelica-Heino M (2015) Transfer and degradation of polyacrylamide-based flocculants in hydrosystems: a review. Environ Sci Pollut Res 22(9):6390–6406
Haveroen ME, MacKinnon MD, Fedorak PM (2005) Polyacrylamide added as a nitrogen source stimulates methanogenesis in consortia from various wastewaters. Water Res 39(14):3333–3341
Grula MM, Huang ML, Sewell G (1994) Interactions of certain polyacrylamides with soil bacteria. Soil Sci 158(4):291–300
Vivas A, Moreno B, Garcia-Rodriguez S, Benítez E (2009) Assessing the impact of composting and vermicomposting on bacterial community size and structure, and microbial functional diversity of an olive-mill waste. Bioresour Technol 100(3):1319–1326
Molina-Muñoz M, Poyatos J, Rodelas B, Pozo C, Manzanera M, Hontoria E, Gonzalez-Lopez J (2010) Microbial enzymatic activities in a pilot-scale MBR experimental plant under different working conditions. Bioresour Technol 101(2):696–704
Li H, Zhang W, Li L, Liu YD, Lin KF, Lu SG, Mu BZ, Du XM, Lu Q, Zhang Q (2012) Effects of 1, 1, 1-trichloroethane on enzymatic activity and bacterial community in anaerobic microcosm form sequencing batch reactors. Ecotoxicology 21(5):1426–1435
Kabelitz N, Machackova J, Imfeld G, Brennerova M, Pieper DH, Heipieper HJ, Junca H (2009) Enhancement of the microbial community biomass and diversity during air sparging bioremediation of a soil highly contaminated with kerosene and BTEX. Appl Microbiol Biotechnol 82(3):565–577
Yang Q, Wang J, Wang H, Chen X, Ren S, Li X, Xu Y, Zhang H, Li X (2012) Evolution of the microbial community in a full-scale printing and dyeing wastewater treatment system. Bioresour Technol 117:155–163
Yao Y, Lu Z, Min H, Gao H, Zhu F (2012) The effect of tetrahydrofuran on the enzymatic activity and microbial community in activated sludge from a sequencing batch reactor. Ecotoxicology 21(1):56–65
Grobicki A, Stuckey D (1992) Hydrodynamic characteristics of the anaerobic baffled reactor. Water Res 26(3):371–378
Zhao LM, Zhang CC, Bao MT, Lu JR (2018) Effects of different electron acceptors on the methanogenesis of hydrolyzed polyacrylamide biodegradation in anaerobic activated sludge systems. Bioresour Technol 247:759–768
Pi Y, Zheng Z, Bao M, Li Y, Zhou Y, Sang G (2015) Treatment of partially hydrolyzed polyacrylamide wastewater by combined Fenton oxidation and anaerobic biological processes. Chem Eng J 273:1–6
Zhao S, Huang G, Cheng G, Wang Y, Fu H (2014) Hardness, COD and turbidity removals from produced water by electrocoagulation pretreatment prior to reverse osmosis membranes. Desalination 344:454–462
Gan S, Ng HK (2012) Modified Fenton oxidation of polycyclic aromatic hydrocarbon (PAH)-contaminated soils and the potential of bioremediation as post-treatment. Sci Total Environ 419:240–249
Yan Y, Feng L, Sun L, Zhou Q (2010) Integration of a coagulation step in a cometabolism sequencing batch reactor for the treatment of DSD acid wastewater from a reduction process. Desalination 250(1):167–171
Lv Z, Yao Y, Lv Z, Min H (2008) Effect of tetrahydrofuran on enzyme activities in activated sludge. Ecotoxicol Environ Saf 70(2):259–265
Bachmeier KL, Williams AE, Warmington JR, Bang SS (2002) Urease activity in microbiologically-induced calcite precipitation. J Biotechnol 93(2):171–181
Joner EJ, Johansen A (2000) Phosphatase activity of external hyphae of two arbuscular mycorrhizal fungi. Mycol Res 104(01):81–86
Podstawczyk D, Witek-Krowiak A, Dawiec A, Bhatnagar A (2015) Biosorption of copper(II) ions by flax meal: empirical modeling and process optimization by response surface methodology (RSM) and artificial neural network (ANN) simulation. Ecol Eng 83:364–379
Ding L, He S, Yan X (2014) Efficient preparation of pseudoalteromone A from marine Pseudoalteromonas rubra QD1-2 by combination of response surface methodology and high-speed counter-current chromatography: a comparison with high-performance liquid chromatography. Appl Microbiol Biotechnol 98(10):4369–4377
Bhatti MS, Reddy AS, Kalia RK, Thukral AK (2011) Modeling and optimization of voltage and treatment time for electrocoagulation removal of hexavalent chromium. Desalination 269(1):157–162
Zhu G, Zheng H, Chen W, Fan W, Zhang P, Tshukudu T (2012) Preparation of a composite coagulant: polymeric aluminum ferric sulfate (PAFS) for wastewater treatment. Desalination 285:315–323
Bao M, Chen Q, Li Y, Jiang G (2010) Biodegradation of partially hydrolyzed polyacrylamide by bacteria isolated from production water after polymer flooding in an oil field. J Hazard Mater 184(1):105–110
Kay-Shoemake JL, Watwood ME, Lentz RD, Sojka RE (1998) Polyacrylamide as an organic nitrogen source for soil microorganisms with potential effects on inorganic soil nitrogen in agricultural soil. Soil Biol Biochem 30(8):1045–1052
Kay-Shoemake JL, Watwood ME, Sojka RE, Lentz RD (1998) Polyacrylamide as a substrate for microbial amidase in culture and soil. Soil Biol Biochem 30(13):1647–1654
Sang G, Pi Y, Bao M, Li Y, Lu J (2015) Biodegradation for hydrolyzed polyacrylamide in the anaerobic baffled reactor combined aeration tank. Ecol Eng 84:121–127
Yu F, Fu R, Xie Y, Chen W (2015) Isolation and characterization of polyacrylamide-degrading bacteria from dewatered sludge. Int J Environ Res Public Health 12(4):4214–4230
Dai X, Luo F, Yi J, He Q, Dong B (2014) Biodegradation of polyacrylamide by anaerobic digestion under mesophilic condition and its performance in actual dewatered sludge system. Bioresour Technol 153:55–61
Wen Q, Chen Z, Zhao Y, Zhang H, Feng Y (2010) Biodegradation of polyacrylamide by bacteria isolated from activated sludge and oil-contaminated soil. J Hazard Mater 175(1):955–959
Muhlbachova G, Sagova-Mareckova M, Omelka M, Szakova J, Tlustos P (2015) The influence of soil organic carbon on interactions between microbial parameters and metal concentrations at a long-term contaminated site. Sci Total Environ 502:218–223
Reddy MV, Srikanth S, Mohan SV, Sarma P (2010) Phosphatase and dehydrogenase activities in anodic chamber of single chamber microbial fuel cell (MFC) at variable substrate loading conditions. Bioelectrochemistry 77(2):125–132
Dai X, Luo F, Zhang D, Dai L, Chen Y, Dong B (2015) Waste-activated sludge fermentation for polyacrylamide biodegradation improved by anaerobic hydrolysis and key microorganisms involved in biological polyacrylamide removal. Sci Rep 5:11675
Wu B, Lan T, Lu D, Liu Z (2014) Ecological and enzymatic responses to petroleum contamination. Environ Sci Process Impact 16(6):1501–1509
Yan M, Zhao L, Bao M, Lu J (2016) Hydrolyzed polyacrylamide biodegradation and mechanism in sequencing batch biofilm reactor. Bioresour Technol 207:315–321
Balezentiene L, Klimas E (2009) Effect of organic and mineral fertilizers and land management on soil enzyme activities. Agronomy Res 7(1):191–197
Mohan SV, Reddy MV (2013) Optimization of critical factors to enhance polyhydroxyalkanoates (PHA) synthesis by mixed culture using Taguchi design of experimental methodology. Bioresour Technol 128:409–416
Le H, Jantarat N, Khanitchaidecha W, Ratananikom K, Nakaruk A (2015) Development of sequencing batch reactor performance for nitrogen wastewater treatment. J Microb Biochem Technol 7:363–366
Li CY, Zhang D, Li XX, Mbadinga SM, Yang SZ, Liu JF, Gu JD, Mu BZ (2016) The biofilm property and its correlationship with high-molecular-weight polyacrylamide degradation in a water injection pipeline of Daqing oilfield. J Hazard Mater 304:388–399
Li J, Hu B, Zheng P, Qaisar M, Mei L (2008) Filamentous granular sludge bulking in a laboratory scale UASB reactor. Bioresour Technol 99(9):3431–3438
Chu ZR, Wang K, Li XK, Zhu MT, Yang L, Zhang J (2015) Microbial characterization of aggregates within a one-stage nitritation–anammox system using high-throughput amplicon sequencing. Chem Eng J 262:41–48
Yang Y, Chen Q, Wall JD, Hu Z (2012) Potential nanosilver impact on anaerobic digestion at moderate silver concentrations. Water Res 46(4):1176–1184
Acknowledgements
This study was financially supported by the Key Research and Development Program of Shandong Province (public welfare special project) (2017GSF217012), the Fundamental Research Funds for the Central Universities (201861016), the National Natural Science Foundation of China (51174181) and the Major Projects of the National High Technology Research and Development Program 863 (2013AA064401).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This is MCTL Contribution No. 196.
Rights and permissions
About this article
Cite this article
Zhao, L., Song, T., Han, D. et al. Hydrolyzed polyacrylamide biotransformation in an up-flow anaerobic sludge blanket reactor system: key enzymes, functional microorganisms, and biodegradation mechanisms. Bioprocess Biosyst Eng 42, 941–951 (2019). https://doi.org/10.1007/s00449-019-02094-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-019-02094-w