Abstract
Patterns of host–parasite association are poorly understood in tropical forests. While we typically observe only snapshots of the diverse assemblages and interactions under variable conditions, there is a desire to make inferences about prevalence and host-specificity patterns. We studied the interaction of ticks with non-volant small mammals in forests of Borneo. We inferred the probability of species interactions from individual-level data in a multi-level Bayesian model that incorporated environmental covariates and advanced estimates for rarely observed species through model averaging. We estimated the likelihood of observing particular interaction frequencies under field conditions and a scenario of exhaustive sampling and examined the consequences for inferring host specificity. We recorded a total of 13 different tick species belonging to the five genera Amblyomma, Dermacentor, Haemaphysalis, Ixodes, and Rhipicephalus from a total of 37 different host species (Rodentia, Scandentia, Carnivora, Soricidae) on 237 out of 1,444 host individuals. Infestation probabilities revealed most variation across host species but less variation across tick species with three common rat and two tree shrew species being most heavily infested. Host species identity explained ca. 75 % of the variation in infestation probability and another 8–10 % was explained by local host abundance. Host traits and site-specific attributes had little explanatory power. Host specificity was estimated to be similarly low for all tick species, which were all likely to infest 34–37 host species if exhaustively sampled. By taking into consideration the hierarchical organization of individual interactions that may take place under variable conditions and that shape host–parasite networks, we can discern uncertainty and sampling bias from true interaction frequencies, whereas network attributes derived from observed values may lead to highly misleading results. Multi-level approaches may help to move this field towards inferential approaches for understanding mechanisms that shape the strength and dynamics in ecological networks.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The quantification of interactions between host and parasite species, which is fundamental for understanding patterns of host specificity and the dynamics of host–parasite associations, is often hindered by insufficient data (Anderson and May 1991). The search for host–parasite associations might be tedious, given generally low parasite prevalence and infestation rates. Using ticks (Acari: Ixodidae), most of which are not rigid host specialists especially in their immature stages (Durden 2006) as an example, inferring host specificity requires a large range of potential host species to be considered in field sampling and analysis. There is considerable uncertainty as to whether any absence of observed tick–host interaction is a true lack of species interaction or simply a matter of not having observed it due to low prevalences or a small number of host individuals being examined (Cumming 2004; Petney et al. 2007). In fact, many tick species may readily switch between host species if their availability changes (Klompen et al. 1996), such as under variable forest and climate conditions. Any concerns about altered host–parasite relationships therefore need an accurate understanding of existing prevalence and species interaction patterns as well as the conditions under which host–parasite relationships may change (Krasnov et al. 2007; LoGiudice et al. 2003). Inevitably, the intricate nature of aspects such as species specificity and complementarity in interactions—be it host–parasite or any other food web or mutualistic interaction—requires consideration of community-level data for proper quantification, since interactions between pairs of species are not independent of other species in the same web (Colwell and Futuyma 1971; Hurlbert 1978). The traditional characterization of host specificity as the sole number of host species approached by a parasite species, for instance, neglects bias by under-sampling rare species or the functional and phylogenetic relationship among members of the host community (Mason et al. 2005; Poulin and Mouillot 2005).
Recently, ecological network approaches have been increasingly used for investigating the interaction of species. These techniques provide helpful frameworks for understanding central aspects of ecology, such as structural patterns in food webs and the functional and complementary roles of species in ecosystems (Bersier et al. 2002; Clauset et al. 2008). Based on adjacency matrices that summarize the presence/absence or frequency of observed interactions among species pairs, however, measures of network attributes may be confounded when species detection and the observation of interactions are less than perfect. In addition, infestation rates and ‘zero-encounters’ of non-infested animals are also meaningful parasitological data, but are only rarely considered in summarizing analyses at the community level. The necessity to account for incomplete detection, sampling and uncertainty in parameters has been well recognized in population and community ecological studies (MacKenzie et al. 2002; Lande et al. 2003; Royle et al. 2005) and for estimating diversity measures (Golicher et al. 2006) but less so in the analysis of species interaction networks (Lafferty and Dunne 2010). Moreover, it has recently been emphasized that future steps in the network analysis of ecological data need to account for environmental variation (Blüthgen and Klein 2011; Olesen et al. 2008). The seminal illustration by Robinson (2009) that the quantitative examination of ecological relations at upper organizational levels of aggregated data may lead to wrong conclusions and that individual-level data are the key to understand upper-level patterns, however, has been little considered in network analysis. In this study we aimed to investigate the host–tick associations of small mammal assemblages at the network level. In view of this aim, we were interested in considering uncertainty in interaction frequencies aggregated into adjacency matrices and how this may bias any inference on derived indices of host specificity. Most efforts in describing host–tick associations in Southeast Asia to date were performed with a taxonomic background (Petney et al. 2007). Consequently, there is considerable uncertainty as to whether host–tick associations are sufficiently well recorded, and whether the lack of host–tick associations is simply a lack of records or a true absence of interaction. Moreover, given the relatively low prevalence of ticks on tropical vertebrates (Durden et al. 2008; Geevarghese and Mishra 2011), detecting a larger range of host–tick associations in the wild is only possible with intensive long-term sampling, but such a sampling scheme would also be more extensive, and so there are likely to be changes in environmental conditions in time and space. Host availability, for example, might change with overall forest conditions (Wells et al. 2007a) or shifts in host abundance and space use with seasonal resource availability, such as during fruiting seasons (Adler 1998). If the individual foraging success of ticks is influenced by environmental factors or host species availability, then variability in the strength of interactions between hosts and parasites at the community level is an emergent consequence of the conditions under which individual interactions take place and we expect such dynamics in interactions to be confounded with host specificity. In this study we illustrate how the application of a multi-level model may overcome problems in the analysis of incompletely sampled data such as in our study.
Materials and methods
Study sites, sampling and tick identification
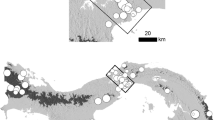
Small mammals were live-captured at six different lowland forest sites in Sabah, Borneo in an effort to understand the effects of forest logging on small mammal ecology (Wells et al. 2007a): unlogged forest at Poring, Kinabalu National Park (06°02′N, 116°42′E), Danum Valley Conservation Area (4°57′N 117°48′E), Tawau Hills National Park (04°23′N, 117°53′E); and logged forests at Luasong Field Centre (4°36′N, 117°23′E), Kg. Monggis (06°13′N, 116°45′E), Kg. Tumbalang (06°08′N, 116°53′E). Each of these study sites was sampled 3 times at equal intervals throughout seasons and years between October 2002 and November 2004. This resulted in a total trapping effort of 40,552 trap-nights in lowland forests. We further conducted small mammal trapping in montane forests at Mesilou, Mount Kinabalu (6°00′N, 116°35′E) and Mount Alap, Crocker Range National Park (5°49′N 116°20′E) at altitudes between 1,875 and 2,230 m a.s.l. with a minor trapping effort of 966 trap nights, following a similar handling protocol as in lowland forests (Wells et al. 2011).
All captured small mammals were lightly anaesthetized with inhalation narcotics (diethylether or isoflurane/forene) before being examined and marked before release at the capture location (Wells et al. 2007a). All encountered ticks were collected either from the container in which the animals were handled or were removed with forceps from the body and stored in 95 % ethanol for later identification in the laboratory. Ticks were identified by L. A. D. and T. N. P. based on available reference collections and various sources of literature. Some larval ticks needed to be cleared in lactophenol and slide-mounted in Hoyer’s medium in order to adequately visualize morphological characters. We could not identify immature stages of Dermacentor sp. ticks because neither larvae nor nymphs of any of the four species known to occur in Borneo (Dermacentor atrosignatus, Dermacentor auratus, Dermacentor compactus, and Dermacentor steini) have been described. Similarly, immature stages have not been described for all of the species of Haemaphysalis known to occur in Borneo and a few immature specimens we collected that belong to this genus did not match any published descriptions; these ticks are listed as Haemaphysalis sp. 1 and sp. 2, respectively in Table 1 and elsewhere. Voucher tick specimens from this study are deposited in the Sabah Parks Museum in Sabah, Malaysia, and the U.S. National Tick Collection at Georgia Southern University, USA.
We characterized all small mammal species by taxonomic family and order (Wilson and Reeder 2005). We further categorized specific patterns of habitat use into terrestrial, scansorial (with climbing activity to 5 m height but species not known to occur in upper canopy levels), and arboreal based on Wells et al. (2006) and personal observations. We categorized forest types into either old-growth or logged for all lowland sites, or as montane old-growth forests. Each sample was further described by sampling month and year. We quantified local host abundance as the number of individuals from the same species investigated within the particular sampling session. Although this is a rather coarse measure of host abundance, we expect this measure, which is directly linked to capture success (i.e. nearly all captured individuals were examined) to be a meaningful indicator of either true abundance or foraging activity of small mammals: both of these factors add up to the exposure of host species to ticks. As some spiny rats (Maxomys rajah and Maxomys surifer) were not distinguishable from one another in the field, we added the unidentified individuals proportionally to the number of identified individuals of both species for sampling efforts. These unidentified individuals were not infested by ticks and were therefore excluded from observation data of host tick associations.
In this study, we use the term ‘infestation probability’ as the probability that a host individual is infested with a tick, whereas we use ‘prevalence’ as a species-level attribute, describing the proportion of host individuals from a population being infected with ticks.
Data analysis
Ecological network data typically consist of two sets of interacting organisms such as hosts and parasites, flowering plants and pollinators, or predators and prey, mostly linked in bipartite relationships, and relational ties are mostly summarized into adjacency matrices at the species-level with a(i,j) given for each pair of interacting species i and j. If a(i,j) is a quantitative measure of individual interactions (i.e. total pair counts), these interactions may take place under variable environmental conditions. Moreover, the relative observation frequencies might be burdened by uncertainty due to sampling efforts and the under-representation of interactions observed for rare species.
With such a background in mind, we established a hierarchical model that accounted for individual-level variation in host–tick associations based on a logistic regression and from which species-level inference can be made. In an individual-level regression model, we assumed the dyad-specific infestation probability of host individual h with tick species j to be a function of tick species identity given as a variable intercept μ(j) and conditional on covariates linked to six host traits (HT: host species identity, family, order, body weight, habitat use, local host abundance) and four environmental attributes (ET: sampling area, forest type, sampling year and month) such that
We linked the model of infestation probability ψ(h,j) to our binary tick presence–absence data through a Bernoulli distribution.
From this individual-level probability ψ(h,j), species-level infestation probability can be modelled as an average infestation probability Φ(i,j) as:
To allow the iterative computation to converge more quickly (see Gelman and Hill 2007), we included redundancy parameters for all multi-level parameter estimates as given here for μ(j):
The variance [termed hereafter “var()”] allocated during Markov chain Monte Carlo (MCMC) updates to each of the different components in the individual-level regression model can be calculated as the squared coefficient estimate multiplied by the variance in the covariate for continuous variables (e.g. \( \text{var} (\alpha_{x} (j)) = \alpha_{2}^{x} \times \text{var} ({\text{HT}}_{x} ) \) for HT x ), for categorical variables the finite sample variance was calculated over redundancy parameters such as var(μ(j)*).
Having estimated infestation probability Φ(i,j), we were able to use this probability to directly calculate indices of host specificity or to generate interaction matrices. We estimated interaction frequency, F(i,j), between host species i and tick species j from the infestation probability Φ(i,j) as a binomial distribution process:
where N(i) is the number of trials, for which we used two scenarios. The first used the number of host individuals sampled in our field study, i.e. N(i) is the total abundance of host species sampled (“FieldScenario”). The second scenario assumed exhaustive and equal sampling of all host species, and used N(i) = 1,000 trials in binomial distributions (“ExhaustiveSample”).
For calculating host specificity, we used a measure derived from Rao’s quadratic entropy (Rao 1982) based on specific infestation probabilities as:
where d(i,s) is the taxonomic distance between the two host species i and s and where Φ(i,j) and Φ(s,j) are the infestation probabilities of host species i and s with tick species j. Taxonomic distance was simply based on the species’ genus, family and order identity, given as half the number of nodes between pairs of species in the resulting taxonomic tree, ranging between 0 and 4. This basic index combines three important aspects of host specificity, namely the number of utilized host species, their specific infestation probability and the taxonomic range of host species utilized.
Prevalence and infestation probability do not necessarily correspond to host availability if it is assumed that specialized species use resources disproportionately more than their availability (Feinsinger et al. 1981). In order to account for overall host availability, we considered the proportion of the total number of individuals from the respective species examined as a coarse but meaningful measure of its relative availability independent of tick prevalence. For simplicity, we have chosen this coarse measure of overall host availability, but local abundance and overall host availability can also be modelled in a hierarchical framework if accurate mark-recapture data are available (Royle et al. 2005; Wenger and Freeman 2008). We thus extended the aforementioned index to account for overall host availability in a similar fashion as taxonomic difference:
and calculated a similar index without including taxonomic distance as:
We calculated the ratio of the sum of indices to the sum of weighting factors, to estimate the relative strength of taxonomic distance and host availability (see Poulin and Mouillot 2005) such as:
for an index that largely accounts for taxonomic distance among hosts and
for an index that largely accounts for the contribution of host availability to species specificity.
We further calculated the specialization index d′ based on Kullback–Leibler distances (Blüthgen and Menzel 2006). We calculated this index for observed counts as well as for random draws from the two scenarios (FieldScenario, ExhaustiveSample). We calculated network indices for interaction networks from the entire data set regardless of forest type, as the effects of forest type and other covariates on infestation probabilities were negligible (see “Results”).
The model was fitted in a Bayesian framework, with the following priors, chosen to be imprecise: for all variance terms U(0,100) and for all sample means N(0,1). We fitted the model with MCMC based on the Gibbs sampler in OpenBUGS 3.1.1 (Lunn et al. 2009). Posteriors were gathered by running ca. 5,000 iterations of two chains after discarding 100,000 iterations. Convergence and mixing was assessed visually. MCMC results are given as either posterior mode and 95 % highest posterior density credible intervals (CI) for Φ(i,j) and variance terms or medians and 5–95 % quantiles for estimates from binomial distributions. The model code is given in the electronic supplementary material (ESM) 1.
Results
We recorded a total of 13 different hard tick species (Ixodidae) belonging to the five genera Amblyomma, Dermacentor, Haemaphysalis, Ixodes, and Rhipicephalus, on 237 out of 1,444 host individuals from a total of 37 different host species (Rodentia, Scandentia, Carnivora, Soricidae) (Table 1). The resulting observed network included 309 host–tick associations, with a total of 597 tick individuals being sampled. The most commonly observed tick species were immature stages (larvae and nymphs) of two species of Dermacentor sp. with 140 and 128 individuals, respectively. Up to four different tick species were sampled from the same host individual. Twenty-three host individuals were infested with ≥5 tick individuals, but most ticks were encountered as singletons.
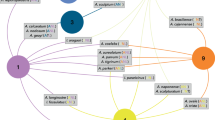
Overall infestation probabilities revealed relatively less variation across tick species but these differed across host species with 75 % (CI 57–89 %) of the variation in infestation probability explained by host species identity compared to only 4 % (CI 1–11 %) of variation in infestation probability explained by tick species identity and <1 % (CI 0–4 %) by host–tick interaction effects. The highest infestation probabilities were estimated to be associated with the rat species Leopoldamys sabanus, Maxomys rajah and the montane Maxomys alticola along with the tree shrews Tupaia longipes and Tupaia gracilis, all of which are relatively common species. For these species, all 13 tick species were likely to have infestation probabilities >1 % (posterior modes >1 %, CI 0.5–9 %) (Fig. 1, ESM 2). For the majority of ca. 78 % of all host–tick associations, however, pair-wise infestation probabilities were likely to be <0. 5 % (based on posterior mode, CI for these estimates: 0–6 %). Notably, there was considerable uncertainty in estimating infestation probabilities with generally large credible intervals: while the overall patterns of a few relatively large infestation probabilities (mostly associated with a few common host species) and mostly low infestation probabilities appears to be a robust result, detailed interpretation of individual infestation probabilities was not feasible. Local host abundance explained another 8–10 % (all CI 1–23 %) of variation in infestation probability with a positive relationship (odds ratios posterior modes 1.08–1.65 for all species). In contrast, forest type, forest site, host body weight, habitat use, year and month of sampling had little effect on infestation probabilities (all posterior mode <1 %, all CI 0–10 %).
The maximum observed number of host species infested was 16 for Dermacentor sp. 1, thirteen for H. papuana, and 12 for Ixodes granulatus, whereas three tick species were only recorded from single host species. Conclusions about host specificity of ticks from these observed numbers, however, would be highly misleading, as the estimated number of host species to be infested by each tick species was larger and likely to be more evenly sized across tick species: if randomly drawn from binomial distributions based on infestation probabilities and the field-based scenario of host individuals examined (“FieldScenario”), all 13 tick species were most likely to be associated with from five to seven host species during single draws (all CI 3–10 species) (Fig. 2). In fact, these estimated numbers of interactions from single random draws were biased by sampling size. If all host species were equally exhaustively sampled (“ExhaustiveSample”), all tick species would be likely to be associated with 28–31 host species per single draw (all CI 21–36 species) (Fig. 2), and to a total of 34–37 host species when all MCMC samples were summarized (Table 1).
Impact of sampling intensity and uncertainty on inferring the number of host species infested by different tick species. Lines indicate the number of observed host species for each tick species when data are aggregated into a single adjacency matrix. Black bars indicate the posterior estimates of the number of host species likely to be infested by each tick species given the estimated infestation probabilities and the numbers of observed host individuals (see “Materials and methods”: “FieldScenario”). Likewise, white bars indicate posterior estimates of the number of host species likely to be infested by each tick species if 1,000 individuals of each species were examined (“ExhaustiveSample”)
Comparing these model-based estimates to the observed data revealed two trends when accounting for uncertainty in observed data, namely lower host specificity (i.e. a larger range of host species) and alignment of relative (spurious) differences in host specificity across tick species.
These trends were also confirmed by host-specificity indices based on Rao’s entropy and the specialization index d′ based on adjacency matrices: calculating these indices of host specificity from observed data included considerably larger ranges of values and differences among the different tick species compared to estimated indices derived from random draws (Figs. 3a, b, 4). The taxonomic distinctness of host species as measured by the index S.TD, for example, were estimated as equally high for all tick species, suggesting that all tick species were likely to infest host species from the full range of taxonomic groups, including tick species observed only from a small range or single host species (Fig. 3a). Likewise, the effect of overall host availability on host specificity (S.AV) is estimated to be similarly low for all tick species, whereas indices derived from observed data were biased by having rarely observed tick species associated by chance with common host species such as the few records of Rhipicephalus haemaphysaloides, Haemaphysalis semermis and Haemaphysalis traguli (Fig. 3b).
Indices of host specificity adapted from Rao’s quadratic entropy for the 13 sampled tick species. Black squares are posterior estimates with 95 % credible interval bars from the model and triangles are calculated values from the observed data, representing a single snapshot. S.TD Accounts for taxonomic distances, S.AV accounts for relative overall host availability
Host specificity of tick species calculated as the specialization index d’ between the frequency of interactions among pairs of species aggregated into matrices. Indices are given for observed data (open triangles) as well as posterior mode estimates with 95 % credible intervals for the two scenarios of random draws based on field data (filled squares) and exhaustive sampling (open squares) (see “Materials and methods”). Grey small dots and lines are plotted for 50 arbitrarily selected random draws and corresponding index values from the field-based scenario, demonstrating the large differences in d’ from different draws under the same scenario
Calculating the specialization index d′ from observed and randomly drawn interaction frequencies under two different scenarios also suggested that host specificity was equally low for all species. Despite the significant bias in estimating this index from observed data or single draws, estimates of host specificity by d′ are lower if species have been exhaustively sampled compared to random draws under a field-based scenario (Fig. 4). Notably, if all host species were equally exhaustively sampled, their representation in the network would largely differ as would their infestation probabilities with ticks.
Discussion
Describing the connection and interaction patterns between host and parasite species with the aid of network analysis has proven useful in elucidating key properties for understanding parasite diversity (Vázquez et al. 2005, 2007; Mouillot et al. 2008). Using a multi-species, multi-level modelling approach, our results emphasize the conceptual benefit of inferring network attributes from individual-based approaches. They show that considering such complexity is critical rather than a negligible detail. Two principal conclusions can be drawn from this study. Firstly, when accounting for incomplete sampling and uncertainty, estimates of interaction frequency may diverge from what is observed, demonstrating that network attributes and indices directly derived from observational data may lead to highly misleading results if there is uncertainty as to whether observations represent true relationships between species. In particular, species specialization might be overestimated for some species if indices are based on observed data. Further, indices derived from observed data may indicate variation and different levels of specialization across the range of examined species that lack verification. Secondly, multi-level models offer a consistent framework to incorporate covariates under which particular interactions take place, providing the basis to understand how possible variation in interactions is partitioned among species-specific and environmental traits.
In its basic hierarchical structure, the model is similar to increasingly used multi-species occupancy models in population and community ecology (MacKenzie et al. 2002; Royle et al. 2005; Wikle 2003) in that it recognizes observed data at the level of individual entities as a random draw out of the true species’ occupancy probabilities. The model thus benefits from considering a wide variety of available information, including the lack of observed interactions (‘zero counts’) during particular sampling events and the relative frequencies of interactions. Excluding such information while analysing network attributes from solely assembling observed data into interaction matrices comprises a considerable loss of information on how intensively species are interacting. Moreover, modelling individual interactions to infer upper-level organization such as interaction networks prevents the assumption that entire communities are observed under constant conditions and circumstances, an assumption that is hardly met in the real world. Highlighting the benefits of our model, we should also admit that an important feature of occupancy models is not considered in our study, namely modelling the state of true presence through a detection model that distinguishes unobserved from truly absent occupancies/interactions through replicated count data (MacKenzie et al. 2002). Replicated examination of host individuals for parasites was simply not feasible in our live-capture and release study, but may be applicable to ecological network studies adopted from monitoring schemes.
We cannot fully exclude that modelling interaction networks from exhaustive sampling scenarios will include some overestimation of interactions and host–parasite associations. Whether such effects are likely to occur needs further study, ideally also including data from host–parasite networks with more species-specific parasites such as lice and helminths.
The real-world network under particular environmental conditions and in a particular time frame would be the state of connectivity relevant for evaluating the meaning of the network, for example in terms of the relative risk of disease transmission in host–parasite networks. Technically, such possible dynamics in networks can be captured if interaction frequencies at the species level [Φ(i,j) in our study] are estimated under different scenarios while accounting for covariance with the environmental variables of interest (see Eq. 2). It would then be possible to estimate site- or forest-type specific interaction probabilities while further considering uncertainty in inferences made at higher levels of organization. In our study, however, this was not feasible as we were not able to trace the effects of environmental covariates such as forest type, area, or season on the infestation probability of different host species. The lack of environmental effects in our study might be attributable to the relatively small sample size of data collected under any particular conditions. Given ongoing forest conversion and destruction in Southeast Asia (Sodhi et al. 2010) and the necessity to understand such massive anthropogenic impact on wildlife, as well as the potential threat to companion animal and human health, understanding the impact of the environment on host–tick relationships in the tropics is desirable despite the large sampling effort required. The species diversity and prevalence of endoparasitic helminths of mammals, for example, have been found to be affected by forest management in Borneo (Wells et al. 2007b) and elsewhere in the tropics (Gillespie and Chapman 2006). Likewise, effects of deforestation and habitat modification have been recorded for mosquito-borne haemosporidian parasites in birds (Sehgal 2010), whereas such effects remain largely unexplored for tropical host–tick associations and those of other ectoparasites (Cumming 2002; Geevarghese and Mishra 2011).
In the light of our results of generally low host specificity, different infestation rates among host species and the effect of local host abundance on infestation probabilities, we expect overall host availability through species diversity, abundance and space use to be an important predictor of tick survival and reproduction. Within the environmental setting of our study, we found a decline of small mammal diversity in logged compared to old-growth forests, mostly due to the decline of relatively rare and more specialized host species such as civets (Wells et al. 2007a). In contrast, for common species, such as the ones found with the highest tick infestation probabilities in this study, there was little obvious effect on their occurrence and abundance in logged versus old-growth forests. Importantly, if these species provide the major source for all tick species considered in this study, there is no evidence that logging at the examined extent has any major effect on tick occurrence or tick assemblage composition. Some caution, however, is necessary here, as we do not know the entire host range of tick species. Larger mammals such as the bearded pig, Sus barbatus, or deer, Tragulus and Muntiacus, among others, may serve as hosts for the same ticks (especially adult ticks) and may thus play a substantial role in their overall population dynamics (Hoogstraal 1964; Kohls 1957).
Viewing our results in a larger context, it is in our opinion an interesting question to critically ask how uncertainty and sampling bias might confound conclusions on the increasing network data available such as those assembled in the Interaction Web Database (http://www.nceas.ucsb.edu/interactionweb/). Comparative studies and meta-analyses are crucial for understanding the general mechanisms or differences among types of ecological interactions, ecosystems or functional and taxonomic groups in network formation (Ings et al. 2009; Thébault and Fontaine 2010). Scrutinizing comparability of network data collected with different sampling protocols and intensity would benefit from going a step backwards and considering the entire range of disaggregated data collected instead of already aggregated interaction matrices. Presumably, in most ecological networks, some portion of the non-interaction of species may be due to the lack of opportunities of contact between species or missed observations rather than due to deliberate species preferences. Discriminating between these aspects can be significant for accurate conclusions and may avoid potential ecological fallacies in network analysis.
References
Adler GH (1998) Impacts of resource abundance on populations of a tropical forest rodent. Ecology 79:242–254
Anderson RM, May RM (1991) Infectious diseases of humans: dynamics and control. Oxford University Press, Oxford
Bersier LF, Banašek-Richter C, Cattin MF (2002) Quantitative descriptors of food-web matrices. Ecology 83:2394–2407
Blüthgen N, Klein A-M (2011) Functional complementarity and specialisation: the role of biodiversity in plant–pollinator interactions. Basic Appl Ecol 12:282–291
Blüthgen N, Menzel F (2006) Measuring specialization in species interaction networks. BMC Ecol 6:9. doi:10.1186/1472-6785-6-9
Clauset A, Moore C, Newman MEJ (2008) Hierarchical structure and the prediction of missing links in networks. Nature 453:98–101
Colwell RK, Futuyma DJ (1971) On the measurement of niche breadth and overlap. Ecology 52:567–568
Cumming GS (2002) Comparing climate and vegetation as limiting factors for species ranges of African ticks. Ecology 83:255–268
Cumming GS (2004) On the relevance of abundance and spatial pattern for interpretations of host–parasite association data. Bull Entomol Res 94:401–409
Durden LA (2006) Taxonomy, host associations, life cycles and vectorial importance of ticks parasitizing small mammals. In: Morand S, Krasnov BR, Poulin R (eds) Micromammals and macroparasites: from evolutionary ecology to management. Springer, Tokyo, pp 91–102
Durden LA, Merker S, Beati L (2008) The tick fauna of Sulawesi, Indonesia (Acari: Ixodoidea: Argasidae and Ixodidae). Exp Appl Acarol 45:85–110
Feinsinger P, Spears EE, Poole RW (1981) A simple measure of niche breadth. Ecology 62:27–32
Geevarghese G, Mishra AC (2011) Haemaphysalis ticks of India. Elsevier, London
Gelman A, Hill J (2007) Data analysis using regression and multilevel/hierarchical models. Cambridge University Press, New York
Gillespie T, Chapman CA (2006) Prediction of parasite infection dynamics in primate metapopulations based on attributes of forest fragmentation. Conserv Biol 20:441–448
Golicher DJ, O’Hara RB, Ruíz-Montoya L, Cayuela L (2006) Lifting a veil on diversity: a Bayesian approach to fitting relative-abundance models. Ecol Appl 16:202–212
Hoogstraal H (1964) Studies of Southeast Asian Haemaphysalis ticks (Ixodidea, Ixodidae). Redescription, hosts, and distribution of H. traguli Oudemans. The larva and nymph of H. vidua W. and N. Identity of H. papuana toxopei Warburton (new combination). J Parasitol 50:765–782
Hurlbert S (1978) The measurement of niche overlap and some relatives. Ecology 59:67–77
Ings TC, et al. (2009) Ecological networks—beyond food webs. J Anim Ecol 78:253–269
Klompen JSH, Black WC IV, Keirans JE, Oliver JH Jr (1996) Evolution of ticks. Annu Rev Entomol 41:141–161
Kohls GM (1957) Malaysian parasites. XVIII. Ticks (Ixodoidea) of Borneo and Malaya. Stud Inst Med Res Malays 28:65–94
Krasnov BR, Stanko M, Morand S (2007) Host community structure and infestation by ixodid ticks: repeatability, dilution effect and ecological specialization. Oecologia 154:185–194
Lafferty KD, Dunne JA (2010) Stochastic ecological network occupancy (SENO) models: a new tool for modeling ecological networks across spatial scales. Theor Ecol 3:123–135
Lande R, Engen S, Sæther B-E (2003) Stochastic population dynamics in ecology and conservation. Oxford University Press, Oxford
LoGiudice K, Ostfeld RS, Schmidt KA, Keesing F (2003) The ecology of infectious disease: effects of host diversity and community composition on Lyme disease risk. Proc Natl Acad Sci USA 100:567–571
Lunn D, Spiegelhalter D, Thomas A, Best N (2009) The BUGS project: evolution, critique and future directions. Stat Med 28:3049–3067
MacKenzie DI, Nichols JD, Lachman GB, Droege S, Royle JA, Langtimm CA (2002) Estimating site occupancy rates when detection probabilities are less than one. Ecology 83:2248–2255
Mason NWH, Mouillot D, Lee WG, Wilson JB (2005) Functional richness, functional evenness and functional divergence: the primary components of functional diversity. Oikos 111:112–118
Mouillot D, Krasnov BR, Poulin R (2008) High intervality explained by phylogenetic constraints in host parasite webs. Ecology 89:2043–2051
Olesen JM, Bascompte J, Elberling H, Jordano P (2008) Temporal dynamics in a pollination network. Ecology 89:1573–1582
Petney TN, Kolonin GV, Robbins RG (2007) Southeast Asian ticks (Acari: Ixodida): a historical perspective. Parasitol Res 101:S201–S205
Poulin R, Mouillot D (2005) Combining phylogenetic and ecological information into a new index of host specificity. J Parasitol 91:511–514
Rao CR (1982) Diversity and dissimilarity coefficients: a unified approach. Theor Pop Biol 21:2443
Robinson WS (2009) Ecological correlations and the behavior of individuals. Int J Epidemiol 38:337–341 [reprinted from (1950) Am Sociol Rev 15(3):351–357]
Royle JA, Nichols JD, Kéry M (2005) Modelling occurrence and abundance of species when detection is imperfect. Oikos 110:353–359
Sehgal RNM (2010) Deforestation and avian infectious diseases. J Exp Biol 213:955–960
Sodhi NS et al (2010) Conserving Southeast Asian forest biodiversity in human-modified landscapes. Biol Conserv 143:2375–2384
Thébault E, Fontaine C (2010) Stability of ecological communities and the architecture of mutualistic and trophic networks. Science 329:853–856
Vázquez DP, Poulin R, Krasnov BR, Shenbrot GI (2005) Species abundance and the distribution of specialization in host–parasite interaction networks. J Anim Ecol 74:946–955
Vázquez DP, Melian CJ, Williams NM, Blüthgen N, Krasnov BR, Poulin R (2007) Species abundance and asymmetric interaction strength in ecological networks. Oikos 116:1120–1127
Wells K, Pfeiffer M, Lakim MB, Kalko EKV (2006) Movement trajectories and habitat partitioning of small mammals in logged and unlogged rain forests on Borneo. J Anim Ecol 75:1212–1223
Wells K, Kalko EKV, Lakim MB, Pfeiffer M (2007a) Effects of rain forest logging on species richness and assemblage composition of small mammals in Southeast Asia. J Biogeogr 34:1087–1099
Wells K, Smales LR, Kalko EKV, Pfeiffer M (2007b) Impact of rain-forest logging on helminth assemblages in small mammals (Muridae, Tupaiidae) from Borneo. J Trop Ecol 23:35–43
Wells K, Lakim MB, Beaucournu JC (2011) Host specificity and niche partitioning in flea–small mammal networks in Bornean rainforests. Med Vet Entomol 25:311–319
Wenger SJ, Freeman MC (2008) Estimating species occurrence, abundance, and detection probability using zero-inflated distributions. Ecology 89:2953–2959
Wikle CK (2003) Hierarchical Bayesian models for predicting the spread of ecological processes. Ecology 84:1382–1394
Wilson DE, Reeder DM (eds) (2005) Mammal species of the world. A taxonomic and geographic reference, 3rd edn. Johns Hopkins University Press, Baltimore
Acknowledgments
We thank the Economic Planning Unit Malaysia for research permission and Sabah Parks and Yayasan Sabah for various kinds of support during field work. We are especially thankful to all staff and people at the different field sites for their warm hospitality. In particular, we thank Alim Biun, Fred Tuh Yit Yu, Jickson Sankin, Awang Matamin, Suati Selimon, Aloysius Mail and Jadda Suhaimi. We thank Brigitte Fiala and K. Eduard Linsenmair for academic and logistic support. We appreciate the mentoring and friendship of the late Elisabeth K. V. Kalko; she sadly passed away, to our disbelief, while this study was being performed. Field work was supported by the German Academic Exchange Service (DAAD).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Wolf Mooij.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wells, K., O’Hara, R.B., Pfeiffer, M. et al. Inferring host specificity and network formation through agent-based models: tick–mammal interactions in Borneo. Oecologia 172, 307–316 (2013). https://doi.org/10.1007/s00442-012-2511-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00442-012-2511-9