Abstract
Alternative pre-mRNA splicing increases the complexity of the proteome that can be generated from the available genomic coding sequences. Dysregulation of the splicing process has been implicated in a vast repertoire of diseases. However, splicing has recently been linked to both the aging process itself and pro-longevity interventions. This review focuses on recent research towards defining RNA splicing as a new hallmark of aging. We highlight dysfunctional alternative splicing events that contribute to the aging phenotype across multiple species, along with recent efforts toward deciphering mechanistic roles for RNA splicing in the regulation of aging and longevity. Further, we discuss recent research demonstrating a direct requirement for specific splicing factors in pro-longevity interventions, and specifically how nutrient signaling pathways interface to splicing factor regulation and downstream splicing targets. Finally, we review the emerging potential of using splicing profiles as a predictor of biological age and life expectancy. Understanding the role of RNA splicing components and downstream targets altered in aging may provide opportunities to develop therapeutics and ultimately extend healthy lifespan in humans.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Aging and RNA homeostasis
Aging is characterized by the progressive decline in physiological function leading to an increased risk of mortality. Advances in public health have increased the proportion of the population that lives into old age, resulting in an increase in the incidence of many chronic age-related diseases including cardiovascular disease, neurodegenerative diseases and cancers (Christensen et al. 2009). Aging is now considered a key risk factor for these chronic diseases and as a result, individuals who reach advanced age are likely to suffer from multiple chronic diseases concurrently (Hung et al. 2011). The current strategy is to treat these co-morbidities in isolation. However, a limit of this approach is that even complete removal of the symptoms of one isolated age-related disease has little impact on remaining conditions. As a result, progress towards increasing the overall disease-free years of life (healthspan) has been marginal (Goldman et al. 2017). A new approach to tackling health and human disease is that of ‘Geroscience’ (Kennedy et al. 2014) which is focused on understanding and then targeting the underlying biology of aging that leads to chronic disease burden, in order to prolong the healthspan of the population. Dietary restriction (DR), a regimen with reduced food intake without malnutrition delays aging and increases healthspan in multiple species. Along with the increase in longevity, DR in organisms from yeast to mammals modulates similar genetic pathways, suggesting that the mechanism of increased lifespan is conserved throughout evolution (Mair and Dillin 2008). Therefore, research has focused on understanding the mechanisms of how DR influences lifespan in order to develop therapeutics that would mimic DR without nutrient restriction in humans.
Research into the mechanisms of aging and longevity to date has largely focused on deterioration of DNA and protein quality control. However, a key intermediary step between transcription and translation in the central dogma is RNA processing, which involves 5′-capping, pre-mRNA splicing, 3′-polyadenylation and RNA editing (Fig. 1). Recently, RNA processing, especially pre-mRNA splicing is increasingly recognized as both an important contributor to the aging process and a causal mediator of pro-longevity interventions.
Regulation of RNA processing and disruption with age and disease. RNA processing fidelity is maintained at multiple points between transcription and translation in normal physiology. With advanced age, disruption of RNA homeostasis can compromise cellular/tissue function and contribute to development of age-related diseases. Depicted are some of the points of dysregulation in RNA homeostasis including splicing factor expression changes and modifications affecting splicing factor localization, spliceosome dysfunction, aberrant splicing, defective RNA surveillance and aberrant mRNA translation
In this review, we summarize key findings and provide our insights on the role of RNA splicing in aging and longevity with a focus on recent studies. Our discussion highlights the current knowledge on splicing and aging across different species including Homo sapiens, Mus musculus, Caenorhabditis elegans, Drosophila melanogaster, and Saccharomyces cerevisiae. Broadly, we have focused on two key areas: (1) age-associated alternative splicing events and splicing factor expression/activity changes and (2) the role of nutrient signaling-mediated longevity on RNA splicing.
Pre-mRNA splicing and alternative splicing
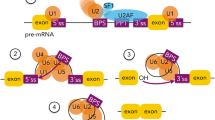
Pre-mRNA splicing is an intricate posttranscriptional process that leads to the removal of introns and joining of exons in a pre-mRNA to form a mature mRNA (House and Lynch 2008). Splicing is a highly dynamic process that is extensively integrated with other gene expression processes such as transcription, mRNA turnover, transport and translation, emphasizing both the importance of splicing precision (Braunschweig et al. 2013), and the potential for detrimental cellular and organismal phenotypes to arise from deregulated splicing. A multi-subunit ribonucleoprotein complex, known as the spliceosome, catalyzes splicing in a two-step trans-esterification reaction. Along with many proteins (> 100), five small nuclear ribonucleoproteins (snRNPs)—U1, U2, U4, U5 and U6 form the constituents of the spliceosome (Wahl et al. 2009). Three essential cis-acting elements: the 5′ splice site (5′ss), the 3′ splice site (3′ss), and the intronic branch-point sequence (BPS) define the core splicing signals necessary for recognition of pre-mRNA by the spliceosome (House and Lynch 2008). In addition to constitutive pre-mRNA splicing, the differential use of these splice sites in a pre-mRNA can result in production of structurally and functionally distinct mRNA and protein variants in a process known as alternative splicing (Nilsen and Graveley 2010). Alternative splicing, therefore, accounts for the complexity of organisms and > 90% of human pre-mRNAs were shown to be alternatively spliced (Wang et al. 2008). Different types of alternative splicing events have been identified, including cassette exon inclusion or skipping, alternative 5′ and 3′ splice sites, mutually exclusive exons, intron retention, alternative promoters and polyA sites (Black 2003) (Fig. 2).
Constitutive and alternative splicing events. Seven types of alternative splicing are cassette exon, alternative 5′ splice site, alternative 3′ splice site, mutually exclusive exons, intron retention, alternative promoter and alternative polyadenylation. Boxes represent exons and lines represent introns
Although alternative splicing can drive functional complexity in higher organisms, this complexity comes with the added potential for dysfunction if the system breaks down. Alternative splicing is regulated by cis-acting elements, trans-acting factors, transcription and chromatin structure (Nilsen and Graveley 2010; Wang et al. 2015). There are additional cis-acting elements known as exonic or intronic enhancers or silencers that provide binding sites for trans-acting factors to either promote or repress exon inclusion. Trans-acting factors comprise splicing activators and repressors. Typically, splicing activators include SR proteins which enhance splice site usage and splicing repressors include hnRNPs, which suppress splice site usage (Busch and Hertel 2012; Wang et al. 2015). However, in many situations, the splicing regulatory functions of SR proteins and hnRNPs are context or position specific (Fu and Ares 2014). For example, SR proteins favor exon inclusion when recruited at the exon, but they repress exon inclusion when bound downstream of the 5′ss. Conversely, hnRNPs mediate exon repression from exonic position while they promote exon inclusion when recruited downstream of the 5′ss (Erkelenz et al. 2013). A combinatorial effect of the activators and repressors determines the final fate of a splicing event (Wang et al. 2015).
Disruptions in the recognition of constitutive splice sites can cause skipping of constitutive exons, intron retention or cryptic splice site activation. This can result in abnormal protein production or mRNA nonsense-mediated decay (NMD) (Faustino and Cooper 2003; Singh and Cooper 2012). In addition, many alternatively spliced transcripts are expressed in a tissue-specific manner, or at a specific developmental stage (Nilsen and Graveley 2010). Therefore, disruptions in alternative splice site recognition can change the ratio of alternatively spliced mRNA transcripts resulting in changes of function and detrimental cellular outcomes (Faustino and Cooper 2003; Singh and Cooper 2012).
Age-associated global changes in alternative splicing events
As longer read RNA deep sequencing becomes commonplace, information about differential isoform usage and RNA processing events with age is increasingly being reported alongside differential gene expression data. Although designating age-related RNA splicing changes as functional/dysfunctional, or even causal to aging is a challenge, what is clear is that age results in increases in isoform variation across species and tissue types. Changes in alternative splicing of select age-related target genes have been reported with advanced aging in human blood (Harries et al. 2011), senescent fibroblasts and endothelial cells (Holly et al. 2013). Similarly, alternative splicing alterations are associated with mouse strain longevity (Lee et al. 2016). Notably, in spleen, alternative splicing changes affect genes involved in cellular senescence, while in muscle, alternative splicing modifications affect inflammatory genes involved in muscle remodeling. In a separate study, age-related increases in alternative exon usage has been demonstrated in a tissue-wide manner across five different mouse tissues: skin, skeletal muscle, bone, thymus, and white adipose tissue (Rodríguez et al. 2016). A subset of these alternatively spliced genes is involved in RNA processing and the spliceosome pathway (Rodríguez et al. 2016). Alternative splicing changes with age in splicing-associated genes have also been observed in Drosophila photoreceptors (Stegeman et al. 2018). Since many components of the RNA processing machinery are themselves regulated by alternative splicing, defects in differential splicing of genes might, therefore, catalyze the aging process via a feed forward mechanism, further impairing RNA processing and translation.
Some of the most striking changes to alternative splicing patterns with age occur in the brain. Since aging results in a decline in cognitive abilities and is the main risk factor for neurodegenerative diseases like Alzheimer’s disease (AD), emphasis has been placed on deciphering genome-wide alternative splicing changes with advanced age in brains from mice (Stilling et al. 2014) and humans (Tollervey et al. 2011; Mazin et al. 2013). A study by Tollervey et al. found splicing changes in 1174 exons with age in human temporal cortex and 95% of these splicing events also occurred in frontotemporal lobar degeneration (FTLD) or AD patients (Tollervey et al. 2011). Age-related alternative splicing changes occurred for genes involved in metabolism and DNA repair (Tollervey et al. 2011). Splicing changes are also observed postnatally in ~ 40% of the genes expressed in two human brain regions: prefrontal cortex and cerebellum. Most of these splicing changes (~ 70%) occur during development while the remaining (~ 30%) occur during aging. A specific increase in inclusion events (particularly intron retention) are observed with age (Mazin et al. 2013). In mice hippocampus, RNA-sequencing analysis revealed significant changes in exon usage at 24 months (436 genes) and 29 months (80 genes) when compared to 3-month-old mice. These changes mainly correspond to genes involved in neuronal plasticity (Stilling et al. 2014). Most of these earlier studies used microarray analysis, exon arrays or qRT-PCR for detecting alternative splicing events, which are targeted approaches and limited by probe design. Moreover, most of the studies focused on exon skipping events. The increasing use of RNA sequencing approaches allow more depth of analysis across the full range of alternative splicing events. For example, in a recent RNA sequencing analysis conducted across 48 different human tissues, Wang et al. identified 49,869 tissue-specific age-associated splicing events, classified into 7 distinct types of alternative splicing categories. These splicing changes broadly correspond to genes involved in aging including DNA repair, DNA damage and apoptosis (Wang et al. 2018). Together, these studies support a potential role of alternative splicing as both a driver and mediator of the aging process.
Intron retention as a signature of the aging process
Increases in intron retention have recently emerged as a common feature in age-related genome-wide splicing analyses. We detected a global increase in intron inclusion in 15-day-old C. elegans compared to young 3-day-old animals, which is suppressed by DR (Heintz et al. 2017). Intron inclusion with age in C. elegans is primarily detected for genes in metabolic processes including lipid catabolism and carbohydrate transport. Intriguingly, increased intron retention has also been shown in young C. elegans on DR (Tabrez et al. 2017; Rollins et al. 2019), as well as in the hippocampus of mice on DR (Tabrez et al. 2017). The effects of retained introns may, therefore, not always be detrimental and could point towards a potential regulatory role for intron inclusion. Similarly, transcriptome analysis of Drosophila heads shows an increase in intron retention with aging at different stages of lifespan (Adusumalli et al. 2019). Drosophila genes with retained introns are linked to TORC1 signaling, protein phosphorylation, memory and longevity. An increase in intron inclusion events with aging is also seen in mouse frontal cortex and hippocampus, human prefrontal cortex as well as in Alzheimer’s disease patients (Adusumalli et al. 2019) in genes involved in mRNA and protein homeostasis. These observations suggest that increase in intron retention is an age-associated event which is conserved across different species.
However, the mechanism leading to intron retention with age remains unclear. Adusumalli et al. speculate that a higher level of nucleosome positioning in older Drosophila might inhibit RNA pol II elongation rate and allow the recruitment of splicing repressors (Adusumalli et al. 2019). Introns are no longer considered genetic junk and a burden to the cell, but rather have important functions in cellular homeostasis (Braunschweig et al. 2014). Two recent studies in yeast demonstrated an accumulation of excised, stable introns and un-spliced transcripts, which are protected from degradation and remain associated with the spliceosome in nutrient-deficient conditions. These stable introns accumulate in a TORC1-dependent manner and protect cells from starvation by specifically downregulating the expression of ribosomal protein genes. This directly contributes to regulating cellular growth depending on nutrient conditions and highlights intron retention as response to nutrient environment (Morgan et al. 2019; Parenteau et al. 2019). Whether age-related intron retention patterns are a response to the loss of cellular homeostasis seen with age or the cause of it remains to be investigated.
In summary, the aging transcriptome has revealed a plethora of age-associated alternative splicing targets using computational tools or qPCR analysis. However, a key question remains: are age-associated splicing events causal drivers of the aging phenotype or a correlative association of functional decline? Hints are beginning to emerge that modulating RNA processing alone is sufficient to prolong healthy aging. For instance, in C. elegans, categorization of lifespan-regulating genes into functional groups revealed an enrichment for RNA binding/processing factors (Curran and Ruvkun 2007). However, future functional studies are still needed to elucidate the causal role of downstream splicing targets in aging and longevity.
Splicing factor changes during aging and longevity
Changes in expression or activity of splicing factors correlate with aging and longevity in humans, mice and C. elegans (Curran and Ruvkun 2007; Holly et al. 2013; Lee et al. 2016; Heintz et al. 2017; Tabrez et al. 2017). However, the causality of splicing factor expression changes remains to be fully demonstrated. In a study profiling gene expression changes with age, ~ 30% of the analyzed splicing factors showed expression changes in human blood in two different cohorts. Some of these splicing factor changes are also associated with cellular senescence in human fibroblasts and endothelial cells. Importantly, age-related expression alterations of transcripts encoding core spliceosomal components such as SF3B1, LSM2, LSM5 and regulatory splicing factors such as SRSF1, SRSF6, SRSF14, HNRNPAB, HNRNPD and HNRNPH3 were identified in both human study cohorts (Holly et al. 2013). In a newer study, splicing factor expression changes were associated with aging phenotypes in humans, suggesting a potential role of splicing factors as drivers of the aging process (Lee et al. 2019b). The expression of HNRNPM and HNRNPA0 correlated with cognitive decline while AKAP17A expression levels correlated with cognitive as well as physical function decline. Age-related changes in expression of other splicing factors including PTBP1/2, RAVER1 and ROD1 were identified in the human temporal cortex (Tollervey et al. 2011).
Splicing factor expression changes are also associated with strain longevity in mice (Lee et al. 2016). These include regulatory splicing factors, Hnrnpa1, Hnrnpa2b1, Hnrnpk, Hnrnpul2, Hnrnpm, Hnrnpd, Hnrnpa0, Hnrnpul1, Srsf3 and Tra2β and core splicing factor, Sf3b1. Notably, two of these splicing factors, Hnrnpa1 and Hnrnpa2b1 also correlate with parental longevity in humans (Lee et al. 2016). However, there is no consensus about which splicing factors correlate with age in different settings. Differences in identity and the direction of expression of splicing factors are seen between different human cohorts, tissues and cell types (Holly et al. 2013; Lee et al. 2016). Therefore, although there is a marked pattern of splicing factor expression changes associated with aging in different settings, functional studies are needed to determine if they are causally linked to longevity. In C. elegans, the branch point binding protein SFA-1 is required for DR and TORC1-mediated extension of lifespan. Overexpression of SFA-1 increases lifespan, supporting a causal and sufficient role in DR longevity. However, both the precise mechanism by which TORC1 modulates SFA-1 activity and how SFA-1 mediates its effects on DR longevity remains unclear (Heintz et al. 2017). In a separate study, the splicing regulator, HRPU-1, was shown to be required for DR-mediated longevity and some of the DR-mediated alternative splicing events in C. elegans. However, the mechanism by which HRPU-1 regulates DR is not known (Tabrez et al. 2017). Much work is, therefore, needed to address the functional mechanisms by which changes to splicing factor activity can modulate the aging process.
Age-associated splicing dysregulation of single genes
Apart from age-associated global splicing changes, splicing pattern changes have been identified in individual genes associated with aging. For instance, in Hutchison Gilford progeria syndrome, a C to T silent mutation in LMNA gene leads to activation of a cryptic 5′splice site in exon 11. This results in production of a truncated lamin A protein, known as progerin with an aberrant function causing progeria (McKenna et al. 2014). The SR proteins, SRSF1 and SRSF6, regulate splicing of the LMNA pre-mRNA, altering the ratio of Lamin A and progerin proteins (Lopez-Mejia et al. 2011). Splicing changes associated with cellular senescence have also been reported. For example, in senescent endothelial cells, an intron retention event during the splicing of endoglin pre-mRNA introduces a premature stop codon that halts translation, resulting in production of the shorter protein isoform (S-endoglin). Binding of SRSF1 near the intronic branchpoint in endoglin pre-mRNA has been shown to inhibit intron removal (Blanco and Bernabeu 2011). Similarly, an exon inclusion event of TP53 pre-mRNA leads to generation of the p53β isoform which promotes cellular senescence (Fujita et al. 2009). The splicing factor SRSF3 represses the inclusion of the p53β-unique exon thereby modulating cellular senescence (Tang et al. 2013).
Nutrient sensing regulation of alternative splicing
Given the changes seen in alternative splicing and spliceosome components with age, a key question is how these changes are regulated. Geroscience research using model organisms has uncovered multiple conserved pathways that act to either exacerbate aging, or can be modulated to reduce age-onset disease incidence and promote longevity (Fontana et al. 2010). One of the most conserved nutrient sensing signaling cascades that modulates aging is insulin/insulin-like signaling (IIS) and the downstream mechanistic target of rapamycin (mTOR). Insulin signaling detects changes in growth factors, while mTOR is activated by amino acids and IIS, and suppression of either increases lifespan in multiple organisms. Recent work has begun to define both how these pathways mechanistically interface with regulation of the RNA splicing machinery and causal roles for RNA splicing in their pro-longevity effects. For example, it was recently discovered that RNA surveillance mechanisms play a role in longevity resulting from mutations in the C. elegans insulin-like receptor, daf-2. Daf-2 mutants have reduced levels of premature termination codon (PTC)-containing transcripts compared to wild type animals (Son et al. 2017). Alternative splicing can produce PTC-containing transcripts that give rise to truncated proteins or are removed by RNA quality control mechanisms. Therefore, it is possible that daf-2 mutation may influence alternative splicing of transcripts. Indeed, insulin signaling in mammalian cells regulates exon inclusion for protein kinase C (PKC)βII mRNA via PI3K signaling and phosphorylation of SR protein SRSF5. This switch in PKC isozyme expression is important for the glucose transport effect of insulin (Patel et al. 2005). Conversely, evidence suggests that splicing of the insulin receptor has an important role in physiology. Different ratios of the insulin receptor isoforms INSR-A and INSR-B are associated with disease states and aging (Belfiore et al. 2017). In rats, advanced age is associated with isoform switching to INSR-A that contributes to insulin resistance observed among aged animals (Serrano et al. 2005). In addition, it was recently found that insulin receptor isoforms are regulated downstream of splicing factor SRSF1 and Ras-MAPK/ERK signaling and have a role in beta cell function and survival (Malakar et al. 2016).
Downstream of insulin signaling, the serine/threonine kinase, AKT, has also been shown to impact splicing both through direct and indirect regulation. AKT can impact splicing indirectly through modulation of splicing factor expression (Latorre et al. 2018), or directly through regulation of SR proteins that bind RNA and impact splicing (Blaustein et al. 2005) or SR protein-specific kinase (SRPK) activity and localization (Zhou et al. 2012). A phosphoproteomic screen of the three AKT isoforms identified 25 RNA processing proteins including multiple splicing factors and splicing regulatory proteins, that are modulated by AKT (Sanidas et al. 2014), suggesting that AKT translates extracellular signals into alternative splicing through post-translational modification of specific splicing factors. Immunoprecipitated AKT phosphorylates SR proteins including SRSF4 and SRSF6 in mammalian cell lines (Jiang et al. 2009). This is potentially through SRPKs, since growth factor-stimulated cells promote SRPK phosphorylation and translocation to the nucleus, SR protein phosphorylation and subsequent changes to alternative splicing through a mechanism involving AKT (Zhou et al. 2012). Recent work also suggests a functional role for AKT regulation of splicing factor activity in age-associated phenotypes such as senescence and cancer. Inhibition of AKT and/or ERK leads to an upregulation of splicing factor expression and alleviation of senescent phenotypes in human fibroblast (Latorre et al. 2018). Furthermore, pharmacological inhibition of PI3K/AKT/mTOR upregulates splicing regulatory protein, hnRNPM,-dependent splicing regulation, which plays a role in cancer treatment (Passacantilli et al. 2017). Further research is needed to determine whether AKT generally acts as a mediator between nutrient signaling, splicing factor activity and age-associated phenotypes.
Active mTOR complex 1 (mTORC1) stimulates protein synthesis by modulating key components of the translation machinery. mTORC1 promotes translation by phosphorylating and inhibiting 4E-binding protein (4EBP), and activating ribosomal subunit S6 kinase (S6K), thereby promoting global translational levels. Conversely, downregulation of mTORC1 inhibits translation and promotes longevity. Additionally, reduced levels of S6 kinase decreases protein synthesis and extends lifespan in yeast, worms, flies and mice (Kennedy and Lamming 2016). Recent data have revealed a connection between mTORC1 signaling and alternative splicing. mTORC1 activation of S6K1 leads to phosphorylation of splicing factor kinase SRPK2, which translocates to the nucleus and activates SR protein binding to the U1–70 K spliceosome component to promote splicing of lipogenesis-related transcripts to fuel cancer metabolism (Lee et al. 2017). mTORC1 activation also influences 3′ splice site utilization and alternative exon usage of components of the spliceosome to regulate the transcriptome, which may serve as another mechanism for mTOR-regulated translational control (Chang et al. 2015, 2019). In particular, mTORC1 controls the expression of U2AF1 isoforms, which then influence 3′ splice site selection. mTOR signaling has also been shown to be regulated by the splicing factor Sam68. The lack of Sam68 leads to mTOR intron 5 retention, introducing a PTC-codon that decreases mTOR protein levels and results in decreased insulin-stimulated S6 and AKT phosphorylation (Huot et al. 2012). Downstream of mTOR signaling, Sam68 also regulates isoform expression of S6K, causing intron 6 inclusion, thereby preventing the expression of p31S6K short isoform (Song and Richard 2015). Expression of the short isoform is frequently upregulated in cancer cell lines (Ben-Hur et al. 2013) Functionally, Sam68 binds to the S6K sequence and prevents alternative splicing by oncogenic splicing factor SRSF1 (Karni et al. 2007). Therefore, alternative splicing is suggested to play a role in various diseases including cancer through regulation of Ras-MAPK and the PI3K-mTOR signaling pathways (Siegfried et al. 2013).
Finally, regulation of alternative splicing through mTOR signaling may be a contributing factor to its impact on longevity. We have shown that an upstream regulator of TORC1 signaling, RAGA-1, is required for maintaining splicing homeostasis in C. elegans (Heintz et al. 2017). In yeast, inhibition of TOR with rapamycin improves splicing efficiency (Munding et al. 2013). Furthermore, TOR-regulated intron accumulation in nutrient-depleted yeast impacts cell growth and survival (Morgan et al. 2019; Parenteau et al. 2019). Altogether, these data support a role for growth factor signaling in regulation of gene expression and alternative splicing through an IIS/AKT/mTOR axis (Fig. 3). What remains to be seen is whether this level of regulation also contributes to the impact of these nutrient sensing pathways on longevity.
Regulation of alternative splicing by nutrient sensing signaling pathways. Examples of nutrient sensing factors, implicated in the mechanisms of aging and longevity, impacting splicing factor regulatory protein phosphorylation, localization and levels. Extracellular signals including insulin/IGF activate growth factor receptors and impact signaling to a number of different pathways. Activated Ras causes a signaling cascade that involves MAPK/ERK signaling to increase SRSF1 expression and subsequent increase in exon inclusion of the insulin receptor (INSR) transcript to form insulin receptor beta (INSR-B). PI3K signaling downstream of insulin increases AKT mediated phosphorylation of SR protein kinases (SRPKs) or SR proteins directly. Phosphorylation induces nuclear localization and can alter splicing of transcripts including PKC exon inclusion to form PKC beta isoform. Also, mTORC1 signaling increases phosphorylation of S6K which phosphorylates SRPK2 and leads to nuclear translocation. SRPK2 then phosphorylates SR proteins. The impact of these splicing regulatory factor-mediated changes in alternative splicing is alterations in isoform expression and cellular functions including lipogenesis and glucose uptake. Finally, nutrient sensing and alternative splicing are important for survival in yeast as nutrient depletion leads to an accumulation of stable introns in a TORC1-dependent manner that protect cells from starvation by downregulating ribosomal biogenesis
RNA splicing as a causal modulator of dietary restriction longevity
Recent work has begun to highlight a role for RNA homeostasis in the effect of dietary restriction. A network-based analysis of the most DR-responsive gene sets in mice across 17 tissues uncovered a role for RNA processing (Swindell 2009). The underlying mechanisms linking DR to changes in exon usage and alternative splicing are beginning to be characterized. Expression of multiple components of the splicing process are shown to be responsive to DR. A recent study identified spliceosome and RNA processing factors to be differentially regulated in the transcriptome, proteome and acetylome analysis in liver of rhesus monkeys (Macaca mulatta) in response to DR. Furthermore, exon analysis identified recruitment of RNA processing mechanisms such as exon skipping and alternate exon usage specifically with DR in the 2-year longitudinal study (Rhoads et al. 2018). Using a combination of polysome profiling and mRNA sequencing, Rollins et al. determined that alternative splicing, leading to intron retention under DR, correlated with diminished translation in C. elegans (Rollins et al. 2019).
Splicing factors previously associated with longevity in humans, mice and worms were shown to have changes in expression in response to DR in mice (Lee et al. 2019a). Now, causal links between DR and RNA processing machinery are also beginning to emerge (Heintz et al. 2017; Tabrez et al. 2017; Rhoads et al. 2018). We have identified two splicing factors, SFA-1 and REPO-1 (SF1/BBP and SF3A2 in mammals), that are specifically required for DR-mediated longevity in C. elegans (Heintz et al. 2017). DR suppresses aging-related deregulation of RNA splicing in a TORC1-dependent manner. Furthermore, genetic perturbations that mimic DR including suppression of TORC1 or activation of the energy sensor AMPK were also suppressed by loss of SFA-1. Further evidence that RNA processing is linked to aging and DR longevity is the association between genes involved in NMD. Knockdown of UPF1 homologue, smg-2 in C. elegans suppresses DR lifespan (Tabrez et al. 2017). In support of a role for NMD, it was found in a separate study that smg-6 and smg-7 were also required for DR-mediated longevity (Rollins et al. 2019). The engagement of NMD is also suggested to be coupled to alternative splicing, since RNAi of splicing factor hrpu-1 downregulates expression of smg-2. The requirement of NMD in longevity is further supported by the observation that mammalian cells exposed to rapamycin, a drug that inhibits mTORC1 and extends lifespan in multiple organisms, augments NMD of certain transcripts in a UPF1 and 4EBP1-dependent manner (Martinez-Nunez et al. 2017). This also includes altered turnover of splicing factors such as SRSF6. Taken together, these results show that longevity paradigms regulate splicing factor expression, splicing and turnover to influence pre-mRNA processing and protein expression. The consequences of altered pre-mRNA processing and expression of mRNA with DR may be reprogramming of the proteome that can directly contribute to longevity.
Conclusions, perspectives and future directions
In summary, a growing body of results from genome-wide transcriptomic analysis has shed light on a previously unappreciated role of RNA splicing in the aging process. Excitingly, work in genetically malleable organisms like C. elegans has also begun to assign these changes as being causally rather than just casually associated with healthy aging and interventions that promote longevity (Fig. 4). Elucidating the role of splicing factors and downstream splicing targets in aging and longevity is now a key area of investigation.
Core and regulatory splicing factors are implicated in longevity. Longevity associated splicing factors participating at different stages of the splicing process are depicted in red. Expression of Hnrnpk, Hnrnpul2, Hnrnpm, Hnrnpd, Hnrnpa0, Hnrnpul1, Sf3b1, Srsf3 and Tra2β are associated with strain longevity in mice (Lee et al. 2016). Hnrnpa1 and Hnrnpa2b1 levels are associated with strain longevity in mice as well as parental longevity in humans (Lee et al. 2016) while expression of PRP-38 is required for mediating longevity in C. elegans (Curran and Ruvkun 2007). Notably, splicing factors SF1/SFA-1, SF3A2/REPO-1 (Heintz et al. 2017) and HNRNPUL1/HRPU-1 (Tabrez et al. 2017) are required for DR-mediated longevity in C. elegans
A remaining challenge is to discriminate whether changes to RNA splicing events seen with age are functional adaptive responses to changes in cellular conditions or aberrant events that induce dysfunction. Age-related, correlative splicing changes have been reported numerously; however, more functional and mechanistic studies to evaluate their effect on health and lifespan are required. Since pre-mRNA splicing occurs co-transcriptionally and is regulated by posttranslational modifications, progress towards this goal can be made using recent technological advances and by combining transcriptomic and proteomic approaches. Studies are beginning to coordinate transcriptional and post-transcriptional changes with DR with alterations at the proteomic level, as illustrated by Rollins et al. (2019) and Rhoads et al. (2018). While these studies contribute to our overall knowledge of the coordination of transcriptional and translational regulation, current insight is limited. The development of high-throughput technologies such as next-generation sequencing and proteomics or ribo-Seq (Calviello and Ohler 2017) and single cell analysis now provide opportunities to integrate systems data at functional regulatory levels and build an interactive network to better understand the contribution of perturbed splicing to the aging process.
Apart from their role in splicing, splicing factors have other cellular functions including chromatin organization, transcription, nucleocytoplasmic transport of RNAs and post-transcriptional regulation (Braunschweig et al. 2013). For example, splicing factor, SF1, was shown to act as a transcriptional repressor (Zhang and Childs 1998) and to interact with transcription factor CA150 affecting RNA polymerase II elongation rates (Goldstrohm et al. 2001). The trans-acting SR protein family functions in coupling transcription to RNA splicing (Braunschweig et al. 2013). The C. elegans RSR-2 splicing protein (mammalian SRRM2/SRm300) is involved in sex determination through its function in transcription (Fontrodona et al. 2013). These are only a few examples to illustrate additional functions of splicing factors that need to be considered and uncoupled in order to define a splicing factor’s mechanistic role in the aging process. To date, longevity paradigms that require specific splicing factors have not been causally linked to changes in alterative splicing specifically.
Splicing factors have also been shown to regulate biogenesis of circRNA, which are a recently characterized type of non-coding RNA species (Kramer et al. 2015). Indeed, circular RNAs have been shown to accumulate during aging in C. elegans, Drosophila and mice, independently of transcriptional upregulation of the host genes (Westholm et al. 2014; Gruner et al. 2016; Cortés-López et al. 2018). The knockdown of individual splicing factors causes alternative splicing changes as well as increased accumulation of circular RNAs in aging Drosophila photoreceptors (Stegeman et al. 2018). Therefore, future goals will be to characterize links between the abundance/activity of individual splicing factors, circular RNAs and alternative splicing events to aging.
The effects of major hallmarks of aging such as loss in protein homeostasis, telomere attrition and genomic instability, likely all affect RNA splicing due to their mechanistic interplay and would also be affected by a loss of RNA homeostasis. Yet, it is currently unknown what initially triggers the age-dependent changes in splicing factor activity and how these triggers impact aging signatures. Age-associated changes to the activity of transcription factors may also lead to differential expression of splicing factors as their downstream target genes (Harries et al. 2011; Deschênes and Chabot 2017). In addition, alternative splicing is also an important regulator of telomerase activity (Sayed et al. 2019). Uncoupling cause and effect for various hallmarks of aging and which if any represents the most treatable intervention remains a challenge.
Heterogeneity in the rate of aging exists between different individuals of a population, i.e. age-matched individuals can have different biological ages. Recently, studies have identified several new biomarkers for biological aging of which epigenetic clocks appears to be the most potent (Jylhävä et al. 2017). Preliminary observations suggest RNA splicing may also act as a predictor of biological age. For instance, we showed that C. elegans of identical chronological age showed differential regulation of a fluorescently labeled alternative splicing reporter mini-gene between individuals. Importantly, young worms that show early deregulation of alternative splicing have a shorter lifespan than controls, demonstrating that a single early splicing event can predict life expectancy and be a potential biomarker of biological age of an organism (Heintz et al. 2017). A similar notion was recently proposed by Wang et al. (2018), following multi-level computational splicing and gene expression analyses across 48 human tissues. They concluded that age-associated splicing profiles can be a better predictor of biological age than gene expression signatures. It will, therefore, be of interest to include in-depth splicing analysis in future omics approaches to generate biomarkers that predict biological age. Such splicing profiles biomarkers of biological age might lead to more targeted therapeutics, along with optimizing the efficacy of current interventions that promote healthy aging.
References
Adusumalli S, Ngian Z, Lin W et al (2019) Increased intron retention is a post-transcriptional signature associated with progressive aging and Alzheimer’s disease. Aging Cell. https://doi.org/10.1111/acel.12928
Belfiore A, Malaguarnera R, Vella V et al (2017) Insulin receptor isoforms in physiology and disease: an updated view. Endocr Rev 38:379–431. https://doi.org/10.1210/er.2017-00073
Ben-Hur V, Denichenko P, Siegfried Z et al (2013) S6K1 alternative splicing modulates its oncogenic activity and regulates mTORC1. Cell Rep 3:103–115. https://doi.org/10.1016/j.celrep.2012.11.020
Black DL (2003) Mechanisms of alternative pre-messenger RNA splicing. Biochemistry 72:291–336. https://doi.org/10.1146/annurev.biochem.72.121801.161720
Blanco FJ, Bernabeu C (2011) Alternative splicing factor or splicing factor-2 plays a key role in intron retention of the endoglin gene during endothelial senescence. Aging Cell 10:896–907. https://doi.org/10.1111/j.1474-9726.2011.00727.x
Blaustein M, Pelisch F, Tanos T et al (2005) Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nat Struct Mol Biology 12:nsmb1020. https://doi.org/10.1038/nsmb1020
Braunschweig U, Gueroussov S, Plocik AM et al (2013) Dynamic integration of splicing within gene regulatory pathways. Cell 152:1252–1269. https://doi.org/10.1016/j.cell.2013.02.034
Braunschweig U, Barbosa-Morais NL, Pan Q et al (2014) Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res 24:1774–1786. https://doi.org/10.1101/gr.177790.114
Busch A, Hertel KJ (2012) Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Rev RNA 3:1–12. https://doi.org/10.1002/wrna.100
Calviello L, Ohler U (2017) Beyond read-counts: ribo-seq data analysis to understand the functions of the transcriptome. Trends Genet 33:728–744. https://doi.org/10.1016/j.tig.2017.08.003
Chang J-W, Zhang W, Yeh H-S et al (2015) mRNA 3′-UTR shortening is a molecular signature of mTORC1 activation. Nat Commun 6:7218. https://doi.org/10.1038/ncomms8218
Chang J-W, Yeh H-S, Park M et al (2019) mTOR-regulated U2af1 tandem exon splicing specifies transcriptome features for translational control. Nucleic Acids Res. https://doi.org/10.1093/nar/gkz761
Christensen K, Doblhammer G, Rau R, Vaupel JW (2009) Ageing populations: the challenges ahead. Lancet 374:1196–1208. https://doi.org/10.1016/s0140-6736(09)61460-4
Cortés-López M, Gruner MR, Cooper DA et al (2018) Global accumulation of circRNAs during aging in Caenorhabditis elegans. Bmc Genomics 19:8. https://doi.org/10.1186/s12864-017-4386-y
Curran SP, Ruvkun G (2007) Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet 3:e56. https://doi.org/10.1371/journal.pgen.0030056
Deschênes M, Chabot B (2017) The emerging role of alternative splicing in senescence and aging. Aging Cell. https://doi.org/10.1111/acel.12646
Erkelenz S, Mueller WF, Evans MS et al (2013) Position-dependent splicing activation and repression by SR and hnRNP proteins rely on common mechanisms. RNA 19:96–102. https://doi.org/10.1261/rna.037044.112
Faustino N, Cooper TA (2003) Pre-mRNA splicing and human disease. Gene Dev 17:419–437. https://doi.org/10.1101/gad.1048803
Fontana L, Partridge L, Longo VD (2010) Extending healthy life span—from yeast to humans. Science 328:321–326. https://doi.org/10.1126/science.1172539
Fontrodona L, Porta-de-la-Riva M, Morán T et al (2013) RSR-2, the Caenorhabditis elegans ortholog of human spliceosomal component SRm300/SRRM2, regulates development by influencing the transcriptional machinery. PLoS Genet 9:e1003543. https://doi.org/10.1371/journal.pgen.1003543
Fu X-D, Ares M (2014) Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet 15:689–701. https://doi.org/10.1038/nrg3778
Fujita K, Mondal AM, Horikawa I et al (2009) p53 isoforms Δ133p53 and p53β are endogenous regulators of replicative cellular senescence. Nat Cell Biol 11:1135–1142. https://doi.org/10.1038/ncb1928
Goldman DP, Cutler D, Rowe JW et al (2017) Substantial health and economic returns from delayed aging may warrant a new focus for medical research. Health Affair 32:1698–1705. https://doi.org/10.1377/hlthaff.2013.0052
Goldstrohm AC, Albrecht TR, Suñé C et al (2001) The transcription elongation factor CA150 interacts with RNA polymerase II and the pre-mRNA splicing factor SF1. Mol Cell Biol 21:7617–7628. https://doi.org/10.1128/mcb.21.22.7617-7628.2001
Gruner H, Cortés-López M, Cooper DA et al (2016) CircRNA accumulation in the aging mouse brain. Sci Rep 6:38907. https://doi.org/10.1038/srep38907
Harries LW, Hernandez D, Henley W et al (2011) Human aging is characterized by focused changes in gene expression and deregulation of alternative splicing. Aging Cell 10:868–878. https://doi.org/10.1111/j.1474-9726.2011.00726.x
Heintz C, Doktor TK, Lanjuin A et al (2017) Splicing factor 1 modulates dietary restriction and TORC1 pathway longevity in C. elegans. Nature 541:102. https://doi.org/10.1038/nature20789
Holly AC, Melzer D, Pilling LC et al (2013) Changes in splicing factor expression are associated with advancing age in man. Mech Ageing Dev 134:356–366. https://doi.org/10.1016/j.mad.2013.05.006
House AE, Lynch KW (2008) Regulation of alternative splicing: more than just the ABCs. J Biol Chem 283:1217–1221. https://doi.org/10.1074/jbc.r700031200
Hung WW, Ross JS, Boockvar KS, Siu AL (2011) Recent trends in chronic disease, impairment and disability among older adults in the United States. BMC Geriatr 11:47. https://doi.org/10.1186/1471-2318-11-47
Huot M-É, Vogel G, Zabarauskas A et al (2012) The Sam68 STAR RNA-binding protein regulates mTOR alternative splicing during adipogenesis. Mol Cell 46:187–199. https://doi.org/10.1016/j.molcel.2012.02.007
Jiang K, Patel NA, Watson JE et al (2009) Akt2 regulation of Cdc2-like kinases (Clk/Sty), serine/arginine-rich (SR) protein phosphorylation, and insulin-induced alternative splicing of PKCβII messenger ribonucleic acid. Endocrinology 150:2087–2097. https://doi.org/10.1210/en.2008-0818
Jylhävä J, Pedersen NL, Hägg S (2017) Biological age predictors. Ebiomedicine 21:29–36. https://doi.org/10.1016/j.ebiom.2017.03.046
Karni R, de Stanchina E, Lowe SW et al (2007) The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol 14:nsmb1209. https://doi.org/10.1038/nsmb1209
Kennedy BK, Lamming DW (2016) The mechanistic target of rapamycin: the grand conductor of metabolism and aging. Cell Metab 23:990–1003. https://doi.org/10.1016/j.cmet.2016.05.009
Kennedy BK, Berger SL, Brunet A et al (2014) Geroscience: linking aging to chronic disease. Cell 159:709–713. https://doi.org/10.1016/j.cell.2014.10.039
Kramer MC, Liang D, Tatomer DC et al (2015) Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Gene Dev 29:2168–2182. https://doi.org/10.1101/gad.270421.115
Latorre E, Ostler EL, Faragher RG, Harries LW (2018) FOXO1 and ETV6 genes may represent novel regulators of splicing factor expression in cellular senescence. Faseb J. https://doi.org/10.1096/fj.201801154r
Lee BP, Pilling LC, Emond F et al (2016) Changes in the expression of splicing factor transcripts and variations in alternative splicing are associated with lifespan in mice and humans. Aging Cell 15:903–913. https://doi.org/10.1111/acel.12499
Lee G, Zheng Y, Cho S et al (2017) Post-transcriptional regulation of de novo lipogenesis by mTORC1-S6K1-SRPK2 signaling. Cell. https://doi.org/10.1016/j.cell.2017.10.037
Lee BP, Mulvey L, Barr G et al (2019a) Dietary restriction in ILSXISS mice is associated with widespread changes in splicing regulatory factor expression levels. Exp Gerontol 128:110736. https://doi.org/10.1016/j.exger.2019.110736
Lee BP, Pilling LC, Bandinelli S et al (2019b) The transcript expression levels of HNRNPM, HNRNPA0 and AKAP17A splicing factors may be predictively associated with ageing phenotypes in human peripheral blood. Biogerontology 20:649–663. https://doi.org/10.1007/s10522-019-09819-0
Lopez-Mejia IC, Vautrot V, Toledo M et al (2011) A conserved splicing mechanism of the LMNA gene controls premature aging. Hum Mol Genet 20:4540–4555. https://doi.org/10.1093/hmg/ddr385
Mair W, Dillin A (2008) Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem 77:727–754. https://doi.org/10.1146/annurev.biochem.77.061206.171059
Malakar P, Chartarifsky L, Hija A et al (2016) Insulin receptor alternative splicing is regulated by insulin signaling and modulates beta cell survival. Sci Rep 6:srep31222. https://doi.org/10.1038/srep31222
Martinez-Nunez RT, Wallace A, Coyne D et al (2017) Modulation of nonsense mediated decay by rapamycin. Nucleic Acids Res 45:3448–3459. https://doi.org/10.1093/nar/gkw1109
Mazin P, Xiong J, Liu X et al (2013) Widespread splicing changes in human brain development and aging. Mol Syst Biol 9:633. https://doi.org/10.1038/msb.2012.67
McKenna T, Rosengardten Y, Viceconte N et al (2014) Embryonic expression of the common progeroid lamin A splice mutation arrests postnatal skin development. Aging Cell 13:292–302. https://doi.org/10.1111/acel.12173
Morgan JT, Fink GR, Bartel DP (2019) Excised linear introns regulate growth in yeast. Nature 565:606–611. https://doi.org/10.1038/s41586-018-0828-1
Munding EM, Shiue L, Katzman S et al (2013) Competition between Pre-mRNAs for the splicing machinery drives global regulation of splicing. Mol Cell 51:338–348. https://doi.org/10.1016/j.molcel.2013.06.012
Nilsen TW, Graveley BR (2010) Expansion of the eukaryotic proteome by alternative splicing. Nature 463:457. https://doi.org/10.1038/nature08909
Parenteau J, Maignon L, Berthoumieux M et al (2019) Introns are mediators of cell response to starvation. Nature 565:612–617. https://doi.org/10.1038/s41586-018-0859-7
Passacantilli I, Frisone P, De Paola E et al (2017) hnRNPM guides an alternative splicing program in response to inhibition of the PI3K/AKT/mTOR pathway in Ewing sarcoma cells. Nucleic Acids Res. https://doi.org/10.1093/nar/gkx831
Patel NA, Kaneko S, Apostolatos HS et al (2005) Molecular and genetic studies imply akt-mediated signaling promotes protein kinase CβII alternative splicing via phosphorylation of serine/arginine-rich splicing factor SRp40. J Biol Chem 280:14302–14309. https://doi.org/10.1074/jbc.m411485200
Rhoads TW, Burhans MS, Chen VB et al (2018) Caloric restriction engages hepatic RNA processing mechanisms in rhesus monkeys. Cell Metab 27:677–688. https://doi.org/10.1016/j.cmet.2018.01.014
Rodríguez SA, Grochová D, McKenna T et al (2016) Global genome splicing analysis reveals an increased number of alternatively spliced genes with aging. Aging Cell 15:267–278. https://doi.org/10.1111/acel.12433
Rollins JA, Shaffer D, Snow SS et al (2019) Dietary restriction induces posttranscriptional regulation of longevity genes. Life Sci Alliance. 2:201800281. https://doi.org/10.26508/lsa.201800281
Sanidas I, Polytarchou C, Hatziapostolou M et al (2014) Phosphoproteomics screen reveals Akt isoform-specific signals linking RNA processing to lung cancer. Mol Cell 53:577–590. https://doi.org/10.1016/j.molcel.2013.12.018
Sayed ME, Yuan L, Robin JD et al (2019) NOVA1 directs PTBP1 to hTERT pre-mRNA and promotes telomerase activity in cancer cells. Oncogene 38:2937–2952. https://doi.org/10.1038/s41388-018-0639-8
Serrano R, Villar M, Martínez C et al (2005) Differential gene expression of insulin receptor isoforms A and B and insulin receptor substrates 1, 2 and 3 in rat tissues: modulation by aging and differentiation in rat adipose tissue. J Mol Endocrinol 34:153–161. https://doi.org/10.1677/jme.1.01635
Siegfried Z, Bonomi S, Ghigna C, Karni R (2013) Regulation of the Ras-MAPK and PI3K-mTOR signalling pathways by alternative splicing in cancer. Int J Cell Biology 2013:568931. https://doi.org/10.1155/2013/568931
Singh RK, Cooper TA (2012) Pre-mRNA splicing in disease and therapeutics. Trends Mol Med 18:472–482. https://doi.org/10.1016/j.molmed.2012.06.006
Son HG, Seo M, Ham S et al (2017) RNA surveillance via nonsense-mediated mRNA decay is crucial for longevity in daf-2/insulin/IGF-1 mutant C. elegans. Nature Commun 8:14749. https://doi.org/10.1038/ncomms14749
Song J, Richard S (2015) Sam68 regulates S6K1 alternative splicing during adipogenesis. Mol Cell Biol 35:1926–1939. https://doi.org/10.1128/mcb.01488-14
Stegeman R, Hall H, Escobedo SE et al (2018) Proper splicing contributes to visual function in the aging Drosophila eye. Aging Cell 17:e12817. https://doi.org/10.1111/acel.12817
Stilling RM, Benito E, Barth J et al (2014) De-regulation of gene expression and alternative splicing affects distinct cellular pathways in the aging hippocampus. Front Cell Neurosci 8:373. https://doi.org/10.3389/fncel.2014.00373
Swindell WR (2009) Genes and gene expression modules associated with caloric restriction and aging in the laboratory mouse. BMC Genomics 10:585. https://doi.org/10.1186/1471-2164-10-585
Tabrez S, Sharma R, Jain V et al (2017) Differential alternative splicing coupled to nonsense-mediated decay of mRNA ensures dietary restriction-induced longevity. Nat Commun 8:306. https://doi.org/10.1038/s41467-017-00370-5
Tang Y, Horikawa I, Ajiro M et al (2013) Downregulation of splicing factor SRSF3 induces p53β, an alternatively spliced isoform of p53 that promotes cellular senescence. Oncogene 32:2792. https://doi.org/10.1038/onc.2012.288
Tollervey JR, Wang Z, Hortobágyi T et al (2011) Analysis of alternative splicing associated with aging and neurodegeneration in the human brain. Genome Res 21:1572–1582. https://doi.org/10.1101/gr.122226.111
Wahl MC, Will CL, Lührmann R (2009) The Spliceosome: design Principles of a Dynamic RNP Machine. Cell 136:701–718. https://doi.org/10.1016/j.cell.2009.02.009
Wang ET, Sandberg R, Luo S et al (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456:470. https://doi.org/10.1038/nature07509
Wang Y, Liu J, Huang B et al (2015) Mechanism of alternative splicing and its regulation. Biomed Rep 3:152–158. https://doi.org/10.3892/br.2014.407
Wang K, Wu D, Zhang H et al (2018) Comprehensive map of age-associated splicing changes across human tissues and their contributions to age-associated diseases. Sci Rep 8:10929. https://doi.org/10.1038/s41598-018-29086-2
Westholm JO, Miura P, Olson S et al (2014) Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep 9:1966–1980. https://doi.org/10.1016/j.celrep.2014.10.062
Zhang D, Childs G (1998) Human ZFM1 protein is a transcriptional repressor that interacts with the transcription activation domain of stage-specific activator protein. J Biol Chem 273:6868–6877. https://doi.org/10.1074/jbc.273.12.6868
Zhou Z, Qiu J, Liu W et al (2012) The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol Cell 47:422–433. https://doi.org/10.1016/j.molcel.2012.05.014
Acknowledgements
We acknowledge Funding from NIA/NIH R01AG051954 and AFAR/Glenn Foundation for Medical Research Breakthroughs in Gerontology Award. P.H. is supported by NIH/NIA Research Supplement to Promote Diversity in Health Related Research R01AG051954-02S1. C.H. is funded by Charles A. King Trust Postdoctoral Fellowship. Figures are created with BioRender.com. We thank the Mair lab members for comments and helpful discussion on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bhadra, M., Howell, P., Dutta, S. et al. Alternative splicing in aging and longevity. Hum Genet 139, 357–369 (2020). https://doi.org/10.1007/s00439-019-02094-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-019-02094-6