Abstract
Nuclear pore complex (NPC) is a fundamental component of the nuclear envelope and is key to the nucleocytoplasmic transport. Mutations in several NUP genes that encode individual components of NPC known as nucleoporins have been identified in recent years among patients with static encephalopathies characterized by developmental delay and microcephaly. We describe a multiplex consanguineous family in which four affected members presented with severe neonatal hypotonia, profound global developmental delay, progressive microcephaly and early death. Autozygome and linkage analysis revealed that this phenotype is linked to a founder disease haplotype (chr9:127,113,732-135,288,807) in which whole exome sequencing revealed the presence of a novel homozygous missense variant in NUP214. Functional analysis of patient-derived fibroblasts recapitulated the dysmorphic phenotype of nuclei that was previously described in NUP214 knockdown cells. In addition, the typical rim staining of NUP214 is largely displaced, further supporting the deleterious effect of the variant. Our data expand the list of NUP genes that are mutated in encephalopathy disorders in humans.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The nucleus is a double membrane-bound organelle that only permits selective transport through the nuclear pore complex (NPC), which together with the lamin and the double nuclear membrane form the nuclear “envelope” (Cronshaw et al. 2002; Rout et al. 2000). NPC is a universal nuclear feature in all eukaryotes (Field et al. 2014). In mammals, NPC comprises around 30 nucleoporins (NUPs), encoded by NUP genes, that form distinct subcomplexes. The 3D structure of the NPC is complex with an eightfold symmetry comprising multiples of eight of each of the subcomplexes, a twofold symmetry comprising pairs of inner and outer ring complexes, and filamentous projections into the cytoplasm and nucleoplasm (Alber et al. 2007; Schwartz 2005). Cryo-electron tomography studies of fully native NPCs have revealed remarkable details about their 3D structure with distinct features observed on the cytoplasmic and nuclear sides (Stoffler et al. 2003). The cytoplasmic side anchors cytoplasmic filaments while the nuclear side anchors the chromatin and has a central ring on top of the central pore, with the latter being partially obstructed by a mobile central plug (Stoffler et al. 2003). This structural conformation endows NPC with the dual function of regulating nucleocytoplasmic transport as well as maintaining structural integrity of the nuclear envelope (Hetzer et al. 2005).
NUP214 forms a subcomplex with NUP88 such that eight subunits of NUP88–NUP214 form the cytoplasmic annual ring that anchors the cytoskeleton to the NPC (Suntharalingam and Wente 2003; Bastos et al. 1997; Kraemer et al. 1994). It was also found to facilitate the export of mRNA through its interaction with DDX19 (Schmitt et al. 1999). Another study demonstrated direct interaction with TTP (Tristetraprolin), a key player in ARE (AU-rich elements)-mediated transcription-level regulation of inflammatory cytokines such as TNFα, and this interaction may regulate TTP subcellular localization (Carman and Nadler 2004). Similarly, NUP214 directly binds SMAD2, SMAD3 and SMAD4 and regulates their nucleocytoplasmic shuttling (Xu et al. 2002, 2003). Knockout of Nup214 in mouse is embryonic lethal (Van Deursen et al. 1996). At the cellular level, NUP214 deficiency results in cell cycle arrest, impaired nucleocytoplasmic transport and accumulation of polyadenylated RNA in the nucleus (Van Deursen et al. 1996). Additionally, nup214 mutant Drosophila larvae display impaired NES nuclear export, most likely as a result of impaired localization of CRM1, a key mediator of NES nuclear export (Sabri et al. 2007; Xylourgidis et al. 2006). Finally, NUP214 is recruited to the mitotic spindles, and its deficiency has been shown to severely impair mitosis, although the latter may also be caused by impaired nucleocytoplasmic shuttling of molecules that are critical for mitosis (Bhattacharjya et al. 2015; Chatel and Fahrenkrog 2011).
Despite the extensive study of NUP214 in cells and in model organisms, our understanding of its medical relevance remains largely limited to the field of cancer. In fact, NUP214 was first identified in the context of cancer where fusion proteins created by translocations involving the N- and C-termini of NUP214 were identified in ALL (acute lymphoblastic leukemia) and AML disease (acute myeloid leukemia), respectively (Graux et al. 2004; Von Lindern et al. 1992). Very recently, an individual with developmental delay and failure to thrive was reported to have deletion of NUP214 in one allele and a deep intronic likely splicing variant in trans (Egloff et al. 2018). In this study, we present data that support NUP214 as a bona fide Mendelian gene, mutation of which causes an autosomal recessive severe form of early infantile encephalopathy.
Materials and methods
Human subjects
The study family was enrolled in an IRB-approved research protocol (KFSHRC RAC#2080006) with informed consent. Venous blood was collected from the patient, healthy siblings and parents. In addition, banked DNA samples were available from two of the three affected siblings. A skin biopsy was also obtained from the proband for functional studies. Phenotypic data were collected from hospital records.
Autozygome and linkage analysis
DNA samples were genotyped on the Axiom SNP Chip platform following the manufacturer’s instructions. Determination of the autozygome was based on regions of homozygosity > 2 Mb as surrogates of autozygosity given the parental consanguinity followed by mapping of the candidate autozygome that is exclusively shared by the affected members using AutoSNPa (http://dna.leeds.ac.uk/autosnpa). Linkage analysis was used to confirm the candidate autozygome and calculate LOD score based on a fully penetrant autosomal recessive model using the EasyLINKAGE package.
Exome sequencing and variant filtering
Exome capture was performed using the TruSeq Exome Enrichment kit (Illumina, San Diego, CA, USA) as per the manufacturer’s instructions. Samples were prepared as an Illumina sequencing library, and in the second step, the sequencing libraries were enriched for the desired target using the Illumina Exome Enrichment protocol. Captured libraries were sequenced using Illumina HiSeq 2000 Sequencer, and the reads mapped against UCSC hg19 (http://genome.ucsc.edu/) by BWA (http://bio-bwa.sourceforge.net/). The SNPs and Indels were detected by SAMTOOLS (http://samtools.sourceforge.net/). The resulting variants were filtered as follows: homozygous → coding/splicing → within candidate autozygome → absent or very rare in Saudi and public exome databases → predicted to be pathogenic by SIFT/PolyPhen/CADD as previously described (Alkuraya 2013).
Cell culture, immunostaining and enveloping surface ratio (ESR) calculation
We followed a standard protocol of establishing a primary fibroblast cell line from a punch skin biopsy and maintained the cells in a standard culture containing MEM + 10% FCS, 1% l-glutamine and 1% penicillin. The following antibodies were used under standard immunofluorescence conditions: Anti-NUP214 antibody (ab70497). Images were taken using Zeiss Axioimager Z2. For ESR (envelope vs surface ratio) calculation, we followed a previously published protocol (Noda et al. 2015). Briefly, exact area of a nucleus was measured as the area surrounded by the contour line, and then a polygon area was divided by the exact area to make the ESR (Noda et al. 2015). Nuclei with a ratio of ≥ 1.05 were defined as dysmorphic.
Results
Clinical report
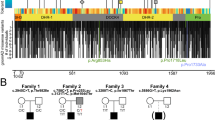
The proband (IV:8) is a 13-month-old girl with severe hypotonia and profound global developmental delay. She was born at term with normal growth parameters. She was admitted at age 2-weeks because of poor weight gain and cyanotic spells and was found to have severe hypotonia and poor suck, but no gross facial dysmorphism. Her severe hypotonia persisted and evolved into profound global developmental delay where she at age 13 months cannot roll over, vocalize or track visually. Abnormal movements were frequently reported and although her EEG was not conclusive for frank epileptiform discharges, she was diagnosed clinically with epilepsy, which remained poorly controlled with therapy. She had frequent lower respiratory tract infections initially thought to be related to aspiration but they persisted even after cessation of oral feeding, placement of gastrostomy tube and undertaking of fundoplication. She had multiple admissions to the PICU for respiratory failure thought to be due to a combination of severe hypotonia and central apnea. Although her somatic growth showed improvement after gastrostomy tube feeding, she had progressive microcephaly. She had extensive metabolic investigations including CSF metabolites, plasma amino acids, acylcarnitines, urine organic acids and mitochondrial sequence analysis, all of which were negative. Her brain MRI was initially normal but later showed evidence of nonspecific atrophy. Nerve conduction studies were normal. Ophthalmologic examination suggested central vision loss. Clinical whole exome sequencing did not reveal any pathogenic variant that explains her phenotype. She died at age 15 months due to respiratory failure. Family history was notable for first cousin healthy parents, two healthy siblings, two siblings who died with an identical clinical presentation at age 1 year and one sister who died within hours of delivery with severe hypotonia (Fig. 1; Table 1).
NUP214 is mutated in a novel recessive form of encephalopathy
By combining the genotypes of the proband, her two deceased affected siblings (no DNA was available from the sister who died shortly after birth), the two healthy siblings and the parents, we were able to identify a candidate autozygous interval chr9:127,113,732–135,288,807 delineated by rs12555646; rs2885495. This was the only autozygous interval exclusively shared by the affected members of the family. Linkage analysis confirmed this interval and assigned a LOD score of 3.2 (Fig. 1c). Although clinical exome sequencing was “negative”, we performed our own exome re-sequencing and analysis since our experience suggested that many such “negative” cases may harbor mutations in novel disease genes (Shamseldin et al. 2017). Indeed, we identified a single novel variant in NUP214 (NM_005085.3:c.461A>G,p.D154G), a gene with no OMIM entry for Mendelian diseases, within the candidate locus, and Sanger sequencing confirmed its full segregation with the phenotype in the family in a strictly autosomal recessive fashion (Fig. 1a). This variant is absent in gnomAD and local database of ethnically matched exomes and is predicted to be pathogenic by SIFT (0.0), PolyPhen (0.9) and CADD (25), and is highly conserved in different species (Fig. 2a, b).
p.D154G impairs the function of NUP214 and leads to abnormal nuclear morphology
Multilobulated, even “flower-like” nuclei have been observed when two individual NUPs were knocked down: NUP153 and NUP214 (Bhattacharjya et al. 2015; Hashizume et al. 2010; Lussi et al. 2010). Thus, we asked whether p.D154G recapitulates the knockdown phenotype of NUP214 in the nucleus of the patient fibroblasts, especially after demonstrating that NUP214 is ubiquitously expressed in different human tissues (Fig. 2c). Indeed, dysmorphic nuclei (defined morphologically by the presence of blebs, hernias, invaginations, and/or lobulations) were far more frequently observed in the patient cells vs controls (Fig. 3a, b). In order to quantify this defect, we measured the ESR, and found that (a) number of nuclei with ESR of > 1.05 in the patient cells is nearly three times that in controls (Fig. 4c) and (b) average ESR in the patient cells is higher than that in controls (Fig. 4D). NUP214 has a characteristic rim-staining pattern (Kinoshita et al. 2012). This pattern was clearly displaced to the nucleoplasm in our patient cells (Figure S1). Taken together, these results show that the p.D154G variant is a deleterious variant that recapitulates the induced NUP214 deficiency described previously.
a Calculation of ESR using polygon area divided by the exact area to calculate the degree of nuclear dysmorphology (see text). b Correlation of increased ESR with nuclear dysmorphology. c Bar graph of the percentage of nuclei with ESR ≥ 1.05 (p value = 0.007). d Average of ESR values in the patient vs control nuclei (error bars are also shown)
Discussion
NUP214 was first studied in the context of cancer where fusion proteins created by translocations involving the N- and C-termini of NUP214 were identified in ALL and AML disease, respectively (Graux et al. 2004; Von Lindern et al. 1992). NUPs share the following major domains in an N- to C- orientation: coiled-coils, FG repeats, α-helical solenoids, β-propellers, and zinc fingers (Devos et al. 2006). The N-terminus has a seven-bladed beta propeller with each blade comprising four antiparallel beta strands that together form a sevenfold axis around a central cavity (Napetschnig et al. 2007). Our variant p.D154 constitutes the third residue of the VIDMKW motif in the A strand of the third blade, which is required for the hydrogen bonding with other beta strands (Napetschnig et al. 2007). This may explain why the p.D154G variant we detected in the study family is associated with such a dramatic disruption of NUP214 localization and function.
The nuclear envelope is a dynamic structure as best seen in its dissolution during cell division (nuclear envelope breakdown, NEBD), which is accompanied by the disassembly of NPC into its individual NUP subcomplexes followed by their reassembly as daughter cells reform their nuclear envelopes (Hetzer et al. 2005). Thus, it is possible to speculate that the pathogenesis of abnormal nuclear morphology we observed in the context of NUP214 mutation is related to impaired dynamics of nuclear envelope stability.
The critical role played by NPC in development is readily evident by the fact that complete knockouts of individual NUPs are usually embryonic lethal in mouse or associated with significant phenotypic abnormalities (Sakuma and D’Angelo 2017). Several aspects of the NPC’s relevance to human diseases have emerged in recent years. In Huntington disease, severe mislocalization and aggregation of NUPs with accompanying abnormal nucleocytoplasmic transport have been observed (Grima et al. 2017). More relevant to the Mendelian disorder we describe in this study is the recent identification of NUP mutations in the context of brain developmental disorders. In 2015, we described a family with a homozygous truncating variant in NUP107 and a combination of microcephaly and chronic kidney disease, also known as Mowat–Galloway syndrome (Alazami et al. 2015). Since then, several studies have similarly shown that biallelic mutations in NUP107 and a number of other NUP genes cause varying combination of microcephaly and glomerular renal disease (Braun et al. 2015, 2018). Very recently, a case with a compound heterozygous mutation in NUP214 was described, albeit with very limited clinical information (developmental delay, failure to thrive and some dysmorphic features) (Egloff et al. 2018).
To our knowledge, the phenotype of NUP214-related encephalopathy we describe here is the most severe early infantile brain-related phenotype observed thus far in the context of NUP mutations. Given the fully lethal phenotype of complete Nup214 deficiency in mouse models, it is likely that our mutation despite its profound deleterious effect is not a complete functional null. The specific involvement of certain body parts in the pathogenesis of NUP-related disorders remains poorly understood although it may be related to tissue-specific differences in the composition of NPCs (Raices and D’angelo 2012). The spatial and temporal variation in NPC composition was first demonstrated for NPU210, but has since been shown for several other NUPs (D’Angelo et al. 2012; Lupu et al. 2008). It is also possible that the demand for normal nucleocytoplasmic transport differs among organs, and that the brain may be particularly vulnerable to impairment of such a fundamental cellular function as has been shown for several others.
In conclusion, we propose that while NUP214 complete deficiency may be lethal in humans, partial deficiency results in a novel autosomal recessive disorder characterized by severe encephalopathy and early death. The pathogenesis of this disorder remains to be fully elucidated although it does seem to involve perturbation of the nuclear envelope integrity. Future cases will shed light on the allelic and phenotypic spectrum of NUP214-related encephalopathy syndrome.
References
Alazami AM, Patel N, Shamseldin HE, Anazi S, Al-Dosari MS, Alzahrani F, Hijazi H, Alshammari M, Aldahmesh MA, Salih MA (2015) Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep 10:148–161
Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT (2007) The molecular architecture of the nuclear pore complex. Nature 450:695
Alkuraya FS (2013) The application of next-generation sequencing in the autozygosity mapping of human recessive diseases. Human Genet 132:1197–1211
Bastos R, De Pouplana LR, Enarson M, Bodoor K, Burke B (1997) Nup84, a novel nucleoporin that is associated with CAN/Nup214 on the cytoplasmic face of the nuclear pore complex. J Cell Biol 137:989–1000
Bhattacharjya S, Roy KS, Ganguly A, Sarkar S, Panda CK, Bhattacharyya D, Bhattacharyya NP, Roychoudhury S (2015) Inhibition of nucleoporin member Nup214 expression by miR-133b perturbs mitotic timing and leads to cell death. Mol Cancer 14:42
Braun DA, Sadowski CE, Kohl S, Lovric S, Astrinidis SA, Pabst WL, Gee HY, Ashraf S, Lawson JA, Shril S (2015) Mutations in nuclear pore genes NUP93, NUP205 and XPO5 cause steroid-resistant nephrotic syndrome. Nat Genet 47:457
Braun DA, Lovric S, Schapiro D, Schneider R, Marquez J, Asif M, Hussain MS, Daga A, Widmeier E, Rao J (2018) Mutations in multiple components of the nuclear pore complex cause nephrotic syndrome. J Clin Investig 128:4313–4328
Carman JA, Nadler SG (2004) Direct association of tristetraprolin with the nucleoporin CAN/Nup214. Biochem Biophys Res Commun 315:445–449
Chatel G, Fahrenkrog B (2011) Nucleoporins: leaving the nuclear pore complex for a successful mitosis. Cell Signal 23:1555–1562
Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ (2002) Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol 158:915–927
D’Angelo MA, Gomez-Cavazos JS, Mei A, Lackner DH, Hetzer MW (2012) A change in nuclear pore complex composition regulates cell differentiation. Dev Cell 22:446–458
Devos D, Dokudovskaya S, Williams R, Alber F, Eswar N, Chait BT, Rout MP, Sali A (2006) Simple fold composition and modular architecture of the nuclear pore complex. Proc Natl Acad Sci 103:2172–2177
Egloff M, Nguyen LS, Siquier-Pernet K, Cormier-Daire V, Baujat G, Attié-Bitach T, Bole-Feysot C, Nitschke P, Vekemans M, Colleaux L, Malan V (2018) Whole-exome sequence analysis highlights the role of unmasked recessive mutations in copy number variants with incomplete penetrance. Eur J Hum Genet 26(6):912–918. https://doi.org/10.1038/s41431-018-0124-4
Field MC, Koreny L, Rout MP (2014) Enriching the pore: splendid complexity from humble origins. Traffic 15:141–156
Graux C, Cools J, Melotte C, Quentmeier H, Ferrando A, Levine R, Vermeesch JR, Stul M, Dutta B, Boeckx N (2004) Fusion of NUP214 to ABL1 on amplified episomes in T-cell acute lymphoblastic leukemia. Nat Genet 36:1084
Grima JC, Daigle JG, Arbez N, Cunningham KC, Zhang K, Ochaba J, Geater C, Morozko E, Stocksdale J, Glatzer JC (2017) Mutant huntingtin disrupts the nuclear pore complex. Neuron 94:93–107.e106
Hashizume C, Nakano H, Yoshida K, Wong RW (2010) Characterization of the role of the tumor marker Nup88 in mitosis. Mol Cancer 9:119
Hetzer MW, Walther TC, Mattaj IW (2005) Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu Rev Cell Dev Biol 21:347–380
Kinoshita Y, Kalir T, Dottino P, Kohtz DS (2012) Nuclear distributions of NUP62 and NUP214 suggest architectural diversity and spatial patterning among nuclear pore complexes. PLoS One 7:e36137
Kraemer D, Wozniak RW, Blobel G, Radu A (1994) The human CAN protein, a putative oncogene product associated with myeloid leukemogenesis, is a nuclear pore complex protein that faces the cytoplasm. Proc Natl Acad Sci 91:1519–1523
Lupu F, Alves A, Anderson K, Doye V, Lacy E (2008) Nuclear pore composition regulates neural stem/progenitor cell differentiation in the mouse embryo. Dev Cell 14:831–842
Lussi YC, Shumaker DK, Shimi T, Fahrenkrog B (2010) The nucleoporin Nup153 affects spindle checkpoint activity due to an association with Mad1. Nucleus 1:71–84
Napetschnig J, Blobel G, Hoelz A (2007) Crystal structure of the N-terminal domain of the human protooncogene Nup214/CAN. Proc Natl Acad Sci 104:1783–1788
Noda A, Mishima S, Hirai Y, Hamasaki K, Landes RD, Mitani H, Haga K, Kiyono T, Nakamura N, Kodama Y (2015) Progerin, the protein responsible for the Hutchinson-Gilford progeria syndrome, increases the unrepaired DNA damages following exposure to ionizing radiation. Genes Environ 37:13
Raices M, D’angelo MA (2012) Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions. Nat Rev Mol Cell Biol 13:687
Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT (2000) The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol 148:635–652
Sabri N, Roth P, Xylourgidis N, Sadeghifar F, Adler J, Samakovlis C (2007) Distinct functions of the Drosophila Nup153 and Nup214 FG domains in nuclear protein transport. J Cell Biol 178:557–565
Sakuma S, D’Angelo MA (2017) The roles of the nuclear pore complex in cellular dysfunction, aging and disease. Semin Cell Dev Biol 68:72–84. https://doi.org/10.1016/j.semcdb.2017.05.006
Schmitt C, von Kobbe C, Bachi A, Pante N, Rodrigues JP, Boscheron C, Rigaut G, Wilm M, Seraphin B, Carmo-Fonseca M (1999) Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J 18:4332–4347
Schwartz TU (2005) Modularity within the architecture of the nuclear pore complex. Curr Opin Struct Biol 15:221–226
Shamseldin HE, Maddirevula S, Faqeih E, Ibrahim N, Hashem M, Shaheen R, Alkuraya FS (2017) Increasing the sensitivity of clinical exome sequencing through improved filtration strategy. Genet Med 19:593
Stoffler D, Feja B, Fahrenkrog B, Walz J, Typke D, Aebi U (2003) Cryo-electron tomography provides novel insights into nuclear pore architecture: implications for nucleocytoplasmic transport. J Mol Biol 328:119–130
Suntharalingam M, Wente SR (2003) Peering through the pore: nuclear pore complex structure, assembly, and function. Dev Cell 4:775–789
Van Deursen J, Boer J, Kasper L, Grosveld G (1996) G2 arrest and impaired nucleocytoplasmic transport in mouse embryos lacking the proto-oncogene CAN/Nup214. EMBO J 15:5574–5583
Von Lindern M, Fornerod M, Van Baal S, Jaegle M, De Wit T, Buijs A, Grosveld G (1992) The translocation (6; 9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, and the expression of a chimeric, leukemia-specific dek-can mRNA. Mol Cell Biol 12:1687–1697
Xu L, Kang Y, Çöl S, Massagué J (2002) Smad2 nucleocytoplasmic shuttling by nucleoporins CAN/Nup214 and Nup153 feeds TGFβ signaling complexes in the cytoplasm and nucleus. Mol Cell 10:271–282
Xu L, Alarcón C, Cöl S, Massagué J (2003) Distinct domain utilization by Smad3 and Smad4 for nucleoporin interaction and nuclear import. J Biol Chem 278(43):42569–42577
Xylourgidis N, Roth P, Sabri N, Tsarouhas V, Samakovlis C (2006) The nucleoporin Nup214 sequesters CRM1 at the nuclear rim and modulates NFκB activation in Drosophila. J Cell Sci 119:4409–4419
Acknowledgements
We thank the study family for their enthusiastic participation. We also thank Mais Hashem for her help as a research coordinator, and the Genotyping and Sequencing Core Facilities at KFSHRC for their technical help. This work was supported in part by King Salman Center for Disability Research (FSA), and the Saudi Human Genome Program (FSA).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
439_2019_1979_MOESM1_ESM.pptx
Figure S1. Displacement of the rim staining of NUP214 to the nucleoplasm of the patient cells compared to control cells (PPTX 966 KB)
Rights and permissions
About this article
Cite this article
Shamseldin, H.E., Makhseed, N., Ibrahim, N. et al. NUP214 deficiency causes severe encephalopathy and microcephaly in humans. Hum Genet 138, 221–229 (2019). https://doi.org/10.1007/s00439-019-01979-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-019-01979-w