Abstract
We report on three unrelated mentally disabled patients, each carrying a de novo balanced translocation that truncates the autism susceptibility candidate 2 (AUTS2) gene at 7q11.2. One of our patients shows relatively mild mental retardation; the other two display more profound disorders. One patient is also physically disabled, exhibiting urogenital and limb malformations in addition to severe mental retardation. The function of AUTS2 is presently unknown, but it has been shown to be disrupted in monozygotic twins with autism and mental retardation, both carrying a translocation t(7;20)(q11.2;p11.2) (de la Barra et al. in Rev Chil Pediatr 57:549–554, 1986; Sultana et al. in Genomics 80:129–134, 2002). Given the overlap of this autism/mental retardation (MR) phenotype and the MR-associated disorders in our patients, together with the fact that mapping of the additional autosomal breakpoints involved did not disclose obvious candidate disease genes, we ascertain with this study that AUTS2 mutations are clearly linked to autosomal dominant mental retardation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mental retardation (MR) is defined by an overall IQ score below 70 and deficits in adaptive behaviour that are recognised in childhood. The disorder affects approximately 2% of the population (e.g. see Ropers et al. 2006). Causes of MR are diverse and often unclear, but in an estimated 60% of severe MR, the cause is genetic. While many genetic forms of MR result from large chromosome imbalances, such as aneuploidies or large duplications and deletions that involve many genes, numerous monogenic causes of MR have also been identified. Several of these disease genes were revealed through characterisation of balanced X chromosome rearrangements, followed by mutation screening in unrelated families with X-linked mental retardation (XLMR) (Billuart et al. 1998; Hagens et al. 2006; Kalscheuer et al. 2003; Kleefstra et al. 2004; Kutsche et al. 2000; Shindo et al. 2002; Shoichet et al. 2003; Tao et al. 2004; Zemni et al. 2000).
When chromosome breakpoints affect autosomal genes, the associated disorder is generally a dominant rather than recessive form. Given that individuals affected with severe MR rarely have children, large multi-generation families with dominant inheritance patterns are uncommon; most mutations causing dominant forms of MR are de novo and therefore difficult to map. For this reason, our current understanding of the genetics of autosomal dominant MR lags far behind our understanding of XLMR. Nonetheless, for several dominant MR-associated disorders, the causative genes have been identified (for comprehensive reviews, see Inlow and Restifo 2004; Chelly et al. 2006). In many cases, these studies involved characterisation of chromosome breakpoints (Borg et al. 2005; Kurotaki et al. 2002; Pichon et al. 2004; Shoichet et al. 2005; Sultana et al. 2002; Tonkin et al. 2004; Vervoort et al. 2002; Yue et al. 2006). The recent advances in our understanding of the aetiology of MR document that defects in diverse pathways result in cognitive impairment. Identification of novel MR genes will broaden our understanding of the pathogenic mechanisms underlying cognitive dysfunction.
As part of our systematic search for causative links between genotype and phenotype in patients with disease-associated balanced chromosome rearrangements we have investigated three unrelated patients with MR and de novo breakpoints in chromosome 7q11.2.
Materials and methods
Cytogenetic analysis and breakpoint mapping by FISH
Samples from patients and parents were obtained after informed consent. Chromosome analysis was performed according to standard high-resolution methods. Breakpoint analyses were performed by fluorescence in situ hybridisation (FISH) experiments on metaphase spreads from the patients’ lymphoblastoid cell lines with biotin-16-dUTP (Boehringer) or digoxigenin-11-dUTP (Boehringer) labelled YAC, BAC or PAC clones from the region of interest. DNA probes were labelled by standard nick translation and detected by immunocytochemistry.
Chromosome isolation and sorting
For chromosome sorting, the lymphoblastoid cell line was cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mM L-glutamine and antibiotics at 37°C in a humidified atmosphere containing 5% CO2. Cells in log phase were treated for 16 h with colcemid (0.05 mg/ml final concentration). Metaphase chromosomes were isolated for flow sorting using buffer containing polyamines as originally described by Sillar and Young (1981), with minor modifications. Chromosome sorts were performed as described previously (Arkesteijn et al. 1999).
Array CGH
Total genomic DNA from the three translocation patients was analysed by array CGH using a sub-megabase resolution whole genome tiling path array, consisting of the human “32k” BAC re-array set, a series of 32,450 overlapping BAC clones obtained from the BAC/PAC Resources Center at Children’s Hospital Oakland Research Institute (Ishkanian et al. 2004; Krzywinski et al. 2004; Osoegawa et al. 2001), the 1 Mb Sanger set (Fiegler et al. 2003a), and a set of 390 sub-telomeric clones (assembled by members of COSTB19: molecular cytogenetics of solid tumors). Hybridisation was done as described previously (Erdogan et al. 2006). In brief, DNA was labelled by random priming (Bioprime array CGH, Invitrogen, Carlsbad, CA) and hybridised overnight at 42°C in a slidebooster (Implen, Munich, Germany). After high stringency washes, slides were scanned using an Agilent scanner (Agilent Technologies, Palo Alto, CA) and images were analysed by Genepix 5.0 (Axon Instruments, Union City, CA). Detailed step-by-step protocols can be found on our website (http://www.molgen.mpg.de/∼abt_rop/molecular_cytogenetics/). Further analysis and visualisation of array CGH data was done using our array CGH software package CGHPRO (Chen et al. 2005). No background subtraction was performed. Normalisation was done by subgrid LOWESS (Cleveland 1979). Copy number gains and losses were determined by a conservative log2 ratio threshold of 0.3 and −0.3, respectively. Profile deviations consisting of three or more neighbouring BAC clones were considered genomic aberrations, unless they coincided with a published polymorphism as listed in the Database of GenomicVariants (http://www.projects.tcag.ca/variation/ version Dec. 13, 2005).
Array painting
For array painting (Fiegler et al. 2003b; Veltman et al. 2003) approximately 6,000 flow-sorted chromosomes in 10 μl of chromosome sorting buffer were used directly for amplification by means of the Genome Plex whole genome amplification (WGA) kit (Rubicon Genomics, Ann Arbor, MI), following the manufacturer’s recommendations. One microgram of PCR product, obtained from derivative chromosomes 7 and 11, was labelled and hybridised as described above. CGHPRO (Chen et al. 2005) was used to visualise the breakpoint-spanning BAC clones, and results were verified by FISH.
Results
Severe MR and multiple congenital anomalies in a patient with a de novo truncation of the AUTS2 gene in intron 5
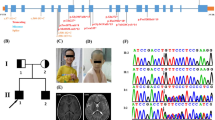
Patient 1 (Fig. 1a) was the first child of healthy non-consanguineous Scottish parents. He was born by spontaneous vaginal delivery at 37 weeks gestation with a birth weight of 2310 g. Dysmorphic features were apparent, and he had poor respiratory effort, requiring immediate admission to the neonatal intensive care unit for conventional mechanical ventilation. At that time, macroglossia, micrognathia, low-set ears, distal arthrogryposis of the hands, talipes equinovarus, and hypospadias were noted. Later, ophthalmological assessment indicated severe bilateral optic nerve hypoplasia with bilaterally absent visual evoked response over the right occipital region and markedly reduced amplitude on the left. Following extubation, he had problems with both central and obstructive apnoea. He had severe esophageal dysmotility and gastroesophageal reflux. Bilateral dislocation of the hip was noted at this time. MRI of the patient’s brain at 2.3 years of age showed a thin corpus callosum, bilateral paucity of white matter with gliosis posteriorly, and a hypoplastic brain stem. He developed a severe kyphoscoliosis with no evidence of vertebral malformation. Moderate to severe sensorineural hearing loss was apparent on analysis of brain stem auditory evoked potentials. At the age of 4 years, he had no discernible speech or language. He now (age 6 years) has a seizure disorder and bilateral vesico-ureteral reflux, and he requires a gastrostomy for enteral feeding. Routine karyotyping indicated a de novo balanced translocation t(3;7)(p21.3;q11.2) (Fig. 2a). Breakpoint mapping by successive fluorescence in situ hybridisation (FISH) experiments led to the identification of breakpoint-spanning clones on both chromosomes 3 and 7. On chromosome 7, clone RP5-910I17 (GenBank accession number AC004927) spans the breakpoint (Fig. 2a). This clone contains part of the AUTS2 gene, which is disrupted by the chromosome rearrangement. Based on sequence comparisons, the breakpoint maps between exons 5 and 6 (UCSC Human Genome Browser, March 2006 update) (Fig. 3). On chromosome 3, clone RP11-241P3 (GenBank accession number AC092048) spans the breakpoint (Fig. 2a). The LYZL4 gene lies at one end of this clone, and may also be disrupted, in which case a fusion protein would be present. LYZL4 is a member of the lysozyme-like gene family with a putative role in human reproduction. Whole genome sub-megabase resolution BAC array CGH did not disclose any other chromosomal abnormalities (data not shown).
Ideograms depicting the rearranged chromosomes with their normal homologues and corresponding FISH-mapping results for critical genomic clones. a Ideogram (left) depicting patient 1 derivative chromosomes and their normal homologues; chromosome 7 overlapping clone RP5-910I17 (left panel) gives a signal on the normal chromosome 7 and split signals on both derivative chromosomes; chromosome 3 overlapping clone RP11-241P3 (right panel) gives a signal on the normal chromosome 3 and split signals on the derivative chromosomes 13 and 7. b Ideogram (left) depicting patient 2 derivative chromosomes and their normal homologues; chromosome 7 overlapping clones RP4-715F13 and RP11-624M10 (upper panels) give signals on the normal chromosomes 7 and split signals on the derivative chromosomes 13 and 7. Chromosome 13 clone RP11-345I22 (lower left panel) maps proximally with respect to the chromosome 13 breakpoint, giving signals on the normal chromosome 13 and the derivative chromosome 13; clone RP11-464D7 (lower right panel) maps distally, giving signals on the normal chromosome 13 and the derivative chromosome 7. c Ideogram (left) depicting patient 3 derivative chromosomes and their normal homologues; chromosome 7 overlapping clone RP11-689B18 (left panel) gives a signal on the normal chromosome 7 and split signals on derivative chromosomes 7 and 11; chromosome 11 overlapping clone RP11-10C6 (right panel) gives a signal on the normal chromosome 11 and split signals on both derivative chromosomes
Borderline MR in a patient with a de novo disruption of the AUTS2 gene between exons 2 and 5
Patient 2 (Fig. 1b) presented with hyperactivity as a child. Milestones were delayed and she was diagnosed with borderline MR. She had feeding problems and severe sleep disturbances during her early years: she slept only for brief periods, and was often awake for much of the night. As a juvenile, she had enuresis. Examination at the age of 27 years revealed mild dysmorphic features including a slight exophthalmia. No malformations were present. Routine chromosome analyses indicated the karyotype 46,XX,t(7;13)(q11.2;q22) de novo (Fig. 2b), and FISH-mapping led to breakpoint localisation on both chromosomes 7 and 13. On chromosome 7, clones RP4-715F13 (GenBank accession number AC006317) and RP11-624M10 (GenBank accession number AC093487) overlap the breakpoint (Fig. 2b), indicating that the AUTS2 gene is also disrupted in this patient. Here the breakpoint is upstream relative to that in the more severely affected patient 1; based on FISH results, it lies between exons 2 and 5 (Fig. 3). Relative to the breakpoint on chromosome 13, clones RP11-345I22 (GenBank accession numbers AQ544047 and AQ544050 for BAC ends) and RP11-464D7 (GenBank accession numbers for BAC ends AQ635412 and AQ586318) mapped proximally and distally, respectively (Fig. 2b). Based on the UCSC Human Genome Browser (March 2006 update), these clones are neighbouring but do not overlap, and the best candidate region (i.e. the 10 kb sequence in between these two clones plus approximately 30 kb on either side) harbours no genes. Two genes, krüppel-like factor 5 (KLF5) and progesterone-induced blocking factor 1 (PIBF or C13orf24), lie further proximally on the proximal clone, and we cannot definitively exclude that KLF5 is disrupted. However, there is no indication that KLF5, which is an essential regulator for vascular remodelling (Shindo et al. 2002) or PIBF, which seems not be expressed in brain tissues (Rozenblum et al. 2002), plays a role in cognition. The distal clone does not contain any known gene. Whole genome sub-megabase resolution BAC array CGH did not disclose any other chromosomal abnormality (data not shown).
Moderate MR in a patient with a de novo disruption of the AUTS2 gene between exons 5 and 7
Patient 3 (Fig. 1c) presented with moderate MR. At the age of ten years, he exhibited a significant speech delay, speaking in five-word sentences. He had a short attention span and was unable to go out alone. This patient has no striking dysmorphic features; however, at the age of 17 he started to develop a bilateral cataract. Like the other two patients studied here, this patient carries a de novo translocation involving 7q11.2: chromosome analysis indicated the karyotype 46,XY,t(7;11)(q11.2;p11.2), depicted in Fig. 2c. In addition, array CGH revealed a duplication of approximately 700 kb at 2p22.3 (data not shown), which we did not investigate further in light of the absence of parental DNA available for array CGH experiments. The translocation breakpoints were localised by array painting using flow-sorted chromosomes (Fig. 4), and results were confirmed by FISH (Fig. 2c). Based on hybridisation results, this translocation also disrupts the AUTS2 gene. Clone RP11-689B18 (no accession number but sequence is available from Ensembl database) from chromosome 7 spans the breakpoint (Fig. 2c), and AUTS2 exon 6 lies close to the distal end of this clone, indicating that the breakpoint lies between AUTS2 exons 5 and 7 (Fig. 3). Clone RP11-10C6 (GenBank accession number AC013602) from chromosome 11 also gives an overlapping signal (Fig. 2c). This clone harbours two genes: PR-domain containing 11 (PRDM11), which has a putative role in cancer, and the membrane synaptotagmin SYT13, which plays a role in vesicular trafficking. It is possible that one of these genes is affected by the breakpoint and thereby plays a causative role, together with the disruption of AUTS2, in the patient’s phenotype. As is the case for patients 1 and 2, if a second gene is disrupted, then the presence of fusion genes may influence the phenotype.
Fine-mapping of patient 3 chromosomal breakpoints by means of array painting. a Flow-sorting of chromosomes enabled the identification and separation of derivative chromosomes 7 and 11 (encircled in the scatterplot). b Array painting result for derivative chromosome 7, with Cy3:Cy5 ratios for each BAC clone indicated (by clone location on the horizontal axis) at the corresponding chromosome position along the chromosome ideogram. The translocation leads to an abrupt ratio shift. The arrow drawn in the zoom-in to the right points to the breakpoint-spanning clone. c Array painting result for derivative chromosome 11, displayed as for derivative chromosome 7 in (b)
The clinical details for patients 1–3 in this study and for the twins with AUTS2 truncations (de la Barra et al. 1986; Sultana et al. 2002) are summarised in Table 1.
Discussion
Based on the current understanding, genes with a wide range of functions may qualify as candidate genes for MR. Identification of multiple patients with mutations in the same gene is arguably one of the most convincing arguments in favor of a causal relationship between gene and disorder. As we show here, for autosomal dominant forms of MR where the predominant common characteristic is MR, matching balanced translocations are a valuable resource in the search for the causative disease genes. In three unrelated individuals with MR-associated balanced translocations involving 7q11.2, breakpoint mapping revealed the disruption of the AUTS2 gene, originally identified as KIAA0442 in a library of large brain-expressed transcripts (Ishikawa et al. 1997) and previously linked to autism and MR through a study on a translocation in identical twins (de la Barra et al. 1986; Sultana et al. 2002). The presence of four unrelated individuals with MR and disruption of AUTS2 (the three described here and the twins described previously) strongly suggests that this gene plays a critical role in human brain function and in the aetiology of autosomal dominant MR.
The AUTS2 gene spans 1.2 Mb of genomic DNA, and the reference sequence comprises 19 exons (mRNA accession number NM_015570). All three patients in this study, as well as those investigated by Sultana et al (2002), have truncations of the AUTS2 gene that lie between exons 2 and 7. Remarkably, this gene region contains very large introns, (e.g. introns 1, 2, 4, and 5 are >218,000 bp each), whereas the rest of the gene contains relatively small introns that are between 936 bp and 4,441 bp long (Sultana et al. 2002). It is also perhaps relevant that AUTS2 lies within a 5 Mb region harbouring relatively few low copy repeats (LCRs), which is flanked by two clusters of LCRs with more than 97% sequence similarity. Aside from this, we identified no specific genomic features that might predispose the region to chromosomal breakage.
The function of AUTS2 (GenBank accession number NP_056385) is unknown, but the protein has a region with homology (39% sequence identity, 60% sequence similarity) to the known human fibrosin 1 protein (FBS1), which is a fibroblast growth factor (Prakash et al. 1995). The reference FBS1 protein sequence (GenBank accession number NP_071897) is only 372 amino acids; however, the Ensembl database (v39 June 2006) features a predicted FBS1 protein with 1323 residues (corresponding to predicted transcript GENSCAN00000033375). Alignment of this predicted protein with the AUTS2 protein sequence reveals an identity of 34% (48% similarity if related amino acid residues are included), which supports the hypothesis that these two proteins may have related functions. Especially in light of the possibility that unique fusion proteins exist, growth factor function for AUTS2 might explain the severe physical anomalies observed in patient 1.
BLAST searches also indicate homology with the XTP9 protein (GenBank accession number AA085466) of unknown function. In addition, the AUTS2 protein contains a PY motif, as pointed out by Sultana et al. (2002), which is a potential WW-domain-binding region involved in protein–protein interactions. Moreover, this motif is present in the activation domain of various transcription factors, suggesting that AUTS2 may play a role in transcriptional regulation. The protein sequence exhibits fairly high cross-species conservation, with 62% amino acid identity between human and zebrafish proteins (based on sequence alignment of zebrafish protein CAD61164 and human protein NP_056385).
All of the patients investigated to date harbour truncations between exons 2 and 7 of AUTS2 (between amino acids 174 and 248 of AUTS2); however, aside from cognitive dysfunction, they present with diverse phenotypes. This clinical variability may in part be due to subtle differences of the breakpoint location within the AUTS2 gene. For example, it is plausible that a disruption of AUTS2 immediately following exon 2, like that in the twins investigated by Sultana et al (2002), might have quite different global effects from one downstream of exon 5, for reasons that we do not fully understand; there are many such genotype–phenotype correlations in other MR-associated disorders. Several XLMR genes, for example, have now been implicated in both non-syndromic and syndromic forms of MR, and for some of these genes, the nature of the mutation seems to play a critical role (Ropers 2006; Ropers and Hamel 2005). It is also noteworthy that there are cases in which identical mutations lead to diverse phenotypes. This phenomenon is poorly understood at the molecular level, but there are numerous MR-associated mutations with this sort of variable expressivity, where considerable intra-familial phenotypic variation is observed (Kwiatkowska et al. 1998; Szudek et al. 2002). The phenotypic differences in the patients we describe here may in fact reflect the non-specific environmental, epigenetic, and/or genetic factors that contribute to such variation.
It is also important to keep in mind that for each of the cases described here, as well as for the twins described previously (de la Barra et al. 1986; Sultana et al. 2002) a contribution of the second breakpoint cannot be ruled out, either through the presence of fusion genes, which may explain the more severe phenotypes observed in patients 1 and 3, or through position effects. Such additional genetic aberrations would likely also have phenotypic effects, but do not diminish the importance of the results presented here. The fact that four unrelated patients with mental retardation carry truncated copies of the AUTS2 gene is highly significant, and it is thus very likely that these AUTS2 aberrations are the primary genetic causes of the observed cognitive disorders. Future investigations will focus on the function of AUTS2 in order to elucidate the pathogenetic mechanisms underlying AUTS2-associated mental retardation.
References
Arkesteijn G, Jumelet E, Hagenbeek A, Smit E, Slater R, Martens A (1999) Reverse chromosome painting for the identification of marker chromosomes and complex translocations in leukemia. Cytometry 35:117–124
Billuart P, Bienvenu T, Ronce N, des Portes V, Vinet MC, Zemni R, Roest Crollius H, Carrie A, Fauchereau F, Cherry M, Briault S, Hamel B, Fryns JP, Beldjord C, Kahn A, Moraine C, Chelly J (1998) Oligophrenin-1 encodes a rhoGAP protein involved in X-linked mental retardation. Nature 392:923–926
Borg I, Freude K, Kubart S, Hoffmann K, Menzel C, Laccone F, Firth H, Ferguson-Smith MA, Tommerup N, Ropers HH, Sargan D, Kalscheuer VM (2005) Disruption of Netrin G1 by a balanced chromosome translocation in a girl with Rett syndrome. Eur J Hum Genet 13:921–927
Chen W, Erdogan F, Ropers HH, Lenzner S, Ullmann R (2005) CGHPRO—a comprehensive data analysis tool for array CGH. BMC Bioinformatics 6:85
Chelly J, Khelfaoui M, Francis F, Cherif B, Bienvenu T (2006) Genetics and pathophysiology of mental retardation. Eur J Hum Genet 14:701–713
Cleveland W (1979) Robust locally weighted regression and smoothing scatterplots. J Am Stat Assoc 74:829–836
de la Barra F, Skoknic V, Alliende A, Raimann E, Cortes F, Lacassie Y (1986) [Twins with autism and mental retardation associated with balanced (7;20) chromosomal translocation]. Rev Chil Pediatr 57:549–554
Erdogan F, Chen W, Kirchhoff M, Kalscheuer VM , Hultschig C, Müller I, Schulz R, Menzel C, Bryndorf T, Ropers HH, Ullmann R (2006) Impact of low copy repeats on the generation of balanced and unbalanced chromosomal aberrations in mental retardation. Cytogenet Genome Res 115
Fiegler H, Carr P, Douglas EJ, Burford DC, Hunt S, Scott CE, Smith J, Vetrie D, Gorman P, Tomlinson IP, Carter NP (2003a) DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer 36:361–374
Fiegler H, Gribble SM, Burford DC, Carr P, Prigmore E, Porter KM, Clegg S, Crolla JA, Dennis NR, Jacobs P, Carter NP (2003b) Array painting: a method for the rapid analysis of aberrant chromosomes using DNA microarrays. J Med Genet 40:664–670
Hagens O, Dubos A, Abidi F, Barbi G, Van Zutven L, Hoeltzenbein M, Tommerup N, Moraine C, Fryns JP, Chelly J, van Bokhoven H, Gecz J, Dollfus H, Ropers HH, Schwartz CE, de Cassia Stocco Dos Santos R, Kalscheuer V, Hanauer A (2006) Disruptions of the novel KIAA1202 gene are associated with X-linked mental retardation. Hum Genet 118:578–590
Inlow JK, Restifo LL (2004) Molecular and comparative genetics of mental retardation. Genetics 166:835–881
Ishikawa K, Nagase T, Nakajima D, Seki N, Ohira M, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O (1997) Prediction of the coding sequences of unidentified human genes. VIII. 78 new cDNA clones from brain which code for large proteins in vitro. DNA Res 4:307–313
Ishkanian AS, Malloff CA, Watson SK, DeLeeuw RJ, Chi B, Coe BP, Snijders A, Albertson DG, Pinkel D, Marra MA, Ling V, MacAulay C, Lam WL (2004) A tiling resolution DNA microarray with complete coverage of the human genome. Nat Genet 36:299–303
Kalscheuer VM, Tao J, Donnelly A, Hollway G, Schwinger E, Kubart S, Menzel C, Hoeltzenbein M, Tommerup N, Eyre H, Harbord M, Haan E, Sutherland GR, Ropers HH, Gecz J (2003) Disruption of the serine/threonine kinase 9 gene causes severe X-linked infantile spasms and mental retardation. Am J Hum Genet 72:1401–1411
Kleefstra T, Yntema HG, Oudakker AR, Banning MJ, Kalscheuer VM, Chelly J, Moraine C, Ropers HH, Fryns JP, Janssen IM, Sistermans EA, Nillesen WN, de Vries LB, Hamel BC, van Bokhoven H (2004) Zinc finger 81 (ZNF81) mutations associated with X-linked mental retardation. J Med Genet 41:394–399
Krzywinski M, Bosdet I, Smailus D, Chiu R, Mathewson C, Wye N, Barber S, Brown-John M, Chan S, Chand S, Cloutier A, Girn N, Lee D, Masson A, Mayo M, Olson T, Pandoh P, Prabhu AL, Schoenmakers E, Tsai M, Albertson D, Lam W, Choy CO, Osoegawa K, Zhao S, de Jong PJ, Schein J, Jones S, Marra MA (2004) A set of BAC clones spanning the human genome. Nucleic Acids Res 32:3651–3660
Kurotaki N, Imaizumi K, Harada N, Masuno M, Kondoh T, Nagai T, Ohashi H, Naritomi K, Tsukahara M, Makita Y, Sugimoto T, Sonoda T, Hasegawa T, Chinen Y, Tomita Ha HA, Kinoshita A, Mizuguchi T, Yoshiura Ki K, Ohta T, Kishino T, Fukushima Y, Niikawa N, Matsumoto N (2002) Haploinsufficiency of NSD1 causes Sotos syndrome. Nat Genet 30:365–366
Kutsche K, Yntema H, Brandt A, Jantke I, Nothwang HG, Orth U, Boavida MG, David D, Chelly J, Fryns JP, Moraine C, Ropers HH, Hamel BC, van Bokhoven H, Gal A (2000) Mutations in ARHGEF6, encoding a guanine nucleotide exchange factor for Rho GTPases, in patients with X-linked mental retardation. Nat Genet 26:247–250
Kwiatkowska J, Jozwiak S, Hall F, Henske EP, Haines JL, McNamara P, Braiser J, Wigowska-Sowinska J, Kasprzyk-Obara J, Short MP, Kwiatkowski DJ (1998) Comprehensive mutational analysis of the TSC1 gene: observations on frequency of mutation, associated features, and nonpenetrance. Ann Hum Genet 62(Pt 4):277–285
Osoegawa K, Mammoser AG, Wu C, Frengen E, Zeng C, Catanese JJ, de Jong PJ (2001) A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res 11:483–496
Pichon B, Vankerckhove S, Bourrouillou G, Duprez L, Abramowicz MJ (2004) A translocation breakpoint disrupts the ASPM gene in a patient with primary microcephaly. Eur J Hum Genet 12:419–421
Prakash S, Robbins PW, Wyler DJ (1995) Cloning and analysis of murine cDNA that encodes a fibrogenic lymphokine, fibrosin. Proc Natl Acad Sci U S A 92:2154–2158
Ropers HH (2006) X-linked mental retardation: many genes for a complex disorder. Curr Opin Genet Dev 16:260–269
Ropers HH, Hamel BC (2005) X-linked mental retardation. Nat Rev Genet 6:46–57
Rozenblum E, Vahteristo P, Sandberg T, Bergthorsson JT, Syrjakoski K, Weaver D, Haraldsson K, Johannsdottir HK, Vehmanen P, Nigam S, Golberger N, Robbins C, Pak E, Dutra A, Gillander E, Stephan DA, Bailey-Wilson J, Juo SH, Kainu T, Arason A, Barkardottir RB, Nevanlinna H, Borg A, Kallioniemi OP (2002) A genomic map of a 6-Mb region at 13q21-q22 implicated in cancer development: identification and characterization of candidate genes. Hum Genet 110:111–121
Shindo T, Manabe I, Fukushima Y, Tobe K, Aizawa K, Miyamoto S, Kawai-Kowase K, Moriyama N, Imai Y, Kawakami H, Nishimatsu H, Ishikawa T, Suzuki T, Morita H, Maemura K, Sata M, Hirata Y, Komukai M, Kagechika H, Kadowaki T, Kurabayashi M, Nagai R (2002) Kruppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nat Med 8:856–863
Shoichet SA, Hoffmann K, Menzel C, Trautmann U, Moser B, Hoeltzenbein M, Echenne B, Partington M, Van Bokhoven H, Moraine C, Fryns JP, Chelly J, Rott HD, Ropers HH, Kalscheuer VM (2003) Mutations in the ZNF41 gene are associated with cognitive deficits: identification of a new candidate for X-linked mental retardation. Am J Hum Genet 73:1341–1354
Shoichet SA, Kunde SA, Viertel P, Schell-Apacik C, von Voss H, Tommerup N, Ropers HH, Kalscheuer VM (2005) Haploinsufficiency of novel FOXG1B variants in a patient with severe mental retardation, brain malformations and microcephaly. Hum Genet 117:536–544
Sillar R, Young BD (1981) A new method for the preparation of metaphase chromosomes for flow analysis. J Histochem Cytochem 1: 747–748
Sultana R, Yu CE, Yu J, Munson J, Chen D, Hua W, Estes A, Cortes F, de la Barra F, Yu D, Haider ST, Trask BJ, Green ED, Raskind WH, Disteche CM, Wijsman E, Dawson G, Storm DR, Schellenberg GD, Villacres EC (2002) Identification of a novel gene on chromosome 7q11.2 interrupted by a translocation breakpoint in a pair of autistic twins. Genomics 80:129–134
Szudek J, Joe H, Friedman JM (2002) Analysis of intrafamilial phenotypic variation in neurofibromatosis 1 (NF1). Genet Epidemiol 23:150–164
Tao J, Van Esch H, Hagedorn-Greiwe M, Hoffmann K, Moser B, Raynaud M, Sperner J, Fryns JP, Schwinger E, Gecz J, Ropers HH, Kalscheuer VM (2004) Mutations in the X-linked cyclin-dependent kinase-like 5 (CDKL5/STK9) gene are associated with severe neurodevelopmental retardation. Am J Hum Genet 75:1149–1154
Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T (2004) NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet 36:636–641
Veltman IM, Veltman JA, Arkesteijn G, Janssen IM, Vissers LE, de Jong PJ, van Kessel AG, Schoenmakers EF (2003) Chromosomal breakpoint mapping by arrayCGH using flow-sorted chromosomes. Biotechniques 35:1066–1070
Vervoort VS, Viljoen D, Smart R, Suthers G, DuPont BR, Abbott A, Schwartz CE (2002) Sorting nexin 3 (SNX3) is disrupted in a patient with a translocation t(6;13)(q21;q12) and microcephaly, microphthalmia, ectrodactyly, prognathism (MMEP) phenotype. J Med Genet 39:893–899
Yue Y, Stout K, Grossmann B, Zechner U, Brinckmann A, White C, Pilz DT, Haaf T (2006) Disruption of TCBA1 associated with a de novo t(1;6)(q32.2;q22.3) presenting in a child with developmental delay and recurrent infections. J Med Genet 43:143–147
Zemni R, Bienvenu T, Vinet MC, Sefiani A, Carrie A, Billuart P, McDonell N, Couvert P, Francis F, Chafey P, Fauchereau F, Friocourt G, des Portes V, Cardona A, Frints S, Meindl A, Brandau O, Ronce N, Moraine C, van Bokhoven H, Ropers HH, Sudbrak R, Kahn A, Fryns JP, Beldjord C, Chelly J (2000) A new gene involved in X-linked mental retardation identified by analysis of an X;2 balanced translocation. Nat Genet 24:167–170
Acknowledgements
We sincerely thank the patients and parents for participation in this study; R. Wegner and H. Neitzel for establishing the karyotype of patient 2; S. Freier and H. Madle for help with cell culture; P. Viertel and M. Schubert for technical assistance; W. Chen for help with LCR analysis; C. Hultschig for spotting the slides; Pieter de Jong and the BACPAC Resources Centre (http://www.bacpac.chori.org/) for providing DNA and clones of the human “32k” BAC re-array set, Nigel Carter and the Mapping Core and Map Finishing groups of the Wellcome Trust Sanger Institute for initial clone supply of the 1 Mb Sanger set and the COST B19 Action “Molecular Cytogenetics of Solid Tumours” for the assembly of the subtelomeric clone set. This work was supported by the National Genome Research Network (NGFN, project numbers 01GR0105 and 01GR0414). The Wilhelm Johannsen Centre for Functional Genome Research was established by the Danish National Research Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kalscheuer, V.M., FitzPatrick, D., Tommerup, N. et al. Mutations in autism susceptibility candidate 2 (AUTS2) in patients with mental retardation. Hum Genet 121, 501–509 (2007). https://doi.org/10.1007/s00439-006-0284-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-006-0284-0